Abstract
Staphylococcus aureus is one of the pathogens most frequently isolated in device-related infections. S. aureus is equipped with surface-associated proteins promoting specific binding to matrix molecules. Clumping factor A (ClfA, encoded by clfA) mediates adhesion to fibrinogen. Whereas the contribution of ClfA to pathogenicity is well documented, the influence of different growth and host parameters on gene activity is unclear. To elucidate this question, we investigated clfA transcript levels in an animal model of device-related infection and in planktonic and sessile bacteria grown in vitro. Specific mRNA from the S. aureus strains Newman, Reynolds, and RN6390 was quantified by LightCycler reverse transcription-PCR. In vitro, clfA transcript levels were low in the early logarithmic growth phase, but a clear increase was observed after the late logarithmic phase. Quantities of clfA transcripts were four to six times higher in the planktonic than in the sessile bacterial subpopulations grown to the stationary phase. During infection, in strains Newman and Reynolds levels of clfA transcripts in exudates accumulating in the infected devices were lower than those in the bacteria grown in vitro to stationary phase. clfA mRNA levels in the exudates increased during the initial phase of infection and remained constant after 96 h postinoculation. In contrast to the in vitro results, quantities of clfA transcripts in the unattached bacteria of the exudates never exceeded the level of clfA transcripts in the sessile bacteria attached to glass beads. However, a clear increase in clfA quantities in the sessile bacteria was observed late in infection after 144 h. In conclusion, maximal clfA transcript levels are reached late during growth in vitro and in vivo.
Staphylococci are the organisms that most commonly cause infections associated with indwelling medical devices (2, 31, 34). Colonization of the device occurs either at the time of implantation or by hematogenous spread during bacteremia (39). After implantation, the sterile surface of the implant is rapidly coated by host proteins. Among them, fibrinogen and fibrin are the major components on newly implanted devices and the dominant ligands mediating binding of Staphylococcus aureus (32, 33). To promote adherence, S. aureus expresses several specific adhesins on its surface, which are called MSCRAMMs, for microbial surface components recognizing adhesive matrix molecules (21). The specific binding of S. aureus to fibrinogen is mainly mediated by clumping factors A (ClfA, encoded by clfA) and B (16, 18). Inactivation of ClfA results in extreme inhibition of S. aureus attachment to fibrinogen-coated surfaces in vitro and ex vivo (15, 18, 36). Furthermore, the role of ClfA as a virulence factor was shown in an endocarditis model, where the clfA-defective mutant produced about 50% less endocarditis than the parent strain (17). The determination of the role of a given gene in pathogenicity by use of site-directed mutants is hampered by the fact that S. aureus harbors redundant surface adhesins, which might be able to take over and compensate for the function of the missing gene. To circumvent this problem, staphylococcal clfA was recently cloned in Streptococcus gordonii (30) and Lactococcus lactis (27), which lack the redundant adhesive molecules. ClfA was expressed as a surface protein in these bacteria, which thereby gained the ability to cause endocarditis in an animal model (26).
Bacteria may specifically alter the expression of certain genes upon binding and replicating on a substrate, possibly via the mediation of various signals such as changes in osmolarity, oxygen tension, and/or cell density (6, 23). S. aureus has indeed been shown to respond with differential gene expression (1) and physiological changes (35) upon binding to a solid substrate. The ability of S. aureus to bind to fibrinogen and fibrin was thought to be an important step in the initiation of device-related infection (32). The postulated ClfA activity early in infection, however, seems inconsistent with the late transcription of clfA during growth in vitro (18, 36). Additionally, the environmental signals leading to clfA expression have not yet been determined. The present study was designed to quantitate clfA transcripts during infection and during in vitro growth. The specific levels of transcripts of clfA in both planktonic and sessile S. aureus were quantified in vitro and in a guinea pig model of device-related infection by using LightCycler reverse transcription-PCR (RT-PCR) (9, 38).
MATERIALS AND METHODS
Bacterial strains and growth condition.
