Abstract
The locus of enterocyte effacement (LEE) is a chromosomal pathogenicity island that encodes the proteins involved in the formation of the attaching and effacing lesions by enterohemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC). The LEE comprises 41 open reading frames organized in five major operons, LEE1, LEE2, LEE3, tir (LEE5), and LEE4, which encode a type III secretion system, the intimin adhesin, the translocated intimin receptor (Tir), and other effector proteins. The first gene of LEE1 encodes the Ler regulator, which activates all the other genes within the LEE. We previously reported that the LEE genes were activated by quorum sensing through Ler (V. Sperandio, J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper, Proc. Natl. Acad. Sci. USA 96:15196-15201, 1999). In this study we report that a putative regulator in the E. coli genome is itself activated by quorum sensing. This regulator is encoded by open reading frame b3243; belongs to the LysR family of regulators; is present in EHEC, EPEC, and E. coli K-12; and shares homology with the AphB and PtxR regulators of Vibrio cholerae and Pseudomonas aeruginosa, respectively. We confirmed the activation of b3243 by quorum sensing by using transcriptional fusions and renamed this regulator quorum-sensing E. coli regulator A (QseA). We observed that QseA activated transcription of ler and therefore of the other LEE genes. An EHEC qseA mutant had a striking reduction of type III secretion activity, which was complemented when qseA was provided in trans. Similar results were also observed with a qseA mutant of EPEC. The QseA regulator is part of the regulatory cascade that regulates EHEC and EPEC virulence genes by quorum sensing.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is responsible for outbreaks of bloody diarrhea in several countries. It has a very low infectious dose (as low as 50 CFU in one outbreak) and colonizes the large intestine, where it causes attaching and effacing (AE) lesions (reviewed in reference 28). The AE lesions are characterized by effacement of the intestinal epithelial cell microvilli and the rearrangement of the cytoskeleton to form pedestal-like structures that cup the bacteria individually. Similar AE lesions are also seen with enteropathogenic E. coli (EPEC), which is a leading cause of infant diarrhea in developing countries (28). The genes involved in the formation of these AE lesions are encoded within a chromosomal pathogenicity island named the locus of enterocyte effacement (LEE) (25). The LEE comprises 41 open reading frames organized in five major operons, LEE1, LEE2, LEE3, tir (LEE5), and LEE4, and contains (i) genes encoding a type III secretion system (13); (ii) the eae gene, encoding the intimin adhesin (14); (iii) tir, which encodes the translocated intimin receptor (15); (iv) the espABD genes, which encode proteins that are secreted by the type III secretion system (6, 16, 23) (including EspA, which forms filamentous-like structures [18] involved in the translocation of EspB and Tir; and (v) ler (for LEE-encoded regulator), which encodes an H-NS-like protein that activates the expression of the LEE genes (26).
We previously reported that the first gene of the LEE1 operon encodes an activator of the LEE genes, which we named Ler (26). Friedberg et al. (10) and Elliott et al. (7) reported that mutations in ler abolished the ability of EPEC and EHEC to produce AE lesions. Sperandio et al. (37) demonstrated that Ler directly binds to the upstream regulatory region of LEE2, which is downstream of LEE3, to activate transcription of both the LEE2 and LEE3 operons. Bustamante et al. (2) reported that Ler acts as an antirepressor for H-NS, probably by competing with H-NS for binding to the regulatory regions of LEE2 and LEE3. These studies demonstrated that Ler directly activates transcription of the LEE genes by competing for binding with H-NS, which represses their expression in the absence of Ler. We previously reported that the LEE genes were regulated by quorum sensing through autoinducer-2 (AI-2), which is synthesized by the product of the luxS gene. Transcription of ler was activated in the presence of AI-2, leading to transcription of the other LEE genes in a cascade fashion (36).
Quorum sensing is a mechanism through which bacteria regulate gene expression by cell density. The bacteria produce hormone-like compounds called autoinducers that interact with regulatory proteins after they reach a certain threshold concentration. Quorum sensing was first described in the regulation of bioluminescence in Vibrio fischeri and Vibrio harveyi (29, 30). The luciferase operon in V. fischeri is regulated by two proteins, LuxI, which is responsible for the production of the acylhomoserine lactone autoinducer, and LuxR, which is activated by this autoinducer to increase transcription of the luciferase operon (8, 9). Since that initial description, homologs of LuxR-LuxI have been identified in other bacteria, and in all of these LuxR-LuxI systems, the bacteria produce an acylhomoserine lactone autoinducer which binds to the LuxR protein and regulates the transcription of several genes involved in a variety of phenotypes, including production of extracellular matrix, antimicrobial agents, flagellation, motility, and other phenotypes (reviewed in references 1 and 31). Gram-positive bacteria have other quorum-sensing systems which employ different components, such as two-component regulatory systems and peptide autoinducers (reviewed in references 1 and 3). One of the most widely distributed systems is the luxS system, first described for V. harveyi. Surette and Bassler (39) showed that V. harveyi produced a second autoinducer, which is not an acylhomoserine lactone, that they named AI-2. AI-2 seems to be a furanone (34), and the enzyme responsible for synthesizing it is encoded by the luxS gene (40). Many bacterial species produce AI-2 and possess the luxS gene, and this quorum-sensing system has been proposed to be involved in interspecies communication. Among these diverse bacterial species that contain the luxS quorum sensing system is E. coli, including EHEC serotype O157:H7 (39).
