Abstract
We identified five different putative wav gene cluster types, which are responsible for the synthesis of the core oligosaccharide (OS) region of Vibrio cholerae lipopolysaccharide. Preliminary evidence that the genes encoded by this cluster are involved in core OS biosynthesis came from analysis of the recently released O1 El Tor V. cholerae genome sequence and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of O1 El Tor mutant strains defective in three genes (waaF, waaL, and wavB). Investigations of 38 different V. cholerae strains by Southern blotting, PCR, and sequencing analyses showed that the O1 El Tor wav gene cluster type is prevalent among clinical isolates of different serogroups associated with cholera and environmental O1 strains. In contrast, we found differences in the wav gene contents of 19 unrelated non-O1, non-O139 environmental and human isolates not associated with cholera. These strains contained four new wav gene cluster types that differ from each other in distinct gene loci, providing evidence for horizontal transfer of wav genes and for limited structural diversity of the core OS among V. cholerae isolates. Our results show genetic diversity in the core OS biosynthesis gene cluster and predominance of the type 1 wav gene locus in strains associated with clinical cholera, suggesting that a specific core OS structure could contribute to V. cholerae virulence.
Vibrio cholerae is a genetically diverse species that persists in aquatic ecosystems and is often associated with plankton and other aquatic organisms (13). V. cholerae is classified on the basis of biochemical tests and DNA homology studies and is further subdivided into serogroups based on the antigenicity of surface polysaccharides (20). Today more than 193 serogroups are known (72). The ability to cause pandemic cholera is mainly restricted to the nonencapsulated serogroup O1, which is further subdivided mainly into two serotypes (Inaba and Ogawa) and biotypes (classical and El Tor). However, during 1992 and 1993, cholera-like outbreaks in Asia were caused by strains of serogroup O139. Molecular and epidemiological analyses, as well as phage typing, revealed that O139 strains are highly related to the O1 El Tor strains. Hence, it is assumed that the epidemic O139 strains were derived from O1 El Tor strains (for a review see reference 20), differing specifically in the genes encoding for the synthesis of a novel type of cell surface polysaccharide (71). In particular, it was found that the genes encoding the O1 antigen had been replaced by a capsule gene locus carrying the genes involved in the synthesis and transport of the O139 antigen and the O139 capsule (reviewed in reference 64). It was also determined that the structures of the O139 antigen and the O139 capsule were identical (36, 38).
There is good evidence that pandemic V. cholerae O1 strains have become adapted to the human intestine by acquisition of virulence factors. Two known factors are cholera toxin (CT), encoded by the filamentous phage CTXΦ, and the toxin-coregulated pilus (TCP), encoded by a pathogenicity island (VPI) (reviewed in reference 20). Most of the environmental V. cholerae strains lack both of these virulence factors; however, there are reports of environmental non-O1, non-O139 strains that are positive for CTXΦ and VPI or variants of VPI (18, 19, 45, 46, 49). Such CT- and TCP-positive non-O1, non-O139 strains can cause severe cholera-like symptoms, but they have been associated only with local outbreaks or isolated cases (18, 45, 60). Two known examples of larger outbreaks of cholera-like diarrheal disease were cases in Sudan in 1968 caused by serogroup O37 (1, 6) and in Czechoslovakia in 1965 caused by a nontyped strain (74). Interestingly, there is also evidence that the pathogenic O37 Sudan strain was derived from an O1 classical strain by genetic exchange of the O-antigen biosynthesis gene cluster (6). It is presently not completely understood what other genetic determinants of the pathogenic O1 strains are responsible for the ability to cause cholera pandemics or if the structure of the surface polysaccharide per se contributes to virulence. CT- and TCP-negative V. cholerae strains can also occasionally cause diarrhea and extraintestinal infections such as bacteremia (17, 47). Not much is known about the virulence mechanism(s) of such strains, although some virulence factors like the RTX toxin (11, 39) or heat-stable enterotoxin (5) are believed to play a role.
Lipopolysaccharide (LPS) consists of three main regions: the lipid A region, the core oligosaccharide (OS), and the O antigen. The O antigen is the immunogenic portion and is known to contribute to the pathogenesis of O1 and O139 V. cholerae strains by facilitating colonization (4, 42, 70). The lipid A region is the highly conserved portion anchored in the outer membrane and is essential for outer membrane integrity (42). The core OS region is also known to have an essential role in maintaining outer membrane stability (27); however, its contribution to V. cholerae virulence has only begun to be investigated (51). The structure of the V. cholerae core OS region has been resolved for two O1 (classical; smooth and rough) strains, two O139 (encapsulated and nonencapsulated) strains, two O22 strains, and one non-O1, non-O139 isolate, H11 (7, 14-16, 37, 38, 68). The O1, O139, and O22 core OS structures are very similar, while the structure of the non-O1, non-O139 isolate H11 differs significantly in side branches (compare Fig. 1A and B). For the core OS of serogroups O1, O139, and O22 it was found that one d-fructose is linked to the d-glucose residue on HepI (15, 16, 37, 38, 68), while in the H11 isolate d-sedoheptulose is found instead at the same position (7, 68). Other basic differences between the O1, O139, and O22 core OS and the H11 core OS are the presence of a terminal heptosyl-IV residue and the linkage position of the O antigen. The attachment site of the O1 antigen to the core OS is unknown, although it is possible that it is linked to the heptosyl-III residue, as has been shown for O22 and O139 isolates (37, 38). In the core OS of strain H11 two additional residues, d-galactose and d-sedoheptulose, are linked to the heptosyl-III residue, providing the acceptor for the O antigen (7). In contrast to the structural analysis, virtually nothing is known about the core OS biosynthetic pathway in V. cholerae; the gene products involved have not been characterized so far, and nothing is known about the genetic organization of the corresponding wav genes.
FIG. 1.
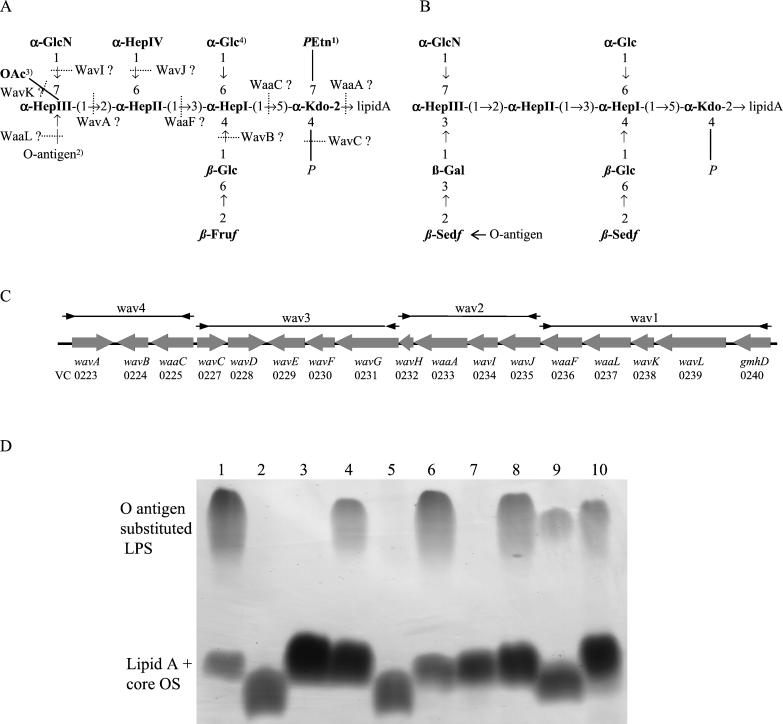
Comparison of the V. cholerae LPS core OS structures and the wav gene cluster. (A) LPS core OS backbone proposed for V. cholerae O1, O139, and O22. The representation is based on the structural analysis of two O1, two O139, and two O22 isolates (15, 31, 37). 1, 2-Aminoethyl phosphate (PEtn) on core OS of one O139 strain (38) and one O22 (37) strain (not reported for the other strains). 2, The O antigen is 1→3 linked in O22 strains (15, 37) and one O139 isolate, whereas a 1→2 linkage was reported for another O139 strain (14, 16). 3, An O-acetyl group in this position was found in O22 strains (15, 37). 4, A second glucose 1→6 linked to this Glc residue was reported for one O22 strain (15) and one O139 strain (16). Hep, l-glycero-d-manno-heptose; Kdo, 3-deoxy-d-manno-octulosonic acid; Sed f, sedoheptulose (D-altro-heptulose); GlcN, N-acetylglucosamine. (B) LPS core OS structure of the non-O1, non-O139 V. cholerae strain H11 (7). (C) Genetic organization of the putative wav gene cluster as deduced from the sequence of the V. cholerae O1 El Tor strain N16961 (26). Southern hybridization probes wav1 to wav4 are indicated by horizontal lines. (D) SDS-PAGE analysis of LPSs from V. cholerae P27459-S (wild type) (lane 1), P27459res118 (lane 2), P27459res118 pACYC waaF (lane 3), P27459res118 pBAD waaLF (lane 4), P27459 waaF::pGP (lane 5), P27459 waaF::pGP pACYC waaF (lane 6), P27459 waaL::pGP (lane 7), P27459 waaL::pGP pBAD waaL (lane 8), P27459 wavB::pGP (lane 9), and P27459 wavB::pGP pAK wavB (lane 10).
In this work, we describe the identification of the putative wav gene cluster of V. cholerae O1, O139, and several non-O1, non-O139 isolates. Mutational and complementation analysis allowed the identification of two core OS biosynthetic enzymes, a putative β-1,4-glucosyl transferase and the putative heptosyl transferase II (encoded by wavB and waaF, respectively), along with the identification of the putative O-antigen ligase (encoded by waaL). Using a genetic approach, the putative wav gene cluster of the recently sequenced strain N16961 (O1 El Tor) (26) was compared with those of several pathogenic and nonpathogenic strains of O1, O139, and non-O1, non-O139 isolates. From this analysis we can deduce that the O1 El Tor wav gene cluster is highly conserved among O1, O139, and non-O1, non-O139 isolates associated with clinical cholera, as well as among O1 environmental strains, whereas distinct wav cluster types can be defined for the environmental and human non-O1, non-O139 isolates. These data show evidence for shuffling of putative wav genes, hence predicting structural diversity of the core OS among V. cholerae strains.