Three different S. aureus strains were used: strain Newman (7), strain Reynolds (13), and strain RN6390 (22). S. aureus was grown to the desired growth phase in Casamino Acids-yeast extract broth (CY) (20) with and without aeration in an Erlenmeyer flask containing 15 to 20 sinter glass beads (Sikug 023/300 A; Schott Schleifer AG, Muttenz, Switzerland).
Animal model of device-related infection.
The guinea pig model of implant infection was used (38). Four perforated Teflon tubes filled with 8 sinter glass beads each were inserted in the flanks of guinea pigs (29). Two weeks after the implantation of the tissue cages, 105 CFU of the test strain were inoculated in the tissue cages. Before inoculation, the interstitial fluid which had accumulated in the tissue cages was checked for sterility. The exudate was aspirated 48, 96, and 144 h after infection. One aliquot of the exudate was immediately stored in liquid nitrogen for subsequent RNA preparation. A second aliquot was used for quantitative bacteriology. Additionally, 96 and 144 h after bacterial challenge one animal infected with each of the three tests strains was sacrificed and the tissue cages were removed. RNA was prepared directly from five glass beads with sessile bacteria; three glass beads were used to count the adherent bacteria.
Bacterial quantification in vitro and in vivo.
Serial dilutions of broth culture and exudates were plated onto blood agar plates (tryptic soy agar containing 5% sheep blood). The number of bacteria adhering to the glass beads was determined as described previously (29). Briefly, for each time point three beads were washed twice with saline, and each bead was placed in a tube with 2 ml of 0.9% NaCl containing EDTA (0.15%) and Triton X-100 (0.1%). The beads were vigorously vortexed in the tubes three times for 15 s. The tubes were then placed in an ultrasonic bath and sonicated for 3 min at 120 W. After an additional mixing step, 100 μl was diluted for quantitative bacterial culture on blood agar plates. With this procedure 97 ± 2% of the adherent bacteria can be removed, as verified with [3H]thymidine-labeled S. aureus (R. B. Spörri et al., unpublished thesis). The plates were incubated at 37°C for 24 h, after which CFU were counted.
RNA isolation.
For RNA isolation from culture, S. aureus was grown in CY to the desired growth phase (8, 12, and 24 h). Approximately 5 × 109 bacteria were pelleted and lysed in 1 ml of Trizol reagent (Gibco BRL, Life Technologies) with 0.5 ml of zirconia-silica beads (0.1 mm in diameter) in a high-speed homogenizer (Savant Instrument, Farmingdale, N.Y.). Total RNA was isolated as directed in the instructions provided by the manufacturer of Trizol. At the same time, approximately 12 glass beads were taken from the same Erlenmeyer flask, washed in sterile saline, and added directly to 1 ml of Trizol reagent. The lysis and RNA isolation of S. aureus on glass beads were performed as described above.
For RNA preparation from exudates, the frozen samples were thawed rapidly and 200-μl aliquots were used. RNA from S. aureus adhering to five glass beads was prepared immediately after sacrificing the animals. RNA was isolated and purified as described previously (11). Contaminating DNA was degraded by digesting RNA samples with DNase as described previously (10).
Slot blot hybridization.
Serial dilutions of sample RNA in 10 mM NaOH-1 mM EDTA were transferred onto a positively charged nylon membrane (Roche Biochemicals, Mannheim, Germany) with a Slot-Blotter (Bio-Rad, Hercules, Calif.). For quantification of sample rRNA, known amounts of a PCR-generated ribosomal DNA fragment were employed as a standard on each blot. Blots were hybridized with an S. aureus-specific rDNA probe as described elsewhere (10), and the concentration of rRNA for each sample was calculated. Equal amounts of sample RNA were serially diluted and probed with a digoxigenin-labeled probe specific for clfA (generated by PCR using primers clfU and clfL; Table 1), and the signals were detected by chemiluminescence.
TABLE 1.
