Abstract
Haemophilus ducreyi, the etiologic agent of chancroid, has been shown to form microcolonies when cultured in the presence of human foreskin fibroblasts. We identified a 15-gene cluster in H. ducreyi that encoded predicted protein products with significant homology to those encoded by the tad (for tight adhesion) locus in Actinobacillus actinomycetemcomitans that is involved in the production of fimbriae by this periodontal pathogen. The first three open reading frames in this H. ducreyi gene cluster encoded predicted proteins with a high degree of identity to the Flp (fimbria-like protein) encoded by the first open reading frame of the tad locus; this 15-gene cluster in H. ducreyi was designated flp. RT-PCR analysis indicated that the H. ducreyi flp gene cluster was likely to be a polycistronic operon. Mutations within the flp gene cluster resulted in an inability to form microcolonies in the presence of human foreskin fibroblasts. In addition, the same mutants were defective in the ability to attach to both plastic and human foreskin fibroblasts in vitro. An H. ducreyi mutant with an inactivated tadA gene exhibited a small decrease in virulence in the temperature-dependent rabbit model for experimental chancroid, whereas another H. ducreyi mutant with inactivated flp-1 and flp-2 genes was as virulent as the wild-type parent strain. These results indicate that the flp gene cluster is essential for microcolony formation by H. ducreyi, whereas this phenotypic trait is not linked to the virulence potential of the pathogen, at least in this animal model of infection.
Haemophilus ducreyi is the etiological agent of the sexually transmitted disease known as chancroid (52). Although the occurrence of this disease is rare in the United States, it is one of the leading causes of genital ulcer disease in some developing countries (49, 52, 53). A number of putative virulence factors of H. ducreyi have been identified, including several outer membrane proteins (17, 45, 47), two toxins (3, 16, 37), lipooligosaccharide (LOS) (8, 15, 20, 48; R. S. Munson, Jr., J. Wang, W. Melaugh, A. A. Campagnari, S. Grass, and B. Gibson, Abstr. Gen. Meet. Am. Soc. Microbiol., abstr. B-451, p. 233, 1996), a copper-zinc superoxide dismutase (42), and a very large protein detected in H. ducreyi culture supernatant fluid (54). To date, isogenic mutants lacking expression of the peptidoglycan-associated lipoprotein (19), the hemoglobin-binding outer membrane protein HgbA (7), and the DsrA outer membrane protein (11) have been shown to exhibit reduced virulence in the human challenge model for experimental chancroid.
The finding that the ability of the H. ducreyi HhdA hemolysin to kill human cells in vitro was contact dependent (3, 36) has stimulated interest in the mechanism(s) used by this pathogen to attach to host components. It has been well documented that H. ducreyi can adhere to human epithelial cells in vitro (1, 2, 14, 30, 51), as well as to extracellular matrix components (9). However, the identities of the gene products responsible for these adherence capabilities have not been determined conclusively. Two different laboratories have indicated that LOS is likely involved in attachment to human cells (1, 20). H. ducreyi also has been shown to express fine tangled pili (13), although it was found that an isogenic H. ducreyi mutant unable to express these pili attached at wild-type levels to extracellular matrix (9) and was fully virulent in the human challenge model (5).
There have been numerous examples of structures that mediate attachment of bacterial cells to target tissue(s). Among the best characterized are the type I (35) and type IV (34, 38, 50) fimbriae, the latter having been identified in a broad spectrum of gram-negative bacteria, including Neisseria gonorrhoeae, Neisseria meningitidis, Moraxella bovis, Pseudomonas aeruginosa, Dichelobacter nodosus, and Vibrio cholerae (44). Recently, a potentially novel class of fimbriae (pili) was identified in both Caulobacter crescentus (43) and Actinobacillus actinomycetemcomitans (26). Fimbriae purified from both organisms were shown to be comprised of a major subunit with an apparent molecular mass of 5 to 6 kDa (25, 43). The amino acid sequences of these small proteins had some similarity to known type IV prepilin proteins, and the A. actinomycetemcomitans protein was designated Flp (for fimbria-like protein) (25). The gene encoding the major fimbrial subunit proved to be part of a large gene cluster in both organisms that may constitute a polycistronic operon.