MATERIALS AND METHODS
V. cholerae strains.
A total of 38 wild-type V. cholerae strains isolated from clinical, human, and environmental sources were used in this study (Table 1). The V. cholerae non-O1, non-O139 strains first described in this study (Table 1) were tested for growth as yellow colonies on thiosulfate citrate bile salt agar (Difco, Heidelberg, Germany) at 37°C and for being oxidase positive (66). The non-O1, non-O139 status was confirmed by absence of slide agglutination with antisera against O1 (Difco) and O139 (see below) and also in Western blot analysis with purified LPS using the same antisera. In addition, we performed PCR analysis for all non-O1, non-O139 strains to confirm the presence of the V. cholerae-specific gene ompW (48) and Southern blot analysis to test the presence of the Vibrio-specific virulence-associated genes ctxAB, tcpA, and toxR. The relationship of all strains was investigated by DNA fingerprinting analysis with IS 1004 as described by Bik et al. (6).
TABLE 1.
Prevalence of virulence-associated genes and wav gene cluster types among V. cholerae isolates
| Isolate no. | Strain | Serogroup, biotype, or serotype | Source of isolationa | Reference or source | tcpAb | ctxb | toxRb | ompWc | wav cluster type |
|---|---|---|---|---|---|---|---|---|---|
| V62 | O395 | O1 classical Ogawa | c, 1964, India | 43 | Pos | Pos | + | + | 1 |
| V14 | C6709 | O1 El Tor Inaba | c, 1991, Peru | 69 | Pos | Pos | Pos | NDd | 1 |
| V19 | CO970 | O1 El Tor Ogawa | c, 1994, India | 51 | Pos | Pos | Pos | ND | 1 |
| V22 | F1873 | O1 El Tor Inaba | c, 1993, Zaire | J. J. Mekalanos | Pos | Pos | Pos | ND | 1 |
| V29 | M799 | O1 El Tor | c, 1989, Hong Kong | 34 | Pos | Pos | Pos | ND | 1 |
| V30 | M804 | O1 El Tor | c, 1962, India | 33 | Pos | Pos | Pos | ND | 1 |
| V31 | M807 | O1 El Tor | c, 1966, Vietnam | 34 | Pos | Pos | Pos | ND | 1 |
| V32 | M817 | O1 El Tor | c, 1974, Chad | 34 | Pos | Pos | Pos | ND | 1 |
| V33 | MAK757 | O1 El Tor Ogawa | c, 1937, Celebes | 43 | Pos | Pos | Pos | ND | 1 |
| V95 | P27459 | O1 El Tor Inaba | c, 1976, Bangladesh | 52 | Pos | Pos | + | + | 1 |
| V241 | 2559-78 | O1 | e, crab, Louisiana | 58 | Pos | Pos | + | + | 1 |
| V243 | 3223-74 | O1 | e, 1974, Guam | 58 | Neg | Neg | ND | + | 1 |
| V2 | AI1837 | O139 | c, 1993, Bangladesh | 28 | Pos | Pos | Pos | ND | 1 |
| V3 | A11838 | O139 | c, 1993, Bangladesh | 28 | Pos | Pos | Pos | ND | 1 |
| V9 | AI4450 | O139 | c, 1993, Bangladesh | 28 | Pos | Pos | Pos | ND | 1 |
| V58 | MO10 | O139 | c, 1993, India | 71 | Pos | Pos | + | + | 1 |
| V61 | MO3 | O139 | c, 1993, India | 71 | Pos | Pos | Pos | ND | 1 |
| V244 | V52 | O37 | c, 1968, Sudan | 6 | Pos | Pos | + | + | 1 |
| V207 | ATCC 25872 | Non-O1, non-O139 | c, 1965, Czechoslovakia | 35 | Pos | Pos | + | + | 1 |
| V215 | Ch18133 | Non-O1, non-O139 | e, 1981, Elbe river, Germany | 8 | − | − | + | + | 2 |
| V194 | A2-2 | Non-O1, non-O139 | e, 2000, Rio, Grande, Texas | K. Klose | − | − | + | + | 3 |
| V196 | B2-2 | O41 | e, 2000, Rio, Grande, Texas | K. Klose | − | − | + | + | 3 |
| V203 | Ch430 | Non-O1, non-O139 | e, 1972, water, Togo | 9 | − | − | + | + | 3 |
| V204 | Ch433 | Non-O1, non-O139 | e, 1972, fish, Togo | 9 | − | − | + | + | 3 |
| V246 | OA2-3 | O5 | e, 2000, Rio, Grande, Texas | K. Klose | − | − | + | + | 3 |
| V247 | CLP-1 | O36 | e, 2000, fish, Spain | K. Klosee | − | − | + | + | 3 |
| V202 | Ch359 | Non-O1, non-O139 | h, 1972, stool, Togo | This study | − | − | + | + | 3 |
| V210 | Ch780 | Non-O1, non-O139 | h, 1985, blood, Germany | This study | − | − | + | + | 3 |
| V211 | Ch821 | Non-O1, non-O139 | h, 1998, stool, Kenya | M. Kist | − | − | + | + | 3 |
| V209 | E8498 | O141 | e, 1978, water, Louisiana | 73 | Pos | Pos | + | + | 4 |
| V213 | Ch18922 | Non-O1, non-O139 | e, 1981, Elbe river, Germany | This study | − | − | + | + | 4 |
| V253 | I0259 | O53 | h, 1984 | 49 | Pos | Pos | ND | + | 4 |
| V208 | Ch762 | Non-O1, non-O139 | h, 1983, blood, Malta | This study | − | − | + | + | 4 |
| V198 | Ch84 | Non-O1, non-O139 | ?, 1970, Japan | R. Sakazaki | − | − | + | + | 4 |
| V192 | O83 | O6 | e, 1993, water, Argentina | K. Klosef | − | − | + | + | 5 |
| V195 | B2-3 | Non-O1, non-O139 | e, 2000, Rio, Grande, Texas | K. Klose | − | − | + | + | 5 |
| V205 | Ch457 | Non-O1, non-O139 | e, 1972, water, Togo | 9 | − | − | + | + | 5 |
| V242 | I528-89 | Non-O1, non-O139 | e, 1979, oyster, Louisiana | 58 | Neg | Neg | + | + | 5 |
c, clinical isolate (the strains were reported to be isolated from patients with cholera symptoms); e, environment; h, human (these strains were isolated from patients; however, there is no information available about clinical manifestations); ?, no information about the source of isolation was available.
Pos and Neg, presence and absence of the indicated virulence-associated genes according to the literature; + and − presence and absence of the indicated gene as determined by Southern blot analysis. The template for amplification of tcpA, ctxAB, and toxR was chromosomal DNA prepared from O1 El Tor strain P27459-S. tcpA was amplified using primers KAR24 and KAR25 (35); ctxAB was amplified using primers ctxA (CTGTTAAACAAAGGGAGCAT) and ctxB (GCAGTAATACATGTTTGGGC), and toxR was amplified using primers PstIToxR (AACTGCAGAGTGTTGGGACAGGGAGATA) and SmaIToxR (TCCCCCGGGCCATGGCGATGTGTCTATTT).
The presence of the V. cholerae-specific gene ompW was shown by PCR analysis as described by Nandi et al. (48).
ND, not determined.
In collaboration with C. Osorio.
In collaboration with M. Waldor.
Escherichia coli strains and growth conditions.
E. coli K-12 strains LE392 (61) and XL-1 (New England Biolabs, Schwalbach, Germany) were utilized for all genetic manipulations, unless the vector being used was a derivative of pGP704, in which case E. coli SM10λpir (44) was used. All strains were grown in Luria broth at 37°C, except as noted otherwise. Antibiotics were used at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 50 or 100 μg/ml; streptomycin, 100 μg/ml; and chloramphenicol, 30 μg/ml (E. coli) and 2 μg/ml (V. cholerae). Strains containing pBAD18-Km (25) derivatives were cultivated under either inducing (with l-arabinose [0.002%, wt/vol]) or repressing (with glucose [0.2%, wt/vol]) conditions. The expression of wavB from plasmid pAK wavB was induced in the presence of 0.3 μg of anhydrotetracycline (Acros Chimica) per ml.
Construction of plasmids.
Internal fragments of waaL, waaF, or wavB were generated by PCR amplification with primers containing EcoRI and SalI restriction sites (underlined below). These fragments were then digested with EcoRI and SalI and ligated into the suicide vector pGP704 (44), which was digested with the same restriction enzymes. Plasmid pGP waaL was constructed using primers waaLintEcoRI (GGAATTCCAACCCGTTCTTGTATACGC) and waaLintSalI (TTACGCGTCGACCAGGCATTCGTGCTCTGTTA), plasmid pGP waaF was generated with primers waaFintEcoRI (GGAATTCCGATGACGAGTTTAGGTCTT) and waaFintSalI (GCAAGTCGACTTTGGAACGCATGCCTGAGG), and plasmid pGP wavB was constructed using primers wavBintEcoRI (GGAATTCGGACGATGCCTTGAGAAAG) and wavBFintSalI (TTACGCGTCGACTCGATTCTGGCAGTCACGA).
To construct complementing plasmid pACYC waaF, the gene-specific oligonucleotides waaF PstI (AAAACTGCAGTACATCGCAGCCAAAAGAGC) and waaF FspI (GAAAATGCGCAGCACCTTTTTCAAACCAGAGG) were designed to introduce PstI and FspI sites (underlined) at the 5′ and 3′ ends of waaF. Following PCR amplification, the product was digested and ligated into the PstI- and FspI-opened plasmid pACYC177 (57). The resulting plasmid, pACYC waaF, expresses waaF from the bla promoter.
For the construction of complementing plasmids pBAD waaL and pBAD waaLF, the waaL gene or the waaL and waaF genes were PCR amplified, using primers waaL NheI (CTAGCAGCTAGCATTAGTTGGAACACGACCCT) and waaL SalI (ACGCTGTCGAC ATATCGCCAACCCAAGAAGG) or primers waaL NheI and waaF SalI (ACGCAAGTCGACAGCACCTTTTTCAAACCAGA). The obtained PCR products containing waaL or waaL and waaF were cloned downstream of the PBAD promoter into plasmid pBAD18-Km (25).
For complementation of wavB in trans a tet promoter-based vector system of pZA31-luc (41) was developed. First, pZA31-luc was modified in order to contain the tetR gene, which was amplified from E. coli strain XL-1 with primerstetR SacI HincII (TTACGTGAGCTCGAGTGTCAACAAAAATTAGG) and tetR SacI HincII (TTACGTGAGCTCAGGGTGGTTAACTCGACATC). The tetR-containing DNA fragment was digested with HincII and inserted into a blunted SacI site of pZA31-luc. Second, the luc gene was replaced with the polylinker multiple cloning site of pBluescript II KS (Stratagene Europe, Amsterdam, The Netherlands) via partial deletion of the luc gene (KpnI-HincII fragment) and subsequent insertion of the polylinker with KpnI and PvuII sites, resulting in plasmid pAKtetR. Third, wavB was PCR amplified with oligonucleotides wavB FspI (GAAAATGCGCATACACCTTTTATACCCAGAT) and wavB HindIII (GAAAGCTTGGGTCGGATTGATATGA), digested with restriction enzymes FspI and HindIII, and subsequently ligated into the HindIII and HincII sites of the expression plasmid pAKtetR. This construction resulted in plasmid pAKwavB.