We proposed that activation of the LEE genes by the AI-2-luxS quorum-sensing system would occur in response to the AI-2 produced by the intestinal flora and that this intraintestinal signaling could be one explanation for the low infectious dose of EHEC (36). The regulatory network of this quorum-sensing system in V. harveyi has been extensively described (reviewed in reference 33); however, very little is known about it in E. coli. In an effort to examine how global quorum-sensing regulation is for E. coli, we hybridized an E. coli gene array with cDNA synthesized from RNA extracted from wild-type EHEC strain 86-24 and its isogenic luxS mutant VS94 (38). The results of that recent study showed that in addition to Shiga toxin and the LEE gene products, quorum sensing regulated ca. 10% of the E. coli genome shared by K-12 and EHEC, including phenotypes such as flagellation and motility that could also play a role in pathogenesis. In the array study we observed 19 putative E. coli transcriptional regulators regulated by the AI-2-luxS quorum-sensing system that might be part of this intricate regulatory network (V. Sperandio and J. B. Kaper, unpublished data).
In the present paper, we describe one of these putative regulators, which is a member of the LysR family of regulators, and demonstrate that it activates transcription of the LEE genes by quorum sensing. This regulator shares homology with AphB from Vibrio cholerae and PtxR from Pseudomonas aeruginosa and was renamed quorum-sensing E. coli regulator A (QseA).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. All E. coli strains were grown aerobically at 37°C in Luria-Bertani medium (LB) or Dulbecco modified Eagle medium (DMEM) (catalog no. 11054; Gibco-BRL). Selective antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and streptomycin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| 86-24 | Stx2+ EHEC strain (serotype O157:H7) | 11 |
| E2348/69 | EPEC wild-type strain | Laboratory stock |
| DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Stratagene |
| SM10λpir | thiL thrL leuB6 supE44 tonA21 lacY1 recA::RP4-2-Tc::Mu Kmr | 32 |
| BB170 | V. harveyi luxN::Tn5 (sensor-1− sensor-2+) | 39 |
| VS94 | 86-24 luxS | 38 |
| VS95 | VS94 with plasmid pVS84 | 38 |
| VS145 | 86-24 qseA | This study |
| VS151 | VS145 with plasmid pVS150 | This study |
| VS193 | E2348/69 qseA | This study |
| VS196 | sepZ1[LEE2]::lacZ chromosomal fusion in E2348/69 | This study |
| VS197 | ler-1[LEE1]::lacZ chromosomal fusion in E2348/69 | This study |
| VS198 | sepZ1[LEE2]::lacZ chromosomal fusion in VS193 | This study |
| VS199 | ler-1[LEE1]::lacZ chromosomal fusion in VS193 | This study |
| VS210 | VS198 with pVS208 | This study |
| VS211 | VS199 with pVS208 | This study |
| Plasmids | ||
| pBluescript KSII | Cloning vector | Stratagene |
| TOPO PCR BLUNT | PCR blunt cloning vector with topoisomerase | Invitrogen |
| pRS551 | lacZ repoter gene fusion vector | 35 |
| pACYC177 | Cloning vector | New England Biolabs |
| pCVD442 | Suicide vector | 5 |
| pVS232Z | ler-1[LEE1]::lacZ in pRS551 | 36 |
| pVS21 | sepZ1[LEE2]::lacZ in pRS551 | 36 |
| pVS26 | orf12[LEE3]::lacZ in pRS551 | 36 |
| pVS24Z | tir::lacZ in pRS551 | 36 |
| pVS130 | qseA into TOPO PCR BLUNT | This study |
| pVS139 | qseA into pBluescript KSII | This study |
| pVS141 | qseA::cat into pBluescript KSII | This study |
| pVS143 | qseA::cat into pCVD442 | This study |
| pVS150 | qseA into pACYC177 | This study |
| pVS208 | pVS150 with tet gene within bla | This study |
| pVS160 | qseA::lacZ into pRS551 | This study |
| pVS194 | sepZ1[LEE2]::lacZ in pJRlacZins | This study |
| pVS195 | ler-1[LEE1]::lacZ in pJRlacZins | This study |
Recombinant DNA techniques.