Oligonucleotide primers and LightCycler hybridization probes
| Target gene | GenBank accession no. | Primer | Sequencea |
|---|---|---|---|
| gyrB | D10489 | gyrU | TTATGGTGCTGGGCAAATACA |
| gyrL | CACCATGTAAACCACCAGATA | ||
| gyrFL1 | ATTTTAACTGTTTTACATGCTGGTGGTAA-F | ||
| gyrLC1 | Red640-TTTGGCGGTGGCGGATACA-ph | ||
| clfA | Z18852 | clfU | GGCGTGGCTTCAGTGCTTGTA |
| clfL | CACCAGTTACCGGCGTTTCTTC | ||
| clfFL1 | GGCGCAAAATCCAGCACAACAGGAA-F | ||
| clfLC1 | Red640-CGACACAATCATCATCAACAAATGC AAC-ph |
F, fluorescein; Red640, LightCycler-Red 640-N-hydroxysuccinimide ester; ph, 3′-phosphate.
Quantification of specific transcripts with LightCycler RT-PCR.
Specific, sequence-modified RNA standards for quantification of gyrB and clfA mRNA (Table 1) were engineered as described elsewhere (10). LightCycler RT-PCR was carried out using the LightCycler RNA amplification kit for hybridization probes (Roche Biochemicals, Basel, Switzerland). Master mixtures were prepared in accordance with the manufacturer's instructions by using the oligonucleotides specific for gyrB and clfA as shown in Table 1. After RT for 20 min at 50°C, the following temperature profile was utilized for amplification: denaturation for 1 cycle at 95°C for 30 s; 45 cycles at 95°C for 1 s (temperature transition, 20°C/s), 55 to 50°C (step size, 1°C; step delay, 1 cycle) for 15 s (temperature transition, 20°C/s), and 72°C for 15 s (temperature transition, 2°C/s); and fluorescence acquisition at 55 to 50°C in single mode. Melting-curve analysis was performed at 45 to 90°C (temperature transition, 0.2°C/s) with step-wise fluorescence acquisition. Sequence-specific standard curves were generated by using 10-fold serial dilutions (104 to 108 copies/μl) of the specific RNA standards. The number of copies of each sample transcript was then determined with the aid of the LightCycler software. The specificity of the PCR was verified by ethidium bromide staining on 3% agarose gels.
RESULTS
Transcription of clfA in planktonic and sessile S. aureus during growth in culture.
For quantitative transcript analysis we developed a method for clfA mRNA determination by LightCycler RT-PCR. Total RNA of S. aureus strain Newman was isolated at various time points during growth, and the specific levels of clfA and gyrB transcripts were quantified. Calculations were carried out with known amounts of synthetic RNA molecules as standards. The level of clfA transcripts was then normalized against the level of gyrB transcripts, which were shown to be constitutively expressed during growth up to early stationary phase (8 h after inoculation) (12). For evaluation of gyrB as the reference transcript in deep stationary phase (24 h after inoculation) total RNA from equivalent numbers of bacteria was independently isolated three times and the quantities of gyrB transcripts were determined by LightCycler RT-PCR. During early logarithmic growth (2.5 h after inoculation) 2.1 copies of gyrB/cell, 4.5 copies of gyrB/cell, and 2.9 copies of gyrB/cell (mean ± standard deviation, 3.2 ± 1.3 copies/cell) were detected, and after 24 h of growth 2.3 copies of gyrB/cell, 2.9 copies of gyrB/cell, and 2.4 copies of gyrB/cell (mean ± standard deviation, 2.5 ± 0.31 copies/cell) were detected. Thus, no significant difference in the amount of gyrB mRNA between the two phases was observed. The ratio of clfA/gyrB transcript significantly increased after 4 h (data not shown), confirming the results obtained by Northern analysis (36).