Construction of mutant strains.
To construct strains containing a mutation in waaL, waaF, or wavB, plasmid pGP waaL, pGP waaF, or pGP wavB was mated by conjugation from E. coli SM10λpir into V. cholerae P27459-S (50) and subsequently selected for streptomycin and ampicillin resistance. The resulting mutant strains had a chromosomal insertion in the gene of interest due to the integration of the plasmid through homologous recombination via the internal gene fragment (44). The correct chromosomal insertion for all mutants was confirmed by Southern blot analysis (data not shown).
LPS analysis.
For screening purposes LPS was isolated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and silver stained or blotted as described previously (50). In other cases, LPS was prepared by the proteinase K digestion method of Hitchcock and Brown (29), separated on SDS-16.5% polyacrylamide gels, and visualized by silver staining as described by Tsai and Frasch (67).
O139 serum.
O139 antiserum was prepared using strain MO45, obtained from Y. Takeda (Tokyo, Japan). A specific-pathogen-free New Zealand White rabbit was immunized by three injections of heated (1 h at 100°C) cell suspensions in normal saline, followed by two injections of cells inactivated in 0.5% formalin-NaCl. The titer against both heated and formalin-inactivated cells of V. cholerae O139 was 1:1,280. The serum then was absorbed with heated (1, h 100°C) antigen of V. cholerae Inaba, strain NIH 35 A3, and tested for reactivity by slide agglutination against three strains of V. cholerae O139. Negative controls included classical and El Tor strains of V. cholerae O1 as well as V. cholerae rough (CA385) and O22; the latter two strains were also received from Y. Takeda.
Southern hybridization.
Southern blotting was performed as described by Southern (63). Briefly, chromosomal DNA was prepared as described by Grimberg et al. (24), digested with appropriate restriction enzymes, fractionated on an agarose gel (0.7%), and transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech, Freiburg, Germany). DNA probe labeling and hybridization were performed by using the ECL direct nucleic acid-labeling and detection system (Amersham Pharmacia Biotech). The hybridization buffer contained NaCl (0.5 M), and high-stringency washing steps were performed for all probes at 42°C in a buffer containing standard saline citrate (0.5%), SDS (0.4%), and urea (6 M).
PCR.
PCRs were performed using an Mastercycler gradient PCR thermocycler (Eppendorf, Hamburg, Germany). Amplifications for the detection of wav genes in the different V. cholerae strains were carried out using Taq polymerase (Supermix; Gibco BRL Life Technologies GmbH, Karlsruhe, Germany). ELONGASE enzyme mix (Gibco BRL Life Technologies GmbH) was used for fragments of >1.5 kb and for cloning. Herculase enzyme mix (Stratagene) was used as the polymerase enzyme in PCRs where products were used for sequencing. Primers specific for each open reading frame (ORF) of the different putative V. cholerae wav gene clusters were designed with Primer3 www primer tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and are listed in Table 2. Primers were purchased from MWG-Biotech (Ebersberg, Germany).
TABLE 2.
Primers used for wav gene amplification
| ORF | Type or straina | Primer (5′→3′)
|
bp | |
|---|---|---|---|---|
| Sense | Antisense | |||
| wavA | 1-5 | GGCAAAATCAACTAACAATCCGGb | GGCGCAAGCTCAGTCAATAC | 1,396 |
| wavB | 1-5 | wavBintSalI | wavBintEcoRI | 538 |
| waaC | 1-5 | TCCCTCTGTATCCTGCGTTT | ACCTGTGTGGCCATGATGT | 1,031 |
| wavC | 1 | CTGCAAAACTACCGGATAACGc | AGCAAACGCTTCAAGACTCC | 713 |
| wavC | 1-5 | CTGCAAAACTACCGGATAACG | TCCATACCTTCTCTTGGTCA | 582 |
| wavD | 1 | ACACCGTCCATTACCTCCAC | TTCTTAATTTTAGCCTAACTCCTTTCC | 880 |
| wavE | 1-4 | TTGATGATGAAGCGATCACC | GCAGCCAATCCTTAAGGTCA | 816 |
| wavF | 1-4 | GGGAATGAGTTCTCGCTTCTT | GGCTGCCTCAAAAAGTCAGT | 714 |
| wavG | 1-4 | ATGTCTGGTGCCTATGTTGG | AAAGGCAACATGGAGAGGAA | 1,487 |
| wavH | 1-4 | GGCACTCTCACCCAAGCTAA | AAATTCGGAAGGGCGCACGG | 294 |
| waaA | 1-4 | GTATGGCCTCTATCGCCGTA | TTTTCGAGAGCTCCACGATT | 1,182 |
| waaA | 5 | TCCGTGGCCTCTATACCTTG | TTTTGGATAGAACCAGACGATTC | 1,237 |
| wavI | 1-4 | GTATTGATCGCCAACCGACT | CGAGATCTTCGGGATTGATG | 757 |
| wavI | 5 | TGATTCGTCTGACTGGCAAC | TTTGAACGAGGGCGATCTAT | 712 |
| wavJd | 1 | CATGAAACACCTCTGGTTTG | CGAAAGGGAACCGTAGCATA | 912 |
| waaF | 1 | waaFPstI | waaFFspI | 1,114 |
| waaF | 2-5 | TGATTGTAGGCCCTTCTTGG | TAGCGCTTCAATGACACGAG | 1,004 |
| waaL | 1 and 2 | waaLNheI | waaLSalI | 1,295 |
| waaL | V194 | TTTCTCCACTAATTGTTCTGCTG | TACAGGACTACTTGAATCAC | 743 |
| waaL | 5 | TGGAATAAACATCGGCATTACA | CATAGCGCCAACAAAGAGAA | 1,104 |
| wavK | 1 and 2 | ATGGCTGGCTTTGGTTTAAT | AATGCAATACTGGCCAATCG | 495 |
| wavL | 1-4 | ATGAATATTTTGATGGCCCT | ATTAAACCAAAGCCAGCCAT | 1,806 |
| wavL | 5 | TTCTAATGGCCCTATCCCAAC | TCTTGGAAAAATCGCAATCC | 1,773 |
| gmhDd | 1-5 | ATGATTATCGTAACTGGCGG | TTACTTACGATTAATCAGCG | 944 |
| wavM | V194 | TGCATCAATTGAACAGCAAA | TAGATTTCTCGCGTCGGTTT | 630 |
| wavN | 4 | GATTGGATCCCTGGAGGATT | TGCTAATACTCCCGCACCTT | 1,881 |
| wavNO | 4 | CCAAAAACGCTTTTTAACTG | GATTGGATCCCTGGAGGATT | 2,800 |
| wavP | 5 | GATGTCGAGGCATTAAACTCG | ATTGGTCGATCCCCTTCTTG | 769 |
| wavQ | 5 | GCAAAAATGGTGGGGTAGTT | ACACACTCTTCTGAACGACGA | 841 |
| wavR | 5 | TCCCATTTAACAAACCACCA | TTTCTTCCCGCTACTGTGCT | 991 |
| wavS | 5 | ACAGATCGGCGATGTCAGTT | GTTGCGTTGCAACAGATAGG | 545 |
| wavT | 5 | ACACAAATGCGTTGAGATGC | TGCTTCTTTGTAGCCATTGA | 776 |
Primers give positive signals for the indicated strain or type of wav gene cluster (Fig. 1).
Primer binds in kdtB (VC0222), downstream of wavA.
Underlined primers are identical.
Annealing at 54°C; all others anneal at 56°C.
DNA sequencing and sequence analysis.
PCR amplification products were prepared for sequencing by using an Amicon Microcon PCR centrifugal filter (Millipore, Eschborn, Germany). DNA sequencing was performed by the dideoxynucleotide chain termination method of Sanger et al. (59) using the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech). Fluorescently labeled DNA primers were purchased from MWG-Biotech. DNA separation and data collection were performed with the LiCor automated sequencing system (MWG-Biotech). DNA sequencing of the PCR fragments derived from strain V192 was done by GATC GmbH (Konstanz, Germany) by use of a primer walking strategy. DNA sequence and protein feature analyses were carried out with tools from the Online Analysis Tools site at http://www.queensu.ca/micr/faculty/kropinski/online.html. Assembly of the DNA sequences was performed with the online program CAP (www.infobiogen.fr/services/analyseq/cgi-bin/cap_in.pl) (32). An ORF search was performed with the National Center for Biotechnology Information (NCBI) Orf Finder, and the ORFs were subsequently subjected to a database search using the BlastX program (version 2.1.2. [2] via the NCBI server). Transmembrane domains were detected using the Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html) (30) and HMMTOP (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (62) online programs. Multiple-sequence alignment was performed at the BCM Search Launcher site (http://searchlauncher.bcm.tmc.edu/) using ClustalW 1.8 and Boxshade (http.//www.ch.embnet.org/software/BOX_form.html). Pairwise sequence alignment was performed using the ALIGN program at the GENESTREAM network server IGH, Montpellier, France (http://www2.igh.cnrs.fr/bin/align-guess.cgi) (53).
Nucleotide sequence accession numbers.
The nucleotide sequences of the waaL genes and the specific DNA fragments for the type 2 to 5 wav gene cluster have been deposited in GenBank. The accession numbers are AF443420 (waaL, strain P27459), AF443421 (waaL, strain O395), AF443422 (waaL, strain V243), AF443423 (waaL, strain MO10), AF443424 (waaL, strain V244), AF443425 (waaL, strain V207), AF443426 (wavC to wavE region of strain V215; type 2), AF443845 (wavI to wavK region of strain V215; type 2), AF444793 (wavC to wavE region of strain V194; type 3), AF444794 (wavI to wavK region of strain V194; type 3), AF443847 (wavC to wavE region of strain V209; type 4), AF444792 (wavI to wavK region of strain V209; type 4), AF443846 (wavI to wavK region of strain V208; type 4), AF449195 (wavC to waaF region of strain V192; type 5), and AF449194 (waaF to gmhD region of strain V192; type 5).
RESULTS
Putative O1 El Tor core OS biosynthesis gene cluster.