In this study, we identified a gene cluster in H. ducreyi that encoded protein products with significant homology to the predicted protein products of the A. actinomycetemcomitans and C. crescentus gene clusters described above. Inactivation of open reading frames (ORFs) within this H. ducreyi gene cluster significantly reduced the ability of H. ducreyi to form microcolonies when cultured in vitro with human foreskin fibroblasts (HFF). Complementation analysis and mutant repair experiments confirmed the involvement of this gene cluster in microcolony formation in vitro. In addition, mutations in this gene cluster affected the ability of H. ducreyi to attach to plastic and to HFF cells in vitro. Finally, one mutation in this gene cluster had a very small but significant deleterious effect on lesion production by H. ducreyi in an animal model for experimental chancroid.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. ducreyi strains 35000 (23) and 35000HP (6) have been described. The mutants constructed in this study are listed in Table 1. H. ducreyi strains were cultivated on chocolate agar (CA) plates prepared from GC medium (Difco Laboratories, Detroit, Mich.) supplemented with 1% (vol/vol) IsoVitaleX (Becton Dickinson, Sparks, Md.) and 2% (wt/vol) hemoglobin or in Columbia broth (Difco) supplemented with hemin (25 μg/ml), 1% IsoVitaleX, and 5% (vol/vol) heat-inactivated fetal bovine serum. Agar plates were incubated in a humidified incubator in an atmosphere of 95% air-5% CO2 at 33°C, whereas liquid cultures were incubated in either a dry-air incubator or a shaking water bath at the same temperature with continuous agitation at 150 rpm. When necessary, chloramphenicol (1 μg/ml), kanamycin (30 μg/ml), or dihydrostreptomycin sulfate (100 μg/ml) was added to culture media. Escherichia coli strains DH5α, HB101, and Top 10 (Invitrogen, Carlsbad, Calif.) were grown at 37°C in Luria-Bertani broth (Difco) or on Luria-Bertani agar with the following concentrations of antibiotics when appropriate: ampicillin, 100 μg/ml; dihydrostreptomycin sulfate, 100 μg/ml; or chloramphenicol, 30 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Host strain for cloning experiments | 41 |
| HB101 | Host strain essential for propagating plasmids carrying mutated H. ducreyi DNA inserts used in electroporation | 41 |
| H. ducreyi | ||
| 35000 | Wild-type strain isolated in Winnipeg, Manitoba, Canada | 22 |
| 35000.400 | Isogenic mutant with the ΔEcat cartridge inserted into the PmeI site of the tadA gene | This study |
| 35000.401 | Isogenic mutant with the Ω-Cm cassette inserted into the AatII site of the rcpA gene | This study |
| 35000HP | Human-passaged variant of strain 35000 | 46 |
| 35000HP.402 | Isogenic mutant containing a deletion of the flp1 and flp2 genes due to replacement of the 0.3-kb SnaBI fragment spanning these two genes with a promoterless cat cartridge | This study |
| 35000HP.402.301 | 35000HP.402 with a kan cartridge inserted into the cdtA ORF | This study |
| 35000HP.402.301REP | 35000HP.402.301 with wild-type flp-1 and flp-2 ORFs in its chromosome | This study |
| Plasmids | ||
| pACYC184 | Broad-host-range cloning vector; Cmr Tetr | New England Biolabs |
| pCRBlunt II-TOPO | E. coli cloning vector; Kanr | Invitrogen |
| pBluescript KS(+) | E. coli cloning vector; Ampr | Stratagene |
| pJL001 | pCRBlunt II-TOPO with a 2.8-kb PCR product containing the tadA ORF | This study |
| pJL002 | pJL001 containing a ΔEcat cartridge at the PmeI site in the tadA ORF | This study |
| pJN009 | pBluescript KS(+) with a 3.7-kb PCR product spanning flp1 through rcpA | This study |
| pJN013 | pJN009 with an Ω-Cm cassette inserted at the AatII site of the rcpA ORF | This study |
| pJN024 | pACYC184 derivative containing a 2.8-kb PCR product that spans from flp1 to orfC | This study |
| pJN025 | pJN024 with the 0.3-kb SnaBI fragment that spans flp1 and flp2 replaced with a promoterless cat gene | This study |
| pJN030 | pJN024 with a kan gene at the NruI site of pACYC184 | This study |
| pJN031 | pJN030 with a 1.2-kb PCR product containing only the intact flp1 and flp2 ORF | This study |
| pJN032 | pACYC184 with a kan gene inserted at the NruI site | This study |
Tissue culture cells and media.