Plasmid purification, PCR, restriction digestion, ligation, transformation, and DNA gel electrophoresis were performed by standard methods (32). DNA sequence analysis was performed at the University of Maryland Biopolymer Facility on an ABI automated sequencer with DNA purified by Qiagen midi-columns. All oligonucleotide primers used are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| CmF | 5′-GTGACGGAAGATCACTTCGCA-3′ |
| CmR | 5′-GCGTTTAAGGTCAACAATAACTGC-3′ |
| K1977 | 5′-CGCGGATCCATAGAGCGCGGTGTTGAACA-3′ |
| K1978 | 5′-CGCGGATCCCGTTGGCACAGGTTTGTACA-3′ |
| K2179 | 5′-CCGGAATTCTCTTCGCGAGACATCGCCTG-3′ |
| K2180 | 5′-CGCGGATCCACGCTCATCTGTAGCTGTCT-3′ |
Plasmid pVS130 was constructed by amplifying the b3243 gene from E. coli K-12 strain MG1655 with Pwo polymerase (Boehringer Mannheim), using primers K1977 and K1978, and cloning into ZERO BLUNT TOPO PCR vector (Invitrogen). Plasmid pVS150 was constructed by cloning b3243 digested with XhoI and HindIII from pVS130 into vector pACYC177 (XhoI/HindIII). Plasmid pVS208 was generated by digesting plasmid pVS150 with ScaI (site within the bla gene) and cloning a blunt tetracycline resistance gene from pBR322. Plasmid pVS139 was constructed by cloning the blunt-ended b3243 PCR product into the EcoRV/SmaI site of vector pBluescript KSII (Stratagene). Plasmid pVS139 was digested with EcoRI (there is a single EcoRI site in the middle of b3243 gene) and blunt ended with the Klenow fragment of DNA polymerase of E. coli. A chloramphenicol resistance cassette (cat) amplified from pACYC184 with Pwo polymerase (Boehringer Mannheim) and primers CmF and CmR was cloned into the blunt-ended EcoRI site of pVS139, generating plasmid pVS141, which has the cloned b3243 gene interrupted by cat. Plasmid pVS141 was digested with XbaI and XhoI, and the b3243::cat cassette was cloned into plasmid pCVD442 (5) digested with XbaI and SalI, generating plasmid pVS143. The EHEC b3243 mutant (named VS145) was generated by allelic exchange using vector pVS143, and the mutants were selected on plates containing chloramphenicol and 5% sucrose as previously described (5). The b3243 mutant (VS145) was complemented with plasmid pVS150, generating strain VS151. The EPEC b3243 mutant (named VS193) was also generated by allelic exchange using vector pVS143 as described above.
Preconditioned media.
To prepare preconditioned media, an initial inoculum of strain 86-24, VS145, or VS151 was grown at 250 rpm in LB for 18 h at 37°C. This culture was diluted 1:1,000 in fresh LB or DMEM and grown to an optical density at 600 nm (OD600) of ca 1.0. Following centrifugation (12,000 × g, 4 min, 25°C) and filtration (0.2-μm-pore-size filter), the pH was adjusted to 7.0.
V. harveyi luminescence assay.
The presence of AI-2 in the preconditioned media was assayed using the V. harveyi BB170 (luxN::Tn5) reporter strain, which responds only to AI-2 (39). The luminescence assays were performed as described (39) and the assays were read in a Wallac 1420 multilabel counter.
Constructions of lacZ operon fusions.
Operon fusions with lacZ were constructed by amplifying the regulatory region of b3243 with Pwo polymerase, using primers K2180 and K2179, and cloning into plasmid pRS551, which contains a promoterless lac operon (35), thereby generating plasmid pVS160. This fusion was transformed into host strains 86-24 (wild-type EHEC), VS94 (luxS mutant of 86-24), and VS95 (VS94 complemented with luxS on a plasmid) (Tables 1 and 2).
We constructed ler-1[LEE1]::lacZ and sepZ1[LEE2]::lacZ fusions in single copy in the chromosome of EPEC strain E2348/69, but for unknown technical reasons we were unable to construct these integrated fusions in EHEC strain 86-24. Plasmids pVS232Z (ler-1[LEE1]::lacZ in pRS551) and pVS21 (sepZ1[LEE2]::lacZ in pRS551) (36) were digested with EcoRV and ScaI, which cut in the middle of the bla and lacZ genes of pRS551, respectively. This digestion will release the LEE::lacZ fusion together with the kanamycin resistance gene. The Km::LEE::lacZ fragment was cloned into plasmid pJRLacZins (21), which is a pCVD442 (5) derivative containing an added lacZ gene, yielding plasmids pVS194 (sepZ1[LEE2]::lacZ) and pVS195 (ler-1[LEE1]::lacZ). During the conjugative transfer of pVS194 and pVS195 by SM10λpir strain to E2348/69 and VS193, the ler-1[LEE1]::lacZ and sepZ1[LEE2]::lacZ fusions were recombined into the lacZ sites of the E2348/69 and VS193 chromosomes, generating the following strains: VS196 (sepZ1[LEE2]::lacZ in E2348/69), VS197 (ler-1[LEE1]::lacZ in E2348/69), VS198 (sepZ1[LEE2]::lacZ in VS193), and VS199 (ler-1[LEE1]::lacZ in VS193). Strains VS198 and VS199 were complemented with the wild-type qseA gene cloned on plasmid pVS208 to generate strains VS210 and VS211, respectively.