Since nothing was known about the organization of the genes responsible for the synthesis of the V. cholerae LPS core OS, we used the recently released genome sequence of the O1 El Tor strain N16961 (26) and analyzed it for the presence of putative core OS biosynthesis genes. Computer analysis suggested that, as in Enterobacteriaceae, the corresponding genes in V. cholerae are clustered together in a region located on chromosome 1, comprising ORFs VC0223 to VC0240 (Fig. 1C). Not shown is ORF VC0222 (left border), whose product shows high similarity to KdtB of E. coli. This protein was recently characterized as being involved in coenzyme A biosynthesis and was renamed CoaD (22). Upstream of VC0240 the O1 antigen biosynthesis gene cluster (rfb) is localized, indicating that in V. cholerae most of the LPS biosynthetic genes are clustered.
According to the new nomenclature system for genes involved in bacterial polysaccharide biosynthesis (54), we designate this putative V. cholerae core OS biosynthesis locus the wav gene cluster. One gene of the wav gene cluster, gmhD, was previously proposed by others to encode the ADP-l-glycero-d-manno-heptose epimerase, involved in the synthesis of the activated heptose precursor (65), and was not renamed by us. All other putative core OS biosynthetic genes were designated wav for genes specific for V. cholerae or waa if homology or experimental data significantly matched already existing Waa protein information or function. Data summarizing the characteristics and proposed functions of the core OS biosynthetic genes are shown in Table 3. The deduced protein sequence of VC0237 did not show high levels of similarity to other proteins in the database. However, it shares typical secondary structure properties, i.e., nine potential membrane-spanning domains, a large periplasmic loop, and a similar hydropathy profile, with several known WaaL enzymes (27) (data not shown). These shared properties and our own experimental characterization (see below) make VC0237 a good candidate to encode the lipid A core:surface polymer ligase, and therefore the corresponding gene was designated waaL. In summary, the putative functions of 10 deduced proteins involved in LPS core assembly could be predicted by the known core OS structure and are summarized in Fig. 1A. At least three additional transferases would be required for completion of the core OS. One is probably encoded by wavL, and it is possible that two others are encoded by the ORFs with no assigned function (wavD, -E, -F, -G, or -H) or that they are encoded outside this locus.
TABLE 3.
Putative core OS biosynthesis gene products of V. cholerae
| ORF straina | New ORF designation | Motifs present in deduced protein sequence | Predicted putative function in core OS assembly | Related sequences (BlastP) | Accession no. or reference | % Identity | wav typesb |
|---|---|---|---|---|---|---|---|
| VC0223 | wavAc | Glycosyl transferase 9 (CDd) | HepIII transferase | PM1294 | AAK03378 | 58 | 1, 2, 3, 4, 5 |
| WaaQ, Haemophilus ducreyi | AAF72875 | 55 | |||||
| VC0224 | wavB | Glycosyl-transferase 2 (CD) | B1,4-Glucosyltransferase | PM1306 | AAK03390 | 68 | 1, 2, 3, 4, 5 |
| LgtF, H. ducreyi | AAF72876 | 67 | |||||
| VC0225 | waaC | Glycosyltransferase 2 + 9 (CD) | HepI transferase | OpsX, Haemophilus influenzae | B64058 | 46 | 1, 2, 3, 4, 5 |
| RfaC, Helicobacter pylori | AAB65778 | 25 | |||||
| VC0227 | wavC | Protein kinase (CD) | Kdo kinase | KdkA, Photobacterium damselae | BAB72027 | 55 | 1, 2, 3, 4, 5 |
| KdkA, H. influenzae | CAC07181 | 48 | |||||
| VC0228 | wavD | Unknown | VC0229 | 31 | 1 | ||
| VC0229 | wavE | Unknown | ORF, P. damselae | BAB72032 | 37 | 1, 2, 3, 4 | |
| VC0228 | 31 | ||||||
| VC0230 | wavF | UPF007 (CD) | Unknown | ORF, P. damselae | BAB72033 | 63 | 1, 2, 3, 4 |
| BcbE, Pasteurella multocida | AAF67267 | 46 | |||||
| VC0231 | wavG | Unknown | ORF, P. damselae | BAB72034 | 53 | 1, 2, 3, 4 | |
| BcbG, P. multocida | AAF67269 | 34 | |||||
| VC0232 | wavH | Unknown | ORF, P. damselae | BAB72026 | 78 | 1, 2, 3, 4 | |
| BcbF, P. multocida | AAF67268 | 64 | |||||
| VC0233 | waaA | Glycosyltransferase I (CD) | Kdo transferase | KdtA, P. damselae | BAB72028 | 51 | 1, 2, 3, 4, 5 |
| WaaA, Salmonella enterica serovar Typhimurium | AAC16417 | 40 | |||||
| VC0234 | wavI | Glycosyltransferase family 32e | Glycosyltransferase | PM1 1,16 | AAK03200 | 51 | 1, 2, 3, 4, 5 |
| VC0235 | wavJ | Glycosyltransferase 9 (CD) | HepIV transferase | WaaC, Bordetella bronchiseptica | CAA07672 | 21 | 1 |
| VC0225 | |||||||
| VC0236 | waaF | Glycosyltransferase 9 (CD) | HepII transferase | RfaF, Pseudomonas aeruginosa | AAG08397 | 53 | 1, 2, 3, 4, 5 |
| RfaF, Salmonella enterica serovar typhimurium | P37421 | 59 | |||||
| VC0237 | waaL | 9 TMHf | O-antigen ligase | 1, 2, 3, 4, 5 | |||
| VC0238 | wavK | O-acetyltransferase family 3g | O-acetyltransferase | LacA, Methanococcus jannaschii | AAB99067 | 30 | 1, 2, 3, 4 |
| VC0239 | wavL | Glycosyltransferase 1 + polysaccharide deacetylase (CD) | Glycosyltransferase | BME11603 | AAL52784 | 32 | 1, 2, 3, 4, 5 |
| RP344 | B71691 | 25 | |||||
| VC0240 | gmhD | Epimerase (CD) | ADP-l-glycero-d-manno-heptose-epimerase | RfaD, H. influenzae | AAC22768 | 73 | 1, 2, 3, 4, 5 |
| RfaD, E. coli | AAA24525 | 74 | |||||
| V194 | wavM | Glycosyltransferase 25 (CD) | Glycosyltransferase (galactosyl?) | Lex2B, H. influenzae HPO826 | AAA6037540 | 34 37 | 3, 4 |
| V209 | wavN | DUF33 (CD) | O-acetyltransferase | SMb20810, Sinorhizobium meliloti | CAC48950 | 28 | 4 |
| O-acetyltransferase family 2h | |||||||
| 9 TMH | PA5238 | E82991 | 28 | ||||
| WbpC, Neisseria meningitidis | AAF42171 | 31 | |||||
| V209 | wavO | O-acetyltransferase family 3g | O-acetyltransferase | ORF11, Campylobacter jejuni | AAF34147 | 42 | 4 |
| V192 | wavP | Glycosyltransferase 2 (CD) | Glycosyltransferase | ORF3, C. jejuni | AAK95997 | 26 | 5 |
| YP00187 | CAC89049 | 29 | |||||
| V192 | wavQ | Unknown | 5 | ||||
| V192 | wavR | pfam 01041 + pfam 00155 + pfam 01053 (CD) | Fuc4Nac pathway (TDP-4-oxo-6-deoxy-d-glucose transaminase) | WecE, S. enterica serovar typhimurium | AAL22774 | 65 | 5 |
| RffA, E. coli | P27833 | 64 | |||||
| YP03859 | CAC93327 | 62 | |||||
| V192 | wavS | Acetyltransferase | Fuc4Nac pathway | WecD, S. enterica serovar typhimurium | AAF33463 | 35 | 5 |
| YP03860 | CAC93328 | 36 | |||||
| V192 | wavT | Glycosyltransferase 2 (CD) | Glycosyltransferase (galactosyl?) | ORF3, C. jejuni | AAK91721 | 30 | 5 |
| CPE0481 | BAB80187 | 28 |
Either the ORF designation of the V. cholerae genome sequence (VC number) or the name of the sequenced strain (V number) used for sequence analysis is indicated.
V. cholerae wav gene cluster types carrying a similar gene.
This ORF was not named waaQ, since characterized enterobacterial WaaQ enzymes link HepIII α-1,7 to HepII (27), whereas the characterized WaaQ enzyme of H. ducreyi links HepIII α-1,2 to HepII (21) as also proposed for V. cholerae (Fig. 1A).
CD, conserved domain database at NCBI via BlastP.
VC0234 is listed in CAZY (http://afmb.cnrs-mrs.fr/|P5cazy/CAZY/index.html). Among the members of the glycosyltransferase family, 32 are characterized N-acetylglucosamine transferases, so it seems possible that WavI is involved in the linkage of α-GlcN to HepIII (Fig. 1A).
Number of predicted transmembrane helices (TMH).
WavK and WavO share conserved regions with the soluble family 3 of O-acetyltransferases (12).
WavN could be assigned to family 2 of large integral membrane O-acetyltransferases (12).
Characterization of the V. cholerae O1 El Tor waaL, waaF, and wavB genes and identification of a spontaneous phage-resistant waaLF mutant.
The recently described spontaneous phage K139.cm9-resistant O1 El Tor mutant strain P27459res118 showed an altered LPS core OS with no attached O antigen (Fig. 1D, lane 2) (50). We previously characterized this mutant as having a deep rough phenotype and as being unable to colonize the small intestine (51). We hypothesized that it may be mutated in the heptosyl transferase gene waaF. To test this hypothesis, we complemented this strain with a plasmid carrying the V. cholerae waaF homologue. In the presence of the waaF-expressing plasmid pACYC waaF, the core OS was restored to full-length core; however, the strain was still unable to ligate O antigen (Fig. 1D, lane 3). To determine the true phenotype of a waaF mutation, we constructed strain P27459 waaF::pGP by plasmid insertion (see Materials and Methods). LPS prepared from P27459 waaF::pGP migrates as far as that from the spontaneous mutant P27459res118 (Fig. 1D, lane 5), but this mutant could be complemented to make wild-type LPS in trans by plasmid pACYC waaF (Fig. 1D, lane 6).