Two HFF cell lines were used in this study. One was described previously (1), and the other (CRL 7014) was obtained from the American Type Culture Collection. HFF were cultivated in RPMI 1640 (Fisher Scientific Co., Pittsburgh, Pa.) supplemented with 2 mM GlutaMAX (GIBCO-BRL, Rockville, Md), 1 mM sodium pyruvate, and 10% heat-inactivated fetal calf serum (HFF medium) in a humidified incubator containing an atmosphere of 95% air-5% CO2 at either 33 or 37°C as indicated. The HaCaT human keratinocyte cell line (12) was maintained in the same medium.
Reverse transcriptase (RT) PCR analysis.
Total RNA was isolated from bacterial cells by using the Totally RNA kit (Ambion, Austin, Tex.) according to the manufacturer's instructions, and residual chromosomal DNA was removed by treatment with the Message Clean kit (GenHunter, Memphis, Tenn.) according to the manufacturer's instructions. RT-PCRs were performed using the Enhanced Avian HS RT-PCR kit (Sigma) according to the manufacturer's instructions with Taq polymerase obtained from Promega substituted for the proofreading thermostable DNA polymerase mixture provided with the kit.
Real-time RT-PCR.
TaqMan primers and probes for H. ducreyi 16S rRNA, tadB, and tadG were designed using the Primer Express software package (PE Applied Biosystems, Novato, Calif.). TaqMan probes (p45 for tadB, p48 for tadG, and p51 for 16S rRNA [Table 2 ]) were synthesized by Biosearch Technologies, Inc. (Novato, Calif.) using 6-carboxyfluorescein for the fluorophore and Black Hole Quencher 1 as the quencher. Primers p43 and p44 (for tadB), p46 and p47 (for tadG), and p49 and p50 (for16S rRNA) (Table 1) were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). Total RNA was isolated as described above. T7 runoff transcripts were generated using the MaxiScript kit (Ambion) according to the manufacturer's instructions. RT-PCRs were performed as described above. All TaqMan reactions were performed using the PE Applied Biosystems GeneAmp 5700 sequence detection system.
TABLE 2.
Primers (5′-3′) used for RT-PCR and cloning procedures
| Primer | Sequence |
|---|---|
| p01 | AATTGCAGTCGCAGTTGC |
| p02 | ATAGCAGTAACACCTTTTTGG |
| p03 | AGTGGATTTTTATTTGCCC |
| p04 | CGAATATCGGTATAAGATAGCC |
| p05 | GAGGGTTGTTTTTCCGCC |
| p06 | GTGTATTTAGTTGGTCATCAGG |
| p07 | ACAAACGATGAAGCAACAG |
| p08 | GATTTCTGTTTTGCCTTCGCC |
| p09 | CACCGGTACGATGGAACG |
| p10 | GGTTTACGTGCGATTTCG |
| p11 | ACTGACTGAAGTGGCTGC |
| p12 | AAGCCGCGACTACGTAGC |
| p13 | ACAAGGTGGGCAATCTGG |
| p14 | GCTTACTCCGTTCATTTTCG |
| p15 | TTAACGCGTTCAGCCGTG |
| p16 | GAAGCCGATTAGTTCAGC |
| p17 | GGTGGTGCGATTATTCTCTAC |
| p18 | CCTTTCGGTTTTTCATCTTC |
| p19 | TCCGCATGTTCTGTACCG |
| p20 | CTAGTGGCTAGCGTTGTT |
| p21 | CGCCACAAAACAGTGAAGC |
| p22 | GAGCGGCTTCTGTAACTG |
| p23 | TCGCAGATAAAACACCAAC |
| p24 | TAGCCTGTAGAATAGCAACG |
| p25 | TCGCAGATAAAACACCAAC |
| p26 | TTTTCTGCGGTAAGGGAG |
| p31 | CGACATGCATGCTTAGCCGCAAAACAGACCGb |
| p32 | AACTGTGATCTTCAGGGTCG |
| p39 | AGAATAATGCGACCACCCG |
| p40 | GCCATAGTGACTCAATCTC |
| p41 | CACGATATCCGCCACACTATCCACCTATCCc |
| p42 | CATGCATGCTAATTATTAAAAGATACACTTb |
| p43 | CTCGTTTAGGCAGAGCGATTG |
| p44 | CATACGGGCTTCTGCTGTCA |
| p45a | TGCGAATAAAATGGAGCAAAAGAAGAAAGCA |
| p46 | CCTATTGTAGCCTTATTGTTTGTTTCA |