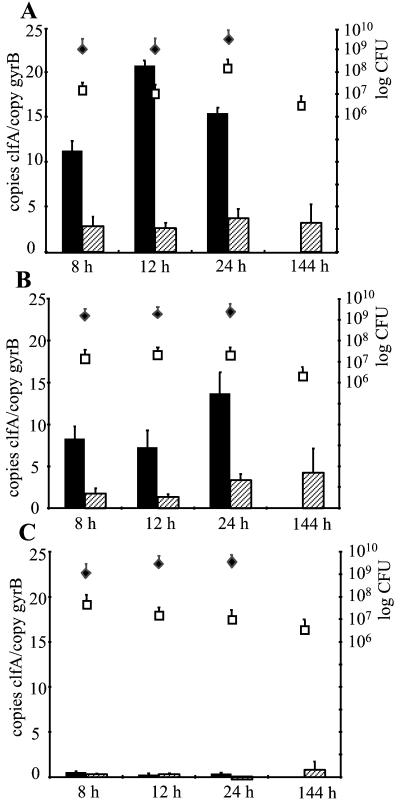
Since bacteria may change their phenotype upon binding to a solid substrate, we investigated the clfA transcript concentrations in planktonic and sessile bacteria grown simultaneously in the same flask. Bacterial RNA was isolated from 1 ml of liquid culture and from the bacteria attached to the glass beads. For the experiments three unrelated, prototypic S. aureus strains, Newman, Reynolds, and RN6390, were used. For all three strains tested, bacterial densities of 1.5 × 109 to 6 × 109 CFU/ml were reached during growth in liquid medium with no slackening in the first 24 h of growth (Fig. 1). After 24 h bacterial numbers declined, requiring changes of growth medium. From this point on, therefore, only sessile bacteria were analyzed. The bacterial counts recovered from the glass beads were similar for all the strains at all time points.
FIG. 1.
Quantitative transcript analysis of S. aureus strains Newman (A), Reynolds (B), and RN6390 (C) by LightCycler RT-PCR in vitro. Levels of clfA transcripts 8, 12, 24, and 144 h after inoculation were quantified in reference to the levels of gyrB transcripts (left y axis). Levels of clfA transcripts were determined in planktonic bacteria (black bars) and in sessile bacteria (hatched bars). At each time point, bacterial densities (right y axis) in planktonic (black diamonds; CFU per milliliter) and in sessile bacteria (white squares; CFU per bead) were determined.
Levels of clfA transcripts were quantified by LightCycler RT-PCR at each scheduled time point during the stationary growth phase. The concentration of clfA mRNA in strain Newman was an average of 6 ± 2 (mean ± standard deviation) times greater in planktonic bacteria than in sessile bacteria at 8, 12, and 24 h of growth (Fig. 1A). Similarly, in strain Reynolds a mean of 4.7 ± 0.7 times more clfA mRNA in planktonic than in sessile bacteria was detected (Fig. 1B). Strain RN6390 showed markedly less clfA transcripts, both in planktonic and in sessile bacteria, than strains Newman and Reynolds (Fig. 1C). Accordingly, no clfA transcript was detectable in RN6390 by using hybridization techniques (data not shown). Prolonged incubation of the sessile bacteria up to 144 h did not result in changes in clfA transcript levels in any of the three strains.
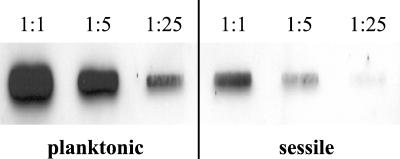
To verify the results obtained by LightCycler RT-PCR, a slot blot hybridization using serial dilutions of equal amounts of total RNA from strain Newman was performed. After 16 h of growth, clfA mRNA quantities were clearly higher in planktonic (Fig. 2, left) than in sessile S. aureus (Fig. 2, right). Thus, comparable results were achieved with the different normalization techniques (gyrB transcript levels in LightCycler RT-PCR, total RNA in slot blot hybridization).
FIG. 2.
Slot blot hybridization for the detection of clfA mRNA in planktonic and sessile bacteria in vitro. Equal amounts of total RNA were serially diluted (1:1, 1:5, 1:25) and hybridized with a digoxigenin-labeled gene probe specific for clfA.
Comparison of the expression of clfA of planktonic and sessile S. aureus during device-related infection.