To investigate the nature of the mutation in strain P27459res118 in more detail, we performed Southern blot analysis. The chromosomal DNA was cut with HindIII and XmnI and probed with a PCR-generated waaF fragment. Compared with the wild type, the restriction fragment generated by both enzymes showed a decrease in fragment length for the mutant res118, indicating a deletion in the waaFL-containing DNA fragment (data not shown). PCR analysis of the waaF-surrounding region with subsequent DNA sequencing confirmed a deletion of 546 bp, affecting both waaF and waaL (data not shown). In the presence of the plasmid pBAD waaLF, encoding both waaF and waaL in trans (see Materials and Methods), the LPS biosynthesis of mutant strain P27459res118 could be restored (Fig. 1D, lane 4). In addition, the waaL gene was inactivated by plasmid integration (see Materials and Methods), and SDS-PAGE analysis with purified LPS from this strain showed no O-antigen ligation, without altering the core OS mobility (Fig. 1D, lane 7). Complementation of the waaL strain with a waaL-carrying plasmid led to restored O-antigen attachment (Fig. 1D, lane 8). The absence of O antigen in the waaL mutants P27459 waaL::pGP and P27459res118 pACYC waaF was also confirmed in Western blot analysis with O1-specific antiserum (data not shown). Taken together, these data along with the computer analysis (see above) suggest that ORF VC0237 encodes the O-antigen ligase WaaL.
To determine the function of VC0224, wavB was inactivated by plasmid integration (see Materials and Methods). SDS-PAGE analysis with purified LPS indicates that the core OS of mutant P27459 wavB::pGP migrates faster (Fig. 1D, lane 9) than the core OS of the wild type. This mutant still ligates O antigen, as is also evident in Western blot analysis (data not shown), indicating that mutant LPS must be deficient in a side branch. The presence of the wavB-expressing plasmid pAK wavB in P27459 wavB::pGP restored the core OS defect (Fig. 1D, lane 10). Along with sequence homology, this mutant LPS phenotype in polyacrylamide gels suggests that wavB most likely encodes the β-1,4-glucosyl transferase. We predict that the LPS from the wavB mutant lacks the β-Fru-βGlc branch on the HepI residue (Fig. 1A), but proof of this awaits structural analysis.
Characterization of V. cholerae strains.
Each of the V. cholerae core OS structures that has been previously investigated shows unique structural features (Fig. 1A and B). Such structural differences should also correlate with genetic variations within the wav gene cluster. However, nothing is known about the extent of the genetic variations or about the distribution of core OS types within the species. To address this issue we investigated 38 different environmental, human, and clinical V. cholerae strains that were isolated at different times (1937 to 2000) and were widespread geographically (Table 1). The genetic relationship of the different isolates was investigated by DNA fingerprinting. It is known that epidemic strains of serogroups O1 and O139 show closely related IS1004 fingerprint patterns (6), which we have also observed for the investigated strains (data not shown). It was also reported that O37 strains isolated from the outbreak in Sudan show fingerprints closely related to those of O1 classical strains, and it was concluded that the toxigenic O37 strains may have been derived from O1 classical strains by genetic exchange of the O-antigen biosynthesis gene cluster (6). We found that strain V207, isolated from an outbreak in Czechoslovakia in 1965 (1), showed an IS1004 fingerprint pattern identical to that of the O37 strain, indicating a close genetic relationship between the two isolates (data not shown). In contrast, the 2 investigated environmental O1 strains and the 19 non-O1, non-O139 strains showed very polymorphic IS1004 fingerprint patterns, indicating that they were unrelated (data not shown). The environmental and human non-O1, non-O139 isolates were further examined for the presence of the virulence genes ctxAB, tcpA, and toxR in Southern blot analysis (summarized in Table 1). According to our hybridization results and previously published data, our collection of V. cholerae strains of different serogroups comprises 17 CT+ TCP+ strains associated with clinical cholera, 4 human CT− TCP− strains, 1 human CT+ TCP+ strain, 2 environmental CT+ TCP+ strains, 13 CT− TCP− environmental strains, and 1 CT− TCP− strain of unknown origin.
The putative O1 El Tor (type 1) wav gene cluster is highly prevalent among epidemic V. cholerae strains.
To determine the distribution of the wav genes, we performed Southern blot experiments. The chromosomal DNAs of all strains were digested with EcoRI, EcoRV, or HindIII and hybridized with the PCR-generated probes wav1 to wav4, which hybridize with the complete O1 El Tor wav region (Fig. 1C). The epidemic strains included nine O1 El Tor strains, one O1 classical strain, and five O139 strains, and the hybridization analyses showed identical restriction patterns with probes wav2 to wav4. Differences in the restriction fragment length between O1 and O139 strains were observed with probe wav1 (data not shown). This observation is due to sequence variations in the different O-antigen biosynthesis gene clusters, which are located immediately upstream of gmhD (64). These results indicate that the epidemic strains of serogroups O1 (El Tor and classical) and O139 are of the same wav gene cluster type. It remains to be established whether the reported minor structural differences between the core OSs of O1 and O139 epidemic strains (Fig. 1A) are due to technical limitations of the structural analysis, differences in the expression of wav genes, or sequence variations not linked with the putative wav gene cluster.
The other V. cholerae strains also hybridized with most of the probes; however, they do show extensive restriction length polymorphism. To further investigate the putative wav gene cluster of these strains, we performed PCR analysis. Based on the DNA sequence obtained from the V. cholerae genome database (The Institute for Genomic Research [TIGR]), we selected DNA primers specific for each ORF of the wav gene cluster (Table 2). The primer wavAsense binds to kdtB, the adjacent ORF upstream of the wav gene cluster, and therefore a positive PCR signal with primer wavAsense verifies the left junction site of the wav gene cluster. Positive signals for all ORFs were obtained in the seven reinvestigated epidemic isolates (four O1 El Tor, one O1 classical, and two O139) and also in four strains which showed differences in the Southern blot analysis (data not shown). The latter strains include the two O1 environmental isolates V241 (CT+ TCP+) and V243 (CT− TCP−) and the two clinical non-O1, non-O139 isolates V207 and V244 (CT+ TCP+), both of which were associated with massive outbreaks of diarrhea.
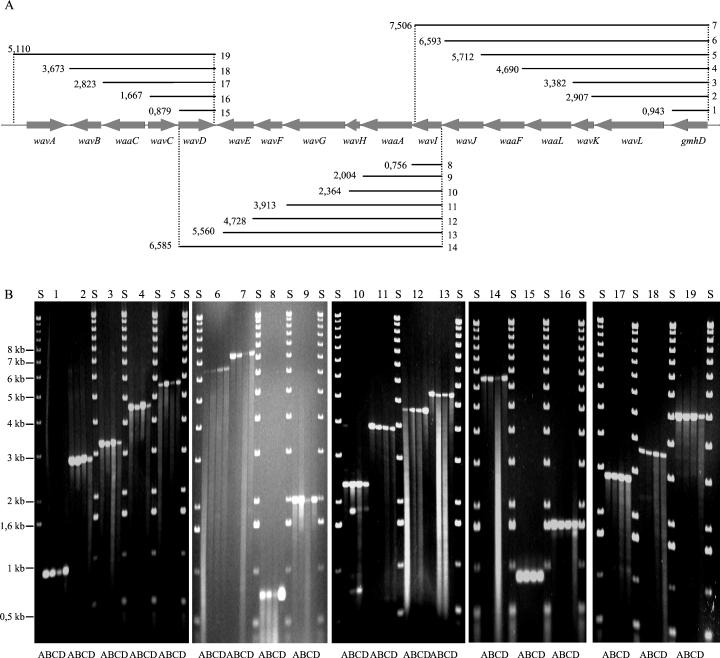
To confirm that the putative wav genes of distantly related strains, i.e., P27459 (O1 El Tor, pandemic), MO10 (O139, epidemic), V244 (O37, clinical), and V243 (O1, environmental, CT− TCP−), are identical in number, order, and orientation, we performed further PCR analysis. In such analyses, the DNA fragment lengths of the ORFs in relation to a chosen starting ORF were measured and compared to the calculated fragment lengths as deduced from the V. cholerae genome database (Fig. 2A). By using one PCR primer specific for each ORF (sense or antisense), it was possible to determine the orientations of all ORFs. Subsequent comparison of the PCR products obtained from the four investigated strains clearly showed that their wav gene clusters were identical (Fig. 2B).
FIG. 2.
Verification of the presence of the type 1 wav gene cluster in four different strains by PCR analysis. (A) Order and orientation of the wav genes and calculated theoretical length of PCR products 1 to 19. The primers used were ORF-specific sense or antisense oligonucleotides (Table 2). (B) Ethidium bromide stained-agarose gel showing the electrophoretic mobilities of PCR products 1 to 19 obtained from strains V243 (lanes A) (O1 environmental isolate), V244 (lanes B) (O37 clinical isolate), P27459 (lanes C) (O1 El Tor pandemic isolate); and MO10 (lanes D) (O139 epidemic isolate). Lanes S, molecular size standard.
Identification of four additional types of putative V. cholerae wav gene cluster.
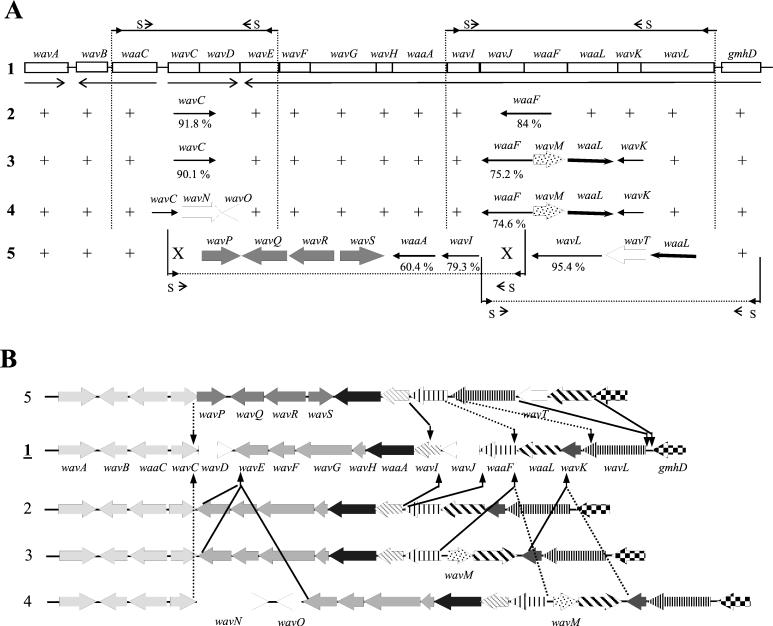
The remaining 19 environmental and human non-O1, non-O139 V. cholerae strains gave positive signals (PCR products) for some of the wav genes (Fig. 3A). The pattern of positive PCR products together with the Southern blot analysis provided evidence that the strains could be subdivided into four additional subtypes of wav gene clusters (designated types 2, 3, 4, and 5). To further investigate the organization of these putative wav gene clusters, we choose one strain from each predicted group and sequenced the unknown regions: strain V215 for type 2, V194 for type 3, V209 for type 4, and V192 for type 5 (Fig. 3A). The sequence data we obtained showed the presence of several type 1 genes that were not detectable in the PCR analysis. They show high identities to the respective O1 El Tor genes (indicated in Fig. 3A). Besides these conserved genes, several new ORFs were identified (Fig. 3A), and the proposed functions of the encoded proteins are described in Table 3. ORFs presumably encoding WaaL enzymes showed no significant homology to VC0237 (Table 4) or other proteins in the database. These genes were identified based on common protein secondary structure features shared with known or proposed O-antigen ligases (data not shown, see above). To verify that the ORFs detected by PCR and sequencing analysis are in the order and orientation proposed (Fig. 3A), we also performed PCR distance analysis with strains V215, V194, V20, and V192. In addition, the PCR data gave no evidence for the presence of additional ORFs (data not shown; summarized in Fig. 3B).