β-Galactosidase assays.
The strains containing the operon::lacZ fusions were grown in DMEM at 37°C to an OD600 of 1.0. These cultures were diluted 1:10 in Z buffer (Na2HPO4, 0.06 M; NaH2PO4, 0.04 M; KCl, 0.01 M; MgSO4, 0.001 M; β-mercaptoethanol, 0.05 M) and assayed for β-galactosidase activity by using o-nitrophenyl-β-d-galactopyranoside as a substrate as previously described (27).
Primer extension.
RNA was extracted from strains grown to an OD600 of 1.0 in DMEM at 37°C according to standard protocols (32). Primer extension was also performed according to standard protocols (32), using primer K983 (36). Primer extension reactions were run in parallel with DNA sequencing reactions of pVS23, which contains the regulatory region of LEE1 (36) and of a λ ladder control (U.S. Biochemicals).
Western blotting.
Total proteins were extracted from strains 86-24, VS94, VS95, VS145, and VS151 grown in DMEM to an OD600 of 1.0. The protein concentration was measured using the assay of Lowry et al. (24). Equal concentrations of total proteins were electrophoresed in sodium dodecyl sulfate-12% polyacrylamide gels (22). Western blotting procedures were performed as previously described (32), and blots were probed with polyclonal antisera directed against the A subunit of Stx2 (kindly provided by Alison O'Brien [Uniformed Services University of the Health Sciences]) and anti-H7 flagellin (J. A. Girón and J. B. Kaper, unpublished data).
Secreted proteins.
Secreted proteins from EHEC strains 86-24, VS145, and VS151 were prepared as described by Jarvis et al. (13) after these strains were grown to an OD600 of 1.0 in DMEM at 37°C.
FAS test.
Fluorescent actin staining (FAS) tests were performed as initially described by Knutton et al. (17). Briefly, bacterial strains were incubated with HeLa cells for 6 h at 37°C 5% CO2, after which epithelial cells were permeabilized with 0.1% Triton and treated with fluorescein isothiocyanate-phalloidin to visualize the accumulation of actin beneath and around the bacteria attached to the HeLa cells. The bacteria were stained with propidium iodide.
RESULTS AND DISCUSSION
E. coli putative regulator b3243 is activated by quorum sensing.
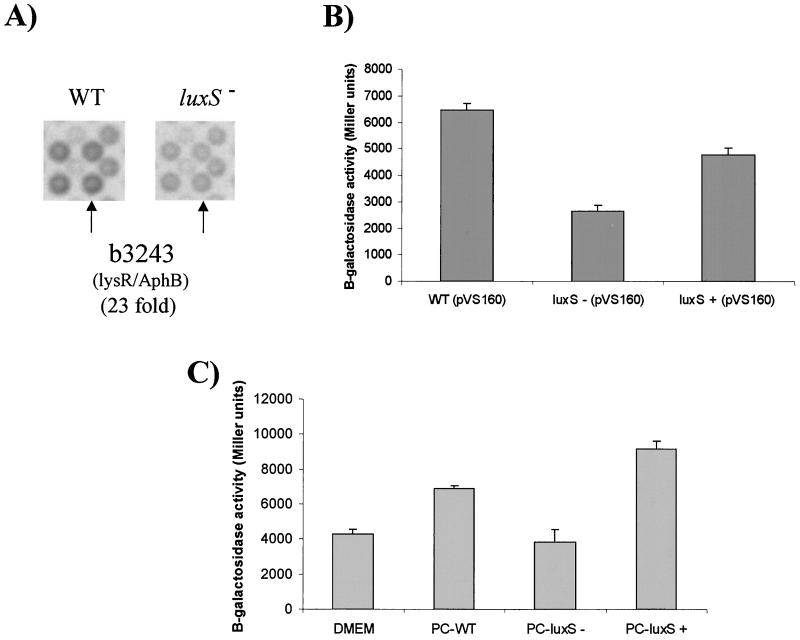
To expand on our initial report that the LEE genes from EHEC were activated by quorum sensing through the luxS system (36), we showed that additional genes in the E. coli genome were also regulated by quorum sensing by using gene array technology (38). Through this array analysis, we found that transcription of 19 putative E. coli transcriptional regulators was either activated or repressed by quorum sensing (data not shown). One of these putative regulators, whose transcription was up-regulated 23-fold in the wild-type strain compared to the luxS mutant, corresponds to the open reading frame referred to as b3243 in the K-12 and the EHEC genomes (GenBank accession numbers AE000403 and AE005552, respectively) (Fig. 1A). This open reading frame encodes a putative regulator of the LysR family, which is characterized by a helix-turn-helix DNA binding domain in its N terminus. The predicted protein encoded by b3243 shares 28% identity and 53% similarity with AphB from V. cholerae, which is involved in regulation of the ToxR regulon, the master virulence regulon of V. cholerae (19, 20). This predicted protein also shares 28% identity and 49% similarity with PtxR from P. aeruginosa, which is an activator of exotoxin A production (12) (Fig. 2). Since this LysR family E. coli regulator shared homology with other virulence regulators and was activated by quorum sensing, we renamed this regulator QseA for quorum-sensing E. coli regulator A.