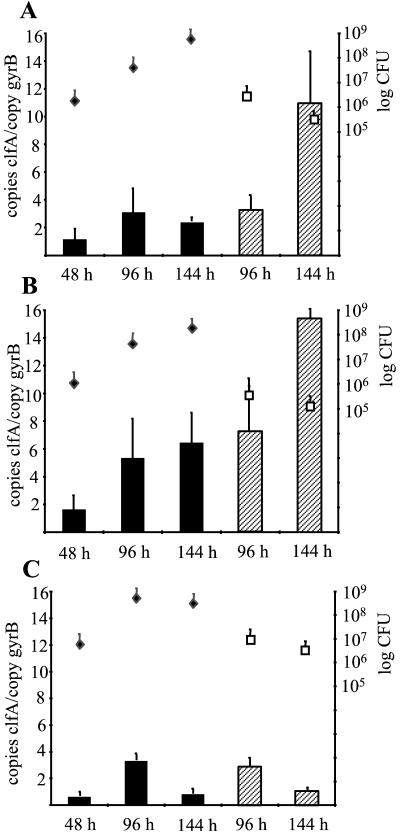
Since ClfA is thought to mediate bacterial attachment to solid surfaces, a well-established animal model for device-related infection was used to analyze clfA transcript quantities during infection. Preimplanted tissue cages containing glass beads were infected with 105 CFU of S. aureus strains Newman, Reynolds, and RN6390. Quantitative analysis of S. aureus transcripts was performed using the aspirated exudates after 48, 96, and 144 h of infection without subculturing the bacteria. Additionally, 96 and 144 h after inoculation animals were sacrificed and the tissue cages containing the glass beads were immediately removed. The bacterial counts for the exudates increased over the course of infection (Fig. 3). On the glass beads, there were no significant differences in the bacterial counts of the three staphylococcal strains after 96 and 144 h of infection (5 × 105 to 106 CFU/bead).
FIG. 3.
Quantitative transcript analysis of S. aureus strains Newman (A), Reynolds (B), and RN6390 (C) by LightCycler RT-PCR during device-related infection. Levels of clfA transcripts in bacteria of the exudates (black bars) and in sessile bacteria attached to glass beads (hatched bars) were quantified in reference to the levels of gyrB transcripts (left y axis). Bacterial densities (right y axis) in the exudates (black diamonds; CFU per milliliter) and on glass beads (white squares; CFU per bead) were determined at each time point.
In the prototypic ClfA-positive strains Newman and Reynolds, the amount of clfA mRNA in the bacteria from the exudates increased from 48 to 96 h and then remained at the same level until 144 h after inoculation (Fig. 3A and B). However, the transcript level remained below that detected in the planktonic bacteria during the stationary growth phase (Fig. 1). In contrast to the in vitro results, levels of clfA transcripts in the unattached bacteria of the exudates never exceeded the levels of clfA transcripts in the sessile bacteria attached to glass beads. In fact, a clear increase of clfA mRNA was observed in the sessile bacteria late in infection, after 144 h.
Strain RN6390 expressed less clfA mRNA than strains Newman and Reynolds during infection, and no increase of clfA mRNA in sessile bacteria 144 h after inoculation could be observed (Fig. 3C). It is worth noting that with this strain expression in the exudates was stronger in vivo than in vitro.
DISCUSSION
ClfA is a cell wall-anchored, fibrinogen-binding protein with the typical characteristics of an MSCRAMM (21). These proteins are responsible for mediating the adherence of S. aureus to matrix proteins, which is thought to be an important step in the establishment of an infection (25). Accordingly, most of the cell wall-anchored MSCRAMMs (protein A [4], fibronectin-binding protein [28, 37], and clumping factor B [18]) analyzed until now were preferentially expressed during the early exponential growth phase. In contrast, clfA transcripts were mainly detected in postexponential growth, indicating specific growth phase-dependent up-regulation of clfA transcription and confirming previous results (36). However, one cannot exclude the possibility that the observed increase in clfA mRNA during growth correlates to an increase in specific transcript stability. The discrimination between de novo synthesis and mRNA decay is a general problem in expression profiling which so far has rarely been addressed.
The expression of some MSCRAMMs in the early growth phase can be explained by the action of global regulatory systems, including agr and sar. Since neither agr (36) nor sarA mutants (unpublished observation) differ from wild-type strains with respect to clfA transcription, the expression of clfA in the late growth cycle must be mediated by regulatory factors independent of agr and sarA. Recently, it was shown that clumping activity is dependent on σB and that a putative σB-dependent promoter is present upstream of clfA (19). σB activity reaches its maximum during the late logarithmic phase and drops toward the stationary phase, as shown by reporter gene fusion experiments using the σB-dependent promoter of asp23 (8). However, in contrast to asp23 promoter activity, we found that clfA transcript levels remained stable during the stationary phase as well. The σB-dependent transcription of clfA is further supported by the down-regulation of clfA in strain RN6390, which was shown to be deficient in σB activity due to a mutation in σB activator gene rsbU (3, 14).