FIG. 3.
Organization of the different wav gene cluster types in environmental and human V. cholerae isolates. ORFs of type 1 wav genes are indicated and named. (A) PCR products were generated with primers waaCas and wavEs from strains V215 (type 2) (yielded a 2.8-kb product), V194 (type 3) (2.8 kb), and V209 (type 4) (5.8 kb) and subsequently sequenced. PCR products were also generated with primers wavIas and wavLs (V215, 5.4-kb fragment; V194, 6.4 kb; V209, 6.4 kb) and subsequently sequenced. DNA fragments for sequencing of the type 5 wav gene cluster were PCR amplified from strain V192 with primer pairs wavCs-waaFs (types 2 to 5) and waaFas (types 2 to 5)-gmhDs (Table 2). +, positive PCR signal with same sizes at gene position for the type 1 wav gene cluster; X, positive PCR signal with primers specific for type 2, 3, and 4 wav genes; S, position of sequencing primer used for initial sequence reactions. New genes found are indicated by large arrows; small arrows indicate a homologue to genes found in the type 1 wav gene cluster. Numbers below the gene name indicate percentage of identity at the nucleotide level for the respective O1 El Tor gene; this is indicated only if the gene was sequenced completely. Relatedness of waaLs is shown in Table 4. (B) Comparison of the five V. cholerae wav gene cluster types and possible rearrangement events. Arrows indicate the gene insertion or deletion regions of types 2 to 5 compared with type 1. The same shading and pattern were assigned to similar wav or waa gene types; new ORFs are indicated with name.
TABLE 4.
Similarity of WaaL enzymes
| WaaL | % Identity witha:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V243 (1) | V62 (1) | V95 (1) | V58 (1) | V244 (1) | V207 (1) | V215 (2) | O22b | V194 (3) | V209 (4) | V208 (4) | V192 (5) | |
| VC0 237 | 99.7 | 99.7 | 100 | 100 | 89.5 | 89.5 | 88 | 89 | 23.7 | 24.9 | 25.7 | 21.8 |
| V244 (1) | 100 | 92.2 | 93.2 | |||||||||
| V215 (2) | 95.7 | |||||||||||
| V194 (3) | 83.4 | 80.4 | 20.9 | |||||||||
| V209 (4) | 86.1 | |||||||||||
The type of the wav gene cluster is indicated in parentheses.
The wav gene cluster of one O22 isolate was partially sequenced along with the O-antigen biosynthesis gene cluster, but those authors did not annotate the wav genes (accession no. AB012957). One of the deduced ORFs shows clear similarity to waaL and therefore is referred to as waaL.
The type 2 wav gene cluster is represented by one isolate.
From our data we can conclude that the type 2 wav locus differ from type 1 only by the absence of two ORFs, wavD, with unknown function, and wavJ, encoding a putative HepIV transferase. None of the other non-O1, non-O139 strains showed a similar PCR product profile, indicating that in our collection only the environmental strain V215 contains a type 2 wav gene cluster.
Comparison of the O-antigen ligases WaaL derived from strains harboring the type 1 and 2 wav gene clusters.
Even though the core OS structures of type 1 and 2 strains seem to be slightly different, we could detect the same waaL gene in PCR analysis. To investigate the relationship of the WaaL enzymes in more detail, we sequenced the waaL genes of seven type 1 strains and V215 (type 2) (Table 4). The deduced protein sequences are of the same length (399 amino acids [aa]) and show a high degree of similarity at the amino acid level (88 to 100%) when compared to VC0237. Among the O1 strains (V243 [environmental], V62 [classical], and V95 [El Tor]) and the O139 strain, the waaL sequences are nearly identical, with the exception of one amino acid substitution in V243 at position 15 and one in V62 at position 348. In contrast, the clinical isolates collected from outbreaks in Sudan and (formerly) Czechoslovakia (V244 and V207) show more sequence diversity (40 amino acid exchanges along the entire sequence) when compared with VC0237. Interestingly, both sequences of V244 and V207 are identical (Table 4), which again, in addition to the IS1004 fingerprint pattern, supports the close genetic relationship of the strains. The putative WaaL enzyme of strain V215 (type 2 gene cluster) also shows several sequence differences compared with VC0237 of O1 El Tor type. However, the V215 subtype of WaaL seems to be closely related to the putative WaaL of an O22 serogroup isolate (95.7% identity [Table 4]) (accession no. AB012957).
The sequence differences among the type 1 and 2 waaL genes indicate a diverse genetic relationship between these strains, but there is evidence that the encoded proteins are functionally related. The WaaL proteins of O22 and O139 strains are 89% identical, but their linkage site to the core OS are identical based on structural data (Fig. 1A); therefore, it seems possible that the other O antigens of type 1 and 2 strains (O1, O37, and those of strains V207 and V215) are also linked to the HepIII residue in a 1,3 linkage.
Most environmental and human isolates contain variants of type 3 and 4 wav gene clusters.
The sequence data we obtained for strains V194 (type 3) and V209 (type 4) revealed that the strains have similar wavI-wavL regions that differ remarkably from type 1 wavI-wavL regions (Fig. 3A, compare rows 3 and 4 with row 1). Instead of the putative HepIV transferase gene wavJ of the type 1 strains, a putative glycosyl transferase gene, wavM, is present. The deduced WavM proteins of strains V194 and V209 share 64.9% sequence identity. The putative WaaL proteins of strain V194 and V209 are 83.4% identical to each other but are unrelated to the type 1 and 2 WaaL enzymes (Table 4).
Initial attempts to detect wavM and waaL among the strains of our collection were only partially successful, although primers specific for each type were used for PCR analysis. To gain more insight into possible sequence divergence, we sequenced the wavI-wavL region for strain V208 (type 4). The results led to the identification of new waaL and wavM alleles with high identity at the DNA and protein levels to those of V194 and V209 (Table 4; Fig. 4). Direct comparison of the three types of WavM proteins revealed that they have a mosaic structure: they share 99% identity in the first 84 aa of the N terminus and lower levels of identity at the C terminus (Fig. 4A). Comparison of the three types of WaaL proteins revealed sequence divergences distributed over the entire length (data not shown). Finally, we performed Southern blot analysis with gene probes (waaL or wavM) derived from strain V194 and found specific detection only of waaL and wavM in strains harboring the type 3 and 4 wav gene clusters. Because of the observed DNA sequence diversity, it remains to be resolved how many subtypes of wavM and waaL do really exist and whether such differences correlate with functional alterations in the encoded enzymes. In addition, the presence of wavK in all type 3 and 4 strains (Table 1) was confirmed in Southern blot analysis, and the presence of waaF was shown in PCR analysis.
FIG. 4.
Comparison of protein sequences, showing multiple alignment of WavM from strains V209 (type 4), V208 (type 4), and V194 (type 3) (A); of the WavC C terminus from strains V215 (type 2), V194 (type 3), V209 (type 4), and V192 (type 5) compared to VC0227 from the V. cholerae genome database (B); and of the C-terminal half of WavE from strains V215 (type 2), V194 (type 3), and V209 (type 4) compared to VC0229 (C). Identical amino acids are shaded black, and conservative changes are shaded gray.
In contrast to the shared wavI-wavL region, strains V194 and V209 showed sequence diversity in the wavC-wavE region. The hypothetical ORF wavD of type 1 strains is absent in both strains, but only V209 contains two additional ORFs, designated wavN and wavO. The sequence analyses provided good evidence that both wavN and wavO could encode O-acetyltransferases (Table 3). To distinguish between type 3 and type 4 wav gene clusters, the waaC-wavE regions of strains from our collection that were positive for wavM and waaL were further analyzed by PCR with primers wavEs and waaCas. We obtained characteristic 2.8-kb PCR products for type 3 strains and 5.8-kb PCR products for type 4 strains, and both PCR products were easily distinguishable from the 3.8-kb PCR products of wavD-containing type 1 strains. In addition, we designed primers to wavC, wavN, and wavNO in order to confirm the presence of these genes. Based on our results, most of the non-O1, non-O139 isolates investigated carry the type 3 or 4 gene cluster or at least variants of them. These include the human CT- and TCP-negative isolates, as well as the human and environmental CT- and TCP-positive strains (Table 1).
V. cholerae isolates harboring the type 5 wav gene cluster.
The additional PCR analyses with primers specific for type 2, 3, and 4 wav genes led also to the detection of wavC and waaF by PCR in the type 5 strain V192 (Fig. 3A). Since only six genes could be detected by PCR, it appeared that the type 5 wav gene cluster is evolutionarily the most unrelated of the wav loci known so far, and indeed the sequence analysis of strain V192 revealed the presence of six new ORFs. Two of the new ORFs, designated wavP and wavT, encode putative glycosyl transferases (Table 3). The gene products of wavR and wavS show significant similarity to different proposed or defined WecE and WecD enzymes (Table 3). In E. coli both proteins are involved in the synthesis of TDP-fucosamine (TDP-4-acetamido-4,6-dideoxy-d-galactose) from TDP-4-keto-6-deoxy-d-glucose (55). It was not possible to determine a role for the hypothetical ORF wavQ because it has no homology to known proteins in the database, whereas another ORF, designated waaL, which also has no homology to other known proteins probably encodes a new variant of V. cholerae O-antigen ligase (see above). Finally PCR and Southern blot analysis with new designed primers for the type 5 genes showed clearly that the environmental strains V195, V242, and V205 also contain the type 5 wav gene locus (data not shown).
Relationship among different wav gene clusters and evidence for horizontal gene transfer.
As summarized in Fig. 3B, our data show evidence of genetic exchange within the V. cholerae wav gene cluster. By comparing the gene clusters (type 1 or 2 versus type 3 or 4) it appears that wavM and waaL (type 3 or 4) replaced waaL (type 1 or 2). In the adjacent wavK, the deduced C-terminal 29 aa of VC0238 show only 44.8% identity to WavK of strains V194, V209, and V208, whereas the remaining portion of the protein sequence is almost identical (data not shown). This could indicate the position of one crossover during genetic exchange; the other seems to be located close to the start codon of waaF. This is suggested by the fact that in O1 El Tor N16961 (type 1) there is an intergenic region of 2 bp between waaF and waaL, while in strains V209, V208, and V194 (type 3 or 4) there is 181 bp between waaF and wavM.