FIG. 1.
Activation of b3243 transcription by quorum sensing. (A) Portion of E. coli K-12 gene array (Sigma-Genosys) containing the b3243 open reading frame hybridized with cDNA synthesized from RNAs extracted from strains 86-24 (wild-type) and VS94 (luxS mutant). (B) β-Galactosidase activities of a qseA::lacZ fusion (pVS160) in 86-24 (wild type [WT]), VS94 (luxS mutant), and VS95 (luxS complemented) strain backgrounds in DMEM at an OD600 of 1.0. (C) β-Galactosidase activity of a qseA::lacZ fusion (pVS160) in VS94 (luxS mutant) in fresh DMEM, DMEM preconditioned by growth of wild-type strain 86-24 (PC-WT), DMEM preconditioned by growth of strain VS94 (PC-luxS −), and DMEM preconditioned by growth of strain VS95 (PC-luxS +). Error bars indicate standard deviations.
FIG. 2.
Alignments of EHEC and K-12 QseA with AphB from V. cholerae and PtxR from P. aeruginosa. Underlined amino acid residues comprise the predicted helix-turn-helix of QseA.
To confirm the regulation of qseA by quorum sensing, we generated a qseA::lacZ transcriptional fusion by amplifying the regulatory region of this gene (from bp 5491 to 6340 of the sequence under GenBank accession number AE000403) and cloning it into a vector containing a promoterless lacZ gene to generate plasmid pVS160 (Table 1). Transcription from the qseA::lacZ fusion was down-regulated 3.5-fold in the EHEC luxS mutant (VS94) compared to the wild-type (86-24) and the complemented (VS95) strains (Fig. 1B). The discrepancy in fold activations between the gene array data and the promoter fusion data is probably due to copy number effects, since multiple copies of the qseA regulatory region in plasmid pVS160 might saturate the effect of other potential regulators involved in the transcriptional activation of qseA. To further confirm that qseA transcription was up-regulated by quorum sensing and not as a result of some metabolic change associated with a luxS mutation, we tested whether transcription of the qseA::lacZ fusion in a luxS mutant (VS94) was activated by exogenous AI-2. Since the final structure of AI-2 is still not reported and this molecule proved to be very difficult to purify (34), we used media preconditioned with the wild-type (86-24) and complemented (VS95) EHEC strains, both of which produce AI-2. As a negative control we used medium preconditioned with the luxS mutant (VS94), which does not produce AI-2. We then observed that transcription of qseA was activated by media preconditioned with both the wild-type and complemented strains but not with medium preconditioned with the luxS mutant (Fig. 1C), indicating that the activation of qseA transcription was due to AI-2 signaling and therefore quorum sensing.
Characterization of a qseA mutant.
Since transcription of qseA was up-regulated by quorum sensing and the predicted QseA protein shared homology with V. cholerae and P. aeruginosa regulators involved in pathogenesis, we generated a qseA isogenic mutant of EHEC strain 86-24, yielding strain VS145. The qseA gene is not in an obvious operon with any other gene in either the K-12 or EHEC chromosomes; hence, the insertion of a chloramphenicol resistance cassette in the middle of the qseA sequence should disrupt only this gene without polar effects.
We initially examined whether a qseA mutation had any effect on AI-2 production by performing the V. harveyi AI-2 luminescence detection assay described by Surette and Bassler (39). Media preconditioned with the wild-type strain (86-24), the qseA mutant (VS145), or VS145 complemented with qseA cloned in a multicopy plasmid (VS151) all induced luminescence in V. harveyi strain BB170 in response to AI-2, indicating that a qseA mutation did not abolish the ability of EHEC to produce AI-2 and that QseA is not involved in the production of AI-2 (data not shown).