We also analyzed whether clfA transcript levels in S. aureus change upon binding to a solid substrate. With the prototypic ClfA-positive strains Newman and Reynolds, we show in vitro that planktonic bacteria yield greater amounts of clfA mRNA than sessile bacteria. In our experimental design, both bacterial populations (planktonic and sessile) were grown simultaneously under identical conditions with aeration in the same flask. The variation that we observed suggests differential gene expression depending on the mode of growth. In a previous study, it was reported that staphylococci express different surface proteins depending on whether they are grown in liquid or on solid media (5). The in vitro signals leading to the observed down-regulation of clfA upon binding to a surface are not yet clear. However, after an analysis of gene expression in planktonic versus sessile Escherichia coli it was concluded that changes in either oxygen tension or osmolarity in the microenvironment of the attached bacteria could account for the changes in gene expression (6, 23). The possible role of aeration in signal transduction is emphasized by our observation that planktonic bacteria grown without aeration (shaking) also expressed less clfA mRNA; amounts were comparable to those found in the fraction of sessile bacteria (data not shown).
Since ClfA of S. aureus mediates adherence to fibrinogen, one of the host proteins coating indwelling material soon after implantation, we compared the clfA mRNA levels in planktonic bacteria and sessile bacteria in an animal model. Strains Newman and Reynolds displayed identical clfA mRNA levels during infection. In these bacteria from the exudates fewer clfA transcripts than in planktonic bacteria during aerated growth in vitro were detected. However, the levels were similar to those found in planktonic bacteria which had been grown without shaking. This is plausible since during infection one would not expect oxygen tensions to be as high as those in an in vitro system with shaking flasks.
We found no differences in clfA transcript levels between bacteria from exudates and those attached to glass beads during the initial course of infection. Surprisingly, however, there was a significant increase in the clfA transcript levels in the sessile bacteria 144 h after infection; a corresponding increase was not detected during prolonged growth in vitro. Thus, infection-specific signals may act on the clfA regulatory pathway. Interestingly, others have been able to show that ClfA expression is induced in small-colony variants (SCV) of S. aureus (P. Vaudeaux, personnel communication). One may speculate that the bacteria may change their phenotype later in infection, mimicking SCV. It was postulated that the SCV phenotype allows the persistence of infection (24). However, after the attached bacteria from our animal model were subcultured, no obvious SCV phenotype was observed. Due to the specific host environment (e.g., energy depletion) in vivo the proposed SCV phenotype may be mediated by regulatory circuits rather than by mutation. Thus, upon subcultivation the phenotype might be lost.
Acknowledgments
We thank Zarko Rajacic for his expert technical assistance in the animal experiments.
This work was supported by grants from fortüne (no. 688-0-0 and 688-0-1) and the Deutsche Forschungsgemeinschaft (Wo 578/3-2). The animal experiments were in part supported by a grant from Bristol-Myers Squibb.