| p47 | GGCTTGCTCTAGTGCATCATTTAA |
| p48a | TAGAGGTAGCAGGCATAATTCAAGATAAGGCCAG |
| p49 | GCGGCAGGCTTAACACATG |
| p50 | TCCCCCTCCATAAGCCAGAT |
| p51a | CCTTGGGTGACGAGTGGCGGA |
Fluorescent probe labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with Black Hole Quencher 1.
SphI site is underlined.
EcoRV site is underlined.
Primer pairs were used in PCR with chromosomal DNA from appropriate wild-type and mutant H. ducreyi strains together with Pfu polymerase. PCR products were inserted into the pCR-BluntII vector (Invitrogen) according to the manufacturer's instructions. The concentrations of all constructs were determined by measuring the optical density at 260 nm (OD260), and this concentration was used to determine the number of DNA molecules per microliter. Tenfold serial dilutions of these constructs were used in PCR with Taq polymerase containing 0.2 μM TaqMan probes, and the dynamic range for all primer-probe pairs was found to span from 108 to 102 molecules/reaction. T7 transcripts were generated as described above, the concentration was determined by measuring the OD260, and 10-fold serial dilutions were made to generate a standard curve for subsequent real-time RT-PCR. For all strains tested, 0.5 μg of total RNA was used to determine the number of transcripts for tadB and tadG, and a 1:1,000 dilution of this solution was used to determine the number of 16S rRNA transcripts.
Mutant construction.
Unless otherwise indicated, Pfu (GIBCO-BRL) was used in PCR to obtain genes or gene fragments. Both DNA strands of cloned PCR products were sequenced using an Applied Biosystems (Foster City, Calif.) model 373A automated sequencer to ensure that no mutations had been introduced by PCR. Each mutant described below was shown to contain the appropriate mutation by both PCR and Southern blot analyses.
(i) tadA mutant.
The oligonucleotide primers p39 and p40 (Table 2) were used in PCR with chromosomal DNA from H. ducreyi strain 35000 to generate a 2.8-kb product containing the tadA ORF. This fragment was cloned into the pCR Blunt II TOPO (Invitrogen) vector to create pJL001. This plasmid was digested with PmeI, which cuts once in the middle of the tadA ORF, and a blunt-ended ΔE cat cartridge from pUC-ΔE cat (provided by Bruce A. Green, Wyeth-Lederle Vaccines and Pediatrics, West Henrietta, N.Y.) was ligated into this site to construct pJL002. The plasmid was used to electroporate E. coli strain HB101 and, after propagation in this strain, was linearized by digestion with EcoRI and used to electroporate H. ducreyi strain 35000 as previously described (24). A chloramphenicol-resistant transformant (35000.400) was selected as the tadA mutant for further study.
(ii) rcpA mutant.
Plasmid pJN009 is pBluescript II KS(+) containing a 3.7-kb H. ducreyi DNA fragment that spans the flp-1 ORF and extends through the rcpA ORF (Fig. 1B). The rcpA ORF was inactivated by ligating a blunt-ended Ω-Cm cartridge (18) into the AatII site within the rcpA ORF, yielding pJN013. This plasmid was used to electroporate E. coli HB101, and the resultant plasmid was digested with EcoRI and used to electroporate H. ducreyi 35000. One of the resultant chloramphenicol-resistant transformants, 35000.401, was selected for further use as an rcpA mutant.