The waaL region of the type 5 wav gene cluster seems to be even more heterogeneous, as we observed that waaL and wavT were inserted between wavL and gmhD. The insertion of waaL (type 5) and wavT seems to have occurred simultaneously with the deletion of waaL and wavK of type 1 strains, leading to small sequence differences in the adjacent ORFs, waaF and wavL. The last C-terminal 7 aa of WavL and the first 5 aa of the N terminus of WaaF from strain V192 (type 5) are different than those of the sequenced O1 El Tor strain (type 1) (data not shown).
Another remarkable divergence can be observed in the 3′ end of the wavC genes, leading to differences in the length and composition of the deduced protein sequence (Fig. 4B) and indicating one end of a likely genetic crossover event (Fig. 3B). The site of the second putative crossover location differs from strain to strain. For strain V192 (type 5), we identified a replacement of the region containing wavDEFGH with four new ORFs (wavPQRS), along with a remarkable divergence in the otherwise highly conserved allele of waaA (57.1% identity to type 1 at the amino acid level). This observed DNA divergence ends at bp 444 of wavI. In strains harboring type 2, 3, and 4 wav gene clusters, wavEFGH are present. In these cluster types the location of a second crossover was found in the 3′ region of wavE, leading also to differences in the deduced C-terminal protein sequence and length of the protein (Fig. 4C). In type 1 strains wavD and in type 4 strains, the putative O-acetyltransferase genes wavNO are inserted between wavC and wavE (Fig. 3B). Finally, another rearrangement was observed between wavI and waaF, which are arranged as wavI waaF in type 2, 3, 4, and 5 strains. However, in type 1 strains wavJ, encoding a putative heptosyl transferase IV, is inserted here. As a consequence, aa 2 to 7 of WavI and the last 38 aa of WaaF in type 1 strains are dissimilar to those of the type 2, 3, and 4 strains, which have otherwise nearly identical sequences.
DISCUSSION
Seven conserved genes (waaA, waaC, waaF, wavA, wavB, wavC, and gmhD) with proposed function in the core OS biosynthesis were found within the whole genome sequence available for V. cholerae O1 El Tor strain N16961 (26); only five of these ORFs (waaA, wavF, wavA, wavB, and gmhD) had been annotated as such (TIGR microbial database, V. cholerae [www.tigr.org]). Knockout mutations and complementation analysis with wavB and waaF confirmed that these genes are indeed involved in core OS biosynthesis. The proposed seven core OS biosynthetic genes are located with other genes of unknown or predicted function on V. cholerae chromosome 1, forming the putative core OS biosynthesis gene cluster comprising ORFs VC0223 to VC0240.
The seven conserved core OS genes were always found to be genetically linked in our investigation of 38 different V. cholerae strains. In addition two genes, wavI and wavL, which probably encode glycosyl transferases, were also found to be conserved among all investigated strains. This indicates a common core OS backbone structure for the V. cholerae strains investigated. We could also provide evidence for the presence of putative waaL (O-antigen ligase) genes in all strains. O-antigen ligase function in the O1 El Tor strain was indirectly proven by confirming the absence of O antigen in SDS-PAGE analysis of waaL knockout mutants. Altogether we identified three different types of putative WaaL proteins of low similarity and by secondary structure prediction; we found similarity in the lengths of the proteins (398 to 403 aa), the numbers of proposed transmembrane helices (at least nine), the locations of the predicted periplasmic loops, and hydrophobicity plots (data not shown). Strains with type 1 or 2 and type 3 or 4 wav gene clusters encode similar WaaL proteins, although such strains are predicted to differ slightly within their core OS structure. This suggests that the WaaL proteins do not seem to recognize the whole structure of the core OS as an acceptor molecule. Interestingly, adjacent to the new identified putative waaL gene alleles of type 3 or 4 and type 5 strains, novel glycosyl transferase genes, wavM and wavT, respectively, were also found. This finding may indicate that WavM and WavT are responsible for adding sugar residues onto the core OS, which in turn could form acceptor molecules for the respective WaaL enzymes.
However, the relationship between the core OS structure and structure of WaaL enzymes has to be addressed in future studies. Recent studies with Salmonella and E. coli WaaL enzymes also indicated that the structure of a core OS is certainly important for O-antigen ligase recognition, but a consistent pattern of WaaL sequences and acceptor structures could not be established (reviewed in reference 27). In contrast, WaaL enzymes seem to be independent from the structure of the ligated polysaccharide, since it was found that one WaaL enzyme can efficiently ligate different O antigens, and therefore it is proposed that they recognize the C55 carrier (27). The V. cholerae ligase enzymes do also not seem to differentiate upon the structure of the ligated polysaccharide; as an example, we show that O1 and O139 strains expressing structurally different O antigens share the same WaaL enzyme.
In general, we assume that the predicted structural differences of the five wav gene cluster types are expected to be located in side branches of the core OS. Such structural differences compared to the type 1 core OS were reported for the environmental isolate H11 (compare Fig. 1A and B). We cannot deduce from our sequence data whether we have strains synthesizing the H11 type of core OS, since strain H11 was not available for this study. Based on our genetic data, we expect that only in strains with the type 1 gene cluster is HepIII replaced with an fourth heptosyl residue, since the predicted corresponding transferase gene wavJ is missing in strains with type 2, 3, 4, and 5 wav loci. The presence of other putative transferase genes, i.e., wavM in type 3 and 4 isolates and wavP and wavT in type 5 isolates, suggests novel modifications of the core OS, which remain to be identified by structural analysis. The presence of WecD and WecE homologues suggests that in type 5 strains fucosamine may be represented in the core OS. Interestingly, we can also predict differences in the presence of O acetylation on the core OS among V. cholerae isolates. O-acetyl groups are rarely found in core OS (12) and seem also not to be present in strains with the type 5 wav gene cluster. In type 1, 2, 3, and 4 strains, probably one O-acetyl group is added by the putative O-acetyltransferase WavK, and the published structural data for two O22 strains (type 1) indicate that it is probably added to the HepIII residue. The presence of two further putative O-acetyltransferases (WavO and WavN) in type 4 strains may lead to additional O acetylation on the core OS.
A common core OS backbone structure with differences in side branches is also known for other gram-negative bacteria. Structural diversity in core OS is thought to be limited by its essential contribution to the maintenance of outer membrane stability (27). We provide evidence that in V. cholerae core OS, variation is due to genetic exchange within the wav gene cluster. Genetic rearrangements occurred in distinct locations between wavC and gmhD and are probably due to insertions, deletions, and/or replacement of DNA fragments rather than to sequence drift. Several DNA exchanges occurred within genes, leading to differences in the deduced amino acid sequences. This could alter the enzymatic activity of the proteins, leading to further core OS structure diversity, e.g., differences in the type of glycosidic linkages. A previous detailed study with LOS of Campylobacter jejuni showed clearly that minor sequence differences within a given LPS biosynthesis gene cluster could, e.g., inactivate or alter the acceptor site recognition of a glycosyl transferase, leading to core OS structure variations (23).
Genetic exchange of wav genes by horizontal gene transfer, leading to structural alteration of the core OS, may be beneficial for V. cholerae by allowing the bacterium to change or improve outer membrane stability and hence to become adapted to different niches. To address this issue, we also looked at the distribution of the wav gene cluster in V. cholerae strains collected over a long period of time from very widespread geographical locations and different environments. Thirteen environmental non-O1, non-O139 V. cholerae isolates were taken from different sources (water, fish, and oyster) and shown to be unrelated in IS1004 fingerprint analysis. Twelve of them were found to be CT negative by ctx hybridization analysis, indicating that they are not lysogenized by the CTXΦ bacteriophage. These strains apparently also lack the VPI, as determined by Southern blot analysis, although it is possible that some of them contain one of the five known variants of the VPI not detectable with our probe (46, 49). These 12 strains were the source for the identification of the new wav gene cluster types 2, 3, 4, and 5, suggesting that among CT- and TCP-negative environmental non-O1, non-O139 V. cholerae strains, the core OS types 2 to 5 are predominant.
Seventeen clinical isolates (ctxA and tcpA positive) derived from different cholera outbreaks were utilized. Among these strains at least three different serogroups were represented: O1 (both biotypes and serotypes), O139, and O37 (the serogroup of strain V207 was not determined). All of these strains had identical type 1 wav gene clusters based on the results of Southern blot and PCR analyses. Based on this observation, it seems interesting to speculate that the corresponding core OS structure could link cell wall physiology as one additional attribute with other virulence factors, making O1 and O139 strains the successor for cholera pathogenesis. It remains to be established whether this could be either direct, e.g., in enhancing colonization or survival in the small intestine, or indirect in facilitating the acquisition of yet-unknown virulence genes. We also obtained evidence that O1 environmental strains, independent of the absence or presence of CT and TCP, carry the type 1 wav gene cluster, which may simply indicate a predominance of type 1 core OS among V. cholerae O1 isolates. There is also evidence that other non-O1, non-O139 environmental strains can harbor the type 1 wav gene cluster, since O22 strains have a similar core OS structure (15, 37), which is not unexpected since we predict horizontal transfer of wav genes.
Another interesting group of strains is represented by the environmental CT- and TCP-positive non-O1, non-O139 isolates. Such strains, and even a V. mimicus isolate, were identified in several recent studies (10, 19, 46, 56), and some of them have been found to colonize infant mice (49), while others do not (10), indicating a heterogeneous group. Moreover, a recent study with O141 strains revealed that such CT- and TCP-positive strains are associated with sporadic severe gastroenteritis (18). Curiously, none of these strains have caused cholera epidemics. Clearly, the mobile elements VPI and CTXΦ are widely distributed among Vibrio spp., but they appear not to be entirely sufficient for causing cholera pandemics, although they are certainly important during human V. cholerae infection. Notably, the O53 human and the O141 environmental CT- and TCP-positive isolates investigated here carry the type 4 gene cluster and not the type 1 gene cluster as found in epidemic strains. It is again tempting to speculate that there may be a link between the core OS structure of type 1 strains and the ability of CT- and TCP-positive strains to cause cholera epidemics. The four CT- and TCP-negative strains used in this study that were isolated from humans possess type 3 or 4 wav gene clusters. These core OS types may also confer some selective advantage in the colonization of humans.