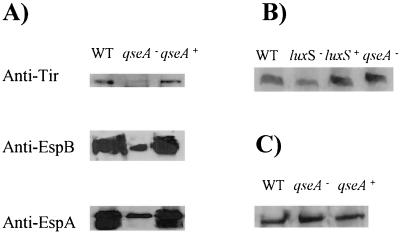
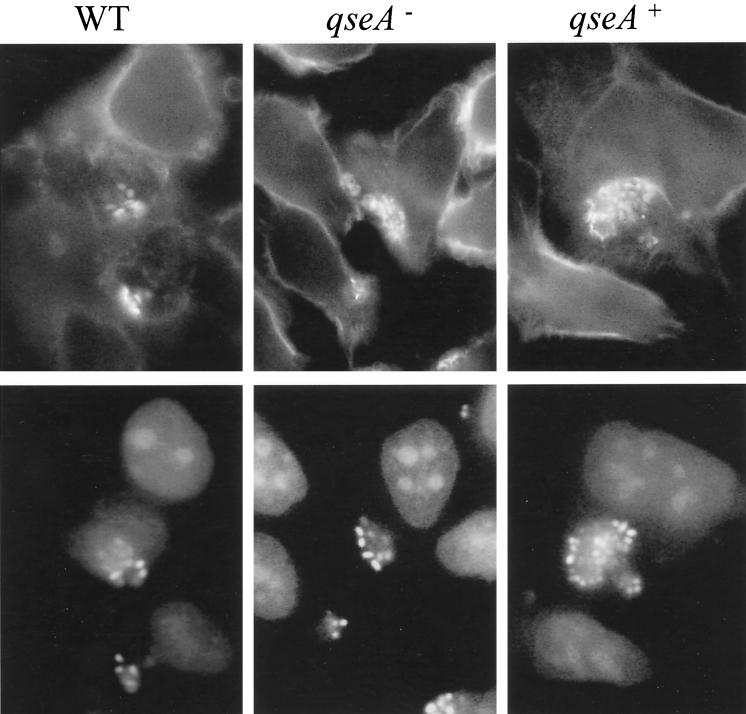
Since we previously reported that quorum sensing activated the LEE genes encoding a type III secretion system (36), we examined whether a qseA mutation had any effect on this secretion system. The qseA mutant secreted strikingly reduced amounts of Tir, EspA, and EspB compared to the wild-type strain (Fig. 3A), and this secretion defect could be complemented when qseA was provided in a multicopy plasmid. Similar effects on secretion of Tir, EspA, and EspB were also observed with the EPEC qseA mutant VS193 (data not shown). The qseA mutant (VS145) is still able to produce AE lesions, which are characterized by the polymerization of actin beneath the bacteria. This phenomenon was observed by the FAS test, in which the actin filaments are labeled by fluorescein (Fig. 4). Although VS145 secretes diminished amounts of Esp proteins and Tir, it would be expected that this strain would still be able to produce AE lesions, since the FAS test is qualitative and not quantitative and this test is positive even when bacteria secrete barely detectable amounts of Esp proteins and Tir (4). The EPEC qseA mutant was also able to produce AE lesions in the FAS test (data not shown).
FIG. 3.
(A) Western blots of secreted proteins from strains 86-24 (wild type [WT]), VS145 (qseA mutant), and VS151 (complemented mutant) probed with anti-Tir, anti-EspA, and anti-EspB polyclonal antisera. (B) Western blot of whole-cell lysates from strains 86-24, VS145, and VS151 with anti-H7 flagellin antiserum. (C) Western blot of whole-cell lysates from strains 86-24, VS145, and VS151 with anti-Stx2A antiserum.
FIG. 4.
FAS test with HeLa cells infected for 6 h at 37°C and 5% CO2 with strains 86-24, VS145, and VS151. The upper panels are the cells stained with fluorescein isothiocyanate-phalloidin, and the lower panels are the bacteria stained with propidium iodide. WT, wild type.
We did not observe any difference in Stx production, flagella production (Fig. 3B and C), or motility (data not shown) between the wild type and the qseA mutant. These results indicate that QseA has a role in regulating the type III secretion LEE genes by quorum sensing but does not appear to regulate genes involved in flagellation, motility, and Stx production.
QseA is an activator of ler.
Since the qseA mutant (VS145) was defective for type III secretion, we examined whether QseA might function as an activator for the LEE genes at the transcriptional level. To address this question, we introduced plasmids containing ler-1[LEE1], sepZ1[LEE2], orf12[LEE3], and tir::lacZ transcriptional fusions in three different genetic backgrounds, the wild-type strain (86-24), the qseA mutant (VS145), and the complemented strain (VS151), and assayed these fusions for β-galactosidase activity. There was a fourfold induction in the expression of LEE1 in VS151 (VS145 complemented with qseA in pACYC177) compared to the expression of this fusion in VS145 (86-24 qseA) (data not shown). We could not observe any significant differences in the expression of ler-1[LEE1]::lacZ between 86-24 and VS145. This might be due to copy number effects, since the ler-1[LEE1]::lacZ fusion is in a multicopy plasmid and qseA is in a single copy in the chromosome of 86-24; when both the ler-1[LEE1]::lacZ fusion and qseA were in multicopy plasmids (VS151), we could observe activation of LEE1 in the presence of qseA. The first gene in the LEE1 operon is ler, which directly activates the LEE2, LEE3, and tir operons (2, 26, 37; J. L. Mellies, unpublished data). There were no significant differences in the expression of β-galactosidase from plasmids containing sepZ1[LEE2], orf12[LEE3], or tir::lacZ fusions in the wild-type, qseA mutant, and complemented strain backgrounds (data not shown). At the present time, we cannot rule out that these results might also be due to copy number effects, since ler was in a single copy in the chromosome in all of these experiments and QseA acts upstream of Ler.