Editor: E. I. Tuomanen
REFERENCES
- 1.Becker, P., W. Hufnagle, G. Peters, and M. Herrmann. 2001. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol 67:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berbari, E. F., A. D. Hanssen, M. C. Duffy, J. M. Steckelberg, D. M. Ilstrup, W. S. Harmsen, and D. R. Osmon. 1998. Risk factors for prosthetic joint infection: case-control study. Clin. Infect. Dis. 27:1247-1254. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björkind, A., and S. Arvidson. 1980. Mutants of Staphylococcus aureus affected in the regulation of exoprotein synthesis. FEMS Microbiol. Lett. 7:203-206. [Google Scholar]
- 5.Cheung, A. L., and V. A. Fischetti. 1988. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infect. Immun. 56:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Duthie, E., and L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 8.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goerke, C., M. G. Bayer, and C. Wolz. 2001. Quantification of bacterial transcripts during infection using competitive reverse transcription-PCR (RT-PCR) and LightCycler RT-PCR. Clin. Diagn. Lab. Immunol. 8:279-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-146. [DOI] [PubMed] [Google Scholar]
- 13.Karakawa, W., and W. Vann. 1982. Capsular polysaccharides of Staphylococcus aureus. Semin. Infect. Dis. 4:285-293. [Google Scholar]
- 14.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 16.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1995. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol. 16:895-907. [DOI] [PubMed] [Google Scholar]
- 17.Moreillon, P., J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. Francois, and P. Vaudaux. 1995. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immun. 63:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni, E. D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 19.Nicholas, R. O., T. Li, D. McDevitt, A. Marra, S. Sucoloski, P. L. Demarsh, and D. R. Gentry. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novick, R. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 21.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 22.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proctor, R. A. 1994. Microbial pathogenic factors: small colony variants, p. 79-90. In A. L. Bisno and F. A. Waldvogel (ed.), Infections associated with indwelling medical devices, 2nd ed. American Society for Microbiology, Washington, D.C.
- 25.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-82. In B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 26.Que, Y. A., P. Francois, J. A. Haefliger, J. M. Entenza, P. Vaudaux, and P. Moreillon. 2001. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect. Immun. 69:6296-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Que, Y. A., J. A. Haefliger, P. Francioli, and P. Moreillon. 2000. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infect. Immun. 68:3516-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saravia-Otten, P., H.-P. Muller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwank, S., Z. Rajacic, W. Zimmerli, and J. Blaser. 1998. Impact of bacterial biofilm formation on in vitro and in vivo activities of antibiotics. Antimicrob. Agents Chemother. 42:895-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stutzmann, M. P., J. M. Entenza, P. Vaudaux, P. Francioli, M. P. Glauser, and P. Moreillon. 2001. Study of Staphylococcus aureus pathogenic genes by transfer and expression in the less virulent organism Streptococcus gordonii. Infect. Immun. 69:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukayama, D. T., R. Estrada, and R. B. Gustilo. 1996. Infection after total hip arthroplasty. A study of the treatment of one hundred and six infections. J. Bone Joint Surg. Am. 78:512-523. [DOI] [PubMed] [Google Scholar]
- 32.Vaudaux, P. E., P. Francois, D. Lew, and F. A. Waldvogel. 2000. Host factors predisposing to and influencing therapy of foreign body infections, p. 1-27. In A. L. Bisno and F. A. Waldvogel (ed.), Infections associated with indwelling medical devices, 3rd ed. American Society for Microbiology, Washington, D.C.
- 33.Vaudaux, P. E., P. Francois, R. A. Proctor, D. McDevitt, T. J. Foster, R. M. Albrecht, D. P. Lew, H. Wabers, and S. L. Cooper. 1995. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 63:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteside, L. A. 1994. Cementless total knee replacement. Nine- to 11-year results and 10-year survivorship analysis. Clin. Orthop. 309:185-192. [PubMed] [Google Scholar]
- 35.Williams, I., F. Paul, D. Lloyd, R. Jepras, I. Critchley, M. Newman, J. Warrack, T. Giokarini, A. J. Hayes, P. F. Randerson, and W. A. Venables. 1999. Flow cytometry and other techniques show that Staphylococcus aureus undergoes significant physiological changes in the early stages of surface-attached culture. Microbiology 145:1325-1333. [DOI] [PubMed] [Google Scholar]
- 36.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 64:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol 36:230-243. [DOI] [PubMed] [Google Scholar]
- 38.Zimmerli, W., F. A. Waldvogel, P. Vaudaux, and U. E. Nydegger. 1982. Pathogenesis of foreign body infection: description and characteristics of an animal model. J. Infect. Dis. 146:487-497. [DOI] [PubMed] [Google Scholar]
- 39.Zimmerli, W., O. Zak, and K. Vosbeck. 1985. Experimental hematogenous infection of subcutaneously implanted foreign bodies. Scand. J. Infect. Dis. 17:303-310. [DOI] [PubMed] [Google Scholar]