This study provides a genetic basis for wav gene cluster typing, and additional data collection with V. cholerae strains should give more information on the core OS distribution in virulent V. cholerae strains not associated with cholera. There is a precedent for specific core OS structures being associated with virulence; for example, recent core OS typing studies with E. coli isolates showed that specific core OS structures are predominant among pathogenic strains (reference 3 and references therein).
Acknowledgments
We thank Emilisa Frirdich for critical reading of the manuscript. M. Waldor, A. Ghose, J. J. Mekalanos, M. Kist, and R. Sakazaki are gratefully acknowledged for providing us with V. cholerae strains. We also thank P. Reeves for the proposed nomenclature for the core OS biosynthetic genes.
This work was funded by BMBF grant 01KI8906.
Editor: J. T. Barbieri
REFERENCES
- 1.Aldova, E., K. Laznickova, E. Stepankova, and J. Lietava. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J. Infect. Dis. 118:25-31. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amor, K., D. Heinrichs, E. Frirdich, K. Ziebell, R. Johnson, and C. Whitfield. 2000. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 68:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attridge, S. R., A. Fazeli, P. A. Manning, and U. H. Stroeher. 2001. Isolation and characterization of bacteriophage-resistant mutants of Vibrio cholerae O139. Microb. Pathog. 30:237-246. [DOI] [PubMed] [Google Scholar]
- 5.Bagchi, K., P. Echeverria, J. D. Arthur, O. Sethabutr, O. Serichantalergs, and C. W. Hoge. 1993. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J. Clin. Microbiol. 31:1315-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock, K., E. V. Vinegradov, O. Holst, and H. Brade. 1994. Isolation and structural analysis of oligosaccharide phosphates containing the complete carbohydrate chain of the lipopolysaccharide from Vibrio cholerae strain H11 (non-O1). Eur. J. Biochem. 225:1029-1039. [DOI] [PubMed] [Google Scholar]
- 8.Bockemühl, J., K. Roch, B. Wohlers, S. Aleksic, V. Aleksic, and R. Wokatsch. 1986. Seasonal distribution of facultatively enteropathogenic vibrios (Vibrio cholerae, Vibrio mimicus, Vibrio parahaemolyticus) in the freshwater of the Elbe river at Hamburg. J. Appl. Bacteriol. 60:435-442. [DOI] [PubMed] [Google Scholar]
- 9.Bockemühl, J., and A. Triemer. 1974. Ecology and epidemiology of Vibrio parahaemolyticus on the coast of Togo. Bull. W. H. O. 51:353-360. [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd, E. F., K. E. Moyer, L. Shi, and M. K. Waldor. 2000. Infectious CTXφ and the Vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, K. H., T. K. Ng, K. Y. Yuen, and W. C. Yam. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, A. J., N. T. Blackburn, and H. Strating. 2000. Pathways for the O-acetylation of bacterial cell wall polysaccharides, p. 187-223. In R. Doyle (ed.), Glycomicrobiology. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 13.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 14.Cox, A. D., J.-R. Brisson, V. Varma, and M. B. Perry. 1996. Structural analysis of the lipopolysaccharide from Vibrio cholerae O139. Carbohydr. Res. 290:43-58. [DOI] [PubMed] [Google Scholar]
- 15.Cox, A. D., J. R. Brisson, P. Thibault, and M. B. Perry. 1997. Structural analysis of the lipopolysaccharide from Vibrio cholerae serotype O22. Carbohydr. Res. 304:191-208. [DOI] [PubMed] [Google Scholar]
- 16.Cox, A. D., and M. B. Perry. 1996. Structural analysis of the O-antigen-core region of the lipopolysaccharide from Vibrio cholerae O139. Carbohydr. Res. 290:59-65. [DOI] [PubMed] [Google Scholar]
- 17.Dalsgaard, A., A. Forslund, L. Bodhidatta, O. Serichantalergs, C. Pitarangsi, L. Pang, T. Shimada, and P. Echeverria. 1999. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol. Infect. 122:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echeverria, P., B. A. Harrison, C. Tirapat, and A. McFarland. 1983. Flies as a source of enteric pathogens in a rural village in Thailand. Appl. Environ. Microbiol. 46:32-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filiatrault, M. J., B. W. Gibson, B. Schilling, S. Sun, R. S. Mundson, Jr., and A. A. Campagnari. 2000. Construction and characterization of Haemophilus ducreyi lipooligosaccharide (LOS) mutants defective in expression of heptosyltransferase III and β1,4-glucosyltransferase: identification of LOS glycoforms containing lactosamine repeats. Infect. Immun. 68:3352-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geerlof, A., A. Lewendon, and W. V. Shaw. 1999. Purification and characterization of phosphopantetheine adenylyltransferase from Escherichia coli. J. Biol. Chem. 274:27105-27111. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, M., M. F. Karwaski, S. Bernatchez, N. M. Young, E. Taboada, J. Michniewicz, A. M. Cunningham, and W. W. Wakarchuk. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327-337. [DOI] [PubMed] [Google Scholar]
- 24.Grimberg, J., S. Maguire, and L. Belluscio. 1989. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 17:8893.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishman, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 28.Higa, N., Y. Honma, J. M. Albert, and M. Iwanaga. 1993. Characterization of Vibrio cholerae O139 synonym bengal isolated from patients with cholera-like disease in Bangladesh. Microbiol. Immunol. 37:971-974. [DOI] [PubMed] [Google Scholar]
- 29.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann, K., and W. Stoffel. 1993. TMbase--a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 31.Holst, O. 1999. Chemical structure of the core region of lipopolysaccharides, p. 115-154. In H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker Inc., New York, N.Y.
- 32.Huang, X. 1992. A contig assembly program based on sensitive detection of fragment overlaps. Genomics 14:237-246. [DOI] [PubMed] [Google Scholar]
- 33.Karaolis, D. K., R. Lan, and P. R. Reeves. 1994. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J. Bacteriol. 176:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karaolis, D. K., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knirel, Y. A., L. Paredes, P.-E. Jansson, A. Weintraub, G. Widmalm, and M. J. Albert. 1995. Structure of the capsular polysaccharide of Vibrio cholerae O139 synonym Bengal containing d-galactose-4,6-cyclophosphate. Eur. J. Biochem. 232:391-396. [DOI] [PubMed] [Google Scholar]
- 37.Knirel, Y. A., S. N. Senchenkova, P. E. Jansson, and A. Weintraub. 1998. More on the structure of Vibrio cholerae O22 lipopolysaccharide. Carbohydr. Res. 310:117-119. [DOI] [PubMed] [Google Scholar]
- 38.Knirel, Y. A., G. Widmalm, S. N. Senchenkova, P.-E. Jansson, and A. Weintraub. 1997. Structural studies on the short-chain lipopolysaccharide of Vibrio cholerae O139 Bengal. Eur. J. Biochem. 247:402-410. [DOI] [PubMed] [Google Scholar]
- 39.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 96:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Logan, S. M., J. W. Conlan, M. A. Monteiro, W. W. Wakarchuk, and E. Altman. 2000. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 35:1156-1167. [DOI] [PubMed] [Google Scholar]
- 41.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manning, P. A., U. H. Stroeher, and R. Morona. 1994. Molecular basis for O-antigen biosynthesis in Vibrio cholerae O1: Ogawa-Inaba switching, p. 77-94. In K. I. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, D.C.
- 43.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 44.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris, J. G. 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179-191. [DOI] [PubMed] [Google Scholar]
- 46.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Namdari, H., C. R. Klaips, and J. L. Hughes. 2000. A cytotoxin-producing strain of V. cholerae non-O1, non-O139 as a cause of cholera and bacteremia after consumption of raw clams. J. Clin. Microbiol. 38:3518-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nandi, B., R. K. Nandy, S. Mukhopadhayay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nandi, B., R. K. Nandy, A. C. P. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nesper, J., D. Kapfhammer, K. E. Klose, H. Merkert, and J. Reidl. 2000. Characterization of Vibrio cholerae O1 antigen as a bacteriophage K139 receptor and identification of IS 1004 insertions aborting O1-antigen biosynthesis. J. Bacteriol. 182:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiβ, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson, G. D. N., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Person, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 54.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. H. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 55.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 56.Riva, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rose, R. E. 1988. The nucleotide sequence of pACYC177. Nucleic Acids Res. 16:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubin, E. J., W. Lin, J. J. Mekalanos, and M. K. Waldor. 1998. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol. Microbiol. 28:1247-1254. [DOI] [PubMed] [Google Scholar]
- 59.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 62.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and S. C. Sensen (ed.), Proceedings of the 6th International Conference on Intelligent Systems and Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 63.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 51:503-517. [DOI] [PubMed] [Google Scholar]
- 64.Stroeher, U. H., K. E. Jedani, and P. A. Manning. 1998. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene 223:269-282. [DOI] [PubMed] [Google Scholar]
- 65.Stroeher, U. H., L. E. Karageorgos, R. Morona, and P. A. Manning. 1995. In Vibrio cholerae serogroup O1, rfaD is closely linked to the rfb operon. Gene 155:67-72. [DOI] [PubMed] [Google Scholar]
- 66.Tison, D. L. 1999. Vibrio, p. 497-506. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology 7th ed. American Society for Microbiology, Washington, D.C.
- 67.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 68.Vinogradov, E. V., K. Bock, O. Holst, and H. Brade. 1995. The structure of the lipid A-core region of the lipopolysaccharides from Vibrio cholerae O1 smooth strain 569B (Inaba) and rough mutant strain 95R (Ogawa). Eur. J. Biochem. 233:152-158. [DOI] [PubMed] [Google Scholar]
- 69.Wachsmuth, I. K., G. M. Evins, P. I. Fields, O. Olsvik, T. Popvic, C. A. Bopp, J. G. Wells, C. Carrillo, and P. A. Blake. 1993. The molecular epidemiology of cholera in Latin America. J. Infect. Dis. 167:621-626. [DOI] [PubMed] [Google Scholar]
- 70.Waldor, M. K., R. Colwell, and J. J. Mekalanos. 1994. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. USA 91:11388-11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamai, S., T. Okitsu, T. Shimada, and Y. Katsube. 1997. Distribution of serogroups of Vibrio cholerae non-O1 non-O139 with specific reference to their ability to produce cholera toxin and addition of novel serogroups. Kansenshogaku Zasshi 10:1037-1045. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto, K., Y. Takeda, T. Miwatani, and J. P. Craig. 1983. Purification and some properties of a non-O1 Vibrio cholerae enterotoxin that is identical to cholera enterotoxin. Infect. Immun. 39:1128-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zinnaka, Y., and C. C. Carpenter, Jr. 1972. An enterotoxin produced by noncholera vibrios. Johns Hopkins Med. J. 131:403-411. [PubMed] [Google Scholar]