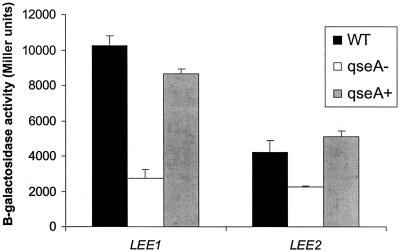
To address potential copy number effects, we generated ler-1[LEE1]::lacZ and sepZ1[LEE2]::lacZ chromosomal single-copy fusions in wild-type EPEC strain E2348/69 and its isogenic qseA mutant (VS193). Transcription of both fusions (ler-1[LEE1] and sepZ1[LEE2]) was decreased in the qseA mutant compared to the wild-type strain (Fig. 5) and was restored when the mutation was complemented with qseA on a plasmid. These results confirm that QseA activates LEE1 (ler transcription) and, through Ler, transcription of LEE2.
FIG. 5.
β-Galactosidase activities of ler-1[LEE1] and sepZ1[LEE2]::lacZ single-copy chromosomal fusions in E2348/69 (wild-type [WT] EPEC), VS193 (EPEC qseA mutant), and VS193 complemented with pVS208 (qseA+). Error bars indicate standard deviations.
QseA acts at the distal LEE1 promoter.
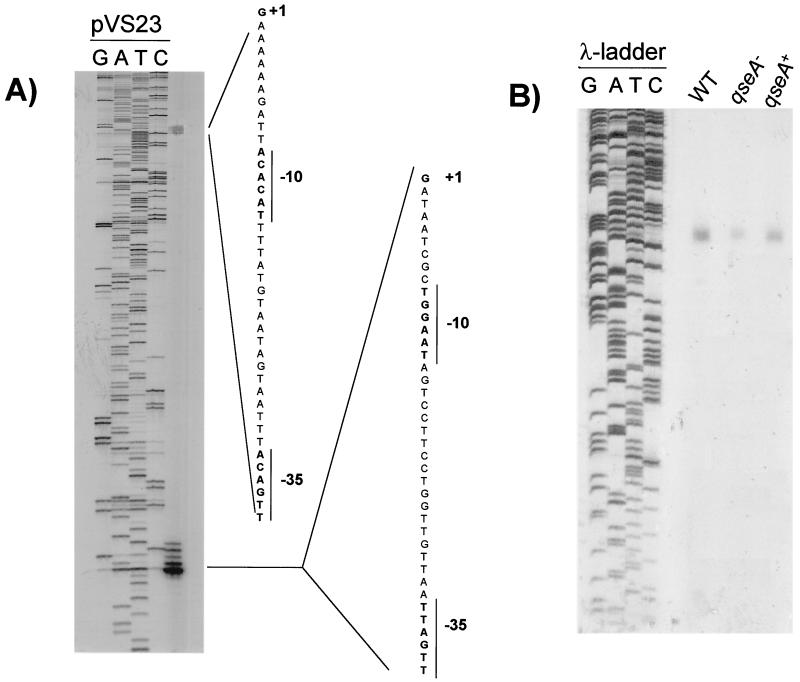
We further confirmed the regulation of LEE1 transcription by QseA by using primer extension analysis. The LEE1 operon in EHEC has two transcriptional start sites and consequently two promoters, one of which is a proximal promoter whose transcriptional start site we previously reported to be located 32 bp upstream from the ler translational start site (36). This promoter is present only in EHEC and is absent in EPEC (7, 36). The second, distal promoter (whose transcriptional start site is located 163 bp upstream from the ler translational start site) is present in both EHEC and EPEC and was previously mapped in EPEC by Mellies et al. (26) (Fig. 6A). In our primer extension analysis, there is a weaker band that mapped to this transcriptional start site and a stronger band that mapped to the previously reported proximal start site (Fig. 6A). Using RNAs extracted from strains 86-24, VS145, and VS151 in the primer extension analysis, we observed a fourfold up-regulation of LEE1 transcription through the distal promoter in 86-24 compared to VS145 (pixel volume intensities of 42,849 and 10,620, respectively) and a threefold up-regulation in VS151 compared to VS145 (pixel volume intensities of 30,261 and 10,620, respectively) (Fig. 6B).
FIG. 6.
(A) Primer extension analysis of the LEE1 promoter region using primer K983 (bp 40912 to 40928 of the EHEC LEE sequence, GenBank accession number AE005593) and RNA from strain 86-24 grown in DMEM to an OD600 of 1.0. The primer extension was run in parallel to a sequencing reaction of plasmid pVS23 (regulatory region of LEE1 in pBluescript). (B) Primer extension analysis of LEE1 using primer K983 and RNAs from strains 86-24, VS145, and VS151 grown in DMEM to an OD600 of 1.0. The primer extensions were run in parallel to a sequencing reaction of a λ ladder control. WT, wild type.
The observation that the promoter involved in QseA activation of LEE1 is the distal promoter, which is also present in EPEC, is consistent with our previous description that LEE1 from both EPEC and EHEC is activated by quorum sensing (36). Taken together, these results suggest that transcription of qseA is activated by quorum sensing, QseA in turn activates transcription of ler, and Ler activates transcription of the LEE2, LEE3, and tir operons in a cascade fashion (Fig. 7).
FIG. 7.
Model of quorum-sensing regulation in EHEC. Quorum sensing activates transcription of qseA, which in turn activates transcription of the LEE genes by quorum sensing. Whether QseA directly binds to the ler promoter is not yet established. There are also additional transcriptional factors, not yet described, involved in the activation of the LEE genes by quorum sensing. For clarity, additional factors that regulate expression of LEE genes by non-quorum-sensing mechanisms, such as integration host factor, H-NS, and Per, are not depicted.
Concluding remarks.
In this study we describe for the first time a regulator that acts upstream of Ler in EHEC and EPEC and is part of the quorum-sensing cascade that regulates the LEE genes through the luxS system. However this regulator is not the sensor for AI-2 in this cascade, i.e., the regulator that binds to AI-2, since a qseA mutant is defective for type III secretion but not for other phenotypes regulated by the luxS quorum-sensing system. We do not know if QseA is directly activating ler transcription or if it is acting through yet another regulator. This question will be addressed in the future by biochemical studies using purified QseA. We also have preliminary data suggesting a role in this quorum-sensing regulatory cascade for three other putative E. coli regulators from the 19 observed through the gene array (data not shown). In Fig. 7 we depict our present model for the quorum-sensing regulatory cascade in EHEC. In the presence of sufficient amounts of AI-2, qseA transcription is activated, and in turn QseA activates the expression of Ler, which activates the LEE genes. Other regulators are involved in the regulation of flagellar, Shiga toxin production, motility, and other metabolic genes by quorum sensing. One two-component regulatory system, which we have named QseBC, is at least partly responsible for regulation of these factors through quorum sensing (V. Sperandio, A. Torres, and J. B. Kaper, submitted for publication). Quorum-sensing regulatory cascades are usually very complex and involve a variety of regulators (reviewed in reference 3). We are currently conducting additional studies to further elucidate this complex quorum-sensing regulatory cascade.
Acknowledgments
We thank Alison O'Brien (Uniformed Services University of the Health Sciences) for the anti-Stx2A antiserum and Kristen Roberts for help with the Western blots.
This work was supported by Public Health Service grants AI41325, AI21657, and DK58957.
Editor: V. J. DiRita
REFERENCES
- 1.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante, V., F. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 3.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devinney, R., I. Nisan, S. Ruschkowski, I. Rosenshine, and B. B. Finlay. 2001. Tir tyrosine phosphorylation and pedestal formation are delayed in enteropathogenic Escherichia coli sepZ::Tn phoA mutant 30-5-1(3). Infect. Immun. 69:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg, M. S., J. Yu, and J. B. Kaper. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175:4670-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. A. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator (Ler) controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engebrecht, J., K. Nealson, and M. Silverman. 1982. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 12.Hamood, A. N., J. A. Colmer, U. A. Ochsner, and M. L. Vasil. 1996. Isolation and characterization of a Pseudomonas aeruginosa gene ptxR, which positively regulates exotoxin A production. Mol. Microbiol. 21:97-110. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis, K. G., J. A. Girón, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a specialized secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jerse, A. E., and J. B. Kaper. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 16.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 17.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutton, S., I. Rosenshine, M. J. Pallen, L. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacikova, G., and K. Skorupski. 1999. The Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraiss, A., J. Blass, and J. Reidl. 1998. A suicide plasmid (pJR lacZins) for targeted integration for non-native genes into the chromosome of Escherichia coli. Technical Tips Online 1:TO1249. [Online.] http://research.bmn.com/tto.
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 34.Schauder, S., S. Kevan, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 35.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53: 85-96. [DOI] [PubMed] [Google Scholar]
- 36.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic E. coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 38.Sperandio, V., A. G. Torres, J. A. Girón, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]