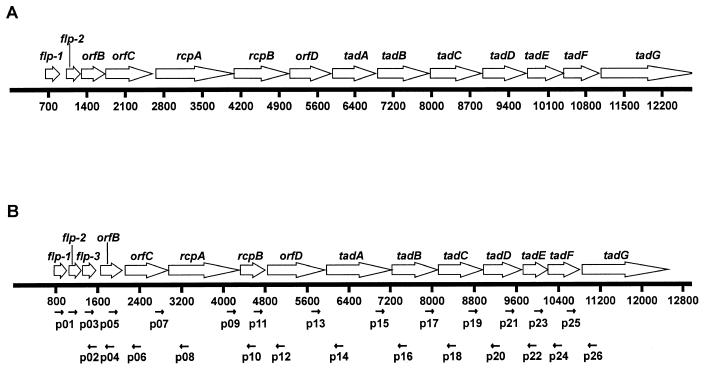
FIG. 1.
Organization of the flp (tad) gene cluster in A. actinomycetemcomitans (A) and the flp gene cluster in H. ducreyi (B). The open arrows represent the directions of transcription of ORFs. Nucleotide position numbers are indicated. (A) ORF map of the 12.4-kb region containing the tad gene cluster in A. actinomycetemcomitans (21, 25, 26). (B) ORF map of the 12.8-kb region containing the flp gene cluster in H. ducreyi. The solid arrows below the scale indicate the relative annealing position of oligonucleotide primers used in PCR and RT-PCR analyses.
(iii) flp-1 flp-2 mutant.
To generate a nonpolar mutation for subsequent use in complementation analysis, the oligonucleotide primers p31 and p32 (Table 2) were used in a PCR to generate a 3.1-kb fragment containing flp-1, flp-2, flp-3, orfB, and most of orfC. Primer p31 contained an SphI site as indicated in Table 2. The PCR product was digested with SphI and EcoRV (which cuts just inside the 5′ end of orfC) and was ligated to SphI/EcoRV-digested pACYC184. This ligation reaction mixture was used to electroporate H. ducreyi 35000, and one of the resultant chloramphenicol-resistant colonies was used as the source of plasmid pJN024. This plasmid was digested with SnaBI, which removed a DNA fragment containing 80% of the flp-1 ORF and 20% of the flp-2 ORF. A 0.8-kb SmaI fragment containing the promoterless chloramphenicol acetyltransferase (cat) gene from pSL1 (33) was then blunt-end ligated into this deletion site, and the ligation reaction mixture was used to electroporate H. ducreyi 35000. The cat cartridge from pSL1 contains a ribosome binding site after the translational stop codon for the cat gene which theoretically allows translation of linked downstream genes (Fig. 1B), thus presumably preventing any polar effects. The resultant chloramphenicol-resistant transformants were screened by restriction digestion and nucleotide sequence analysis to identify a construct with the promoterless cat cartridge oriented in the proper direction in the H. ducreyi DNA insert. This plasmid, pJN025, was digested with AgeI to excise the pACYC184-derived cat gene and the origin of replication. The 5.4-kb AgeI fragment containing the mutated flp-1 flp-2 DNA was purified by preparative agarose gel electrophoresis using the Gene Clean II system (Bio 101, Vista, Calif.) and used to electroporate H. ducreyi strain 35000HP. A chloramphenicol-resistant transformant, 35000HP.402, was selected as the flp-1 flp-2 mutant for further study.
Complementation analysis.
The recombinant plasmid pJN024 was digested with NruI, and a kan cartridge (Amersham Pharmacia) was inserted by blunt-end ligation into this NruI site. The resultant plasmid, pJN030, was then digested with both SphI and EcoRV to excise the entire H. ducreyi DNA insert. Primers p41 (containing an EcoRV site) and p42 (containing an SphI site) (Table 2) were used in PCR as before to generate a 1.2-kb product that contained only the intact flp-1 and flp-2 ORFs and 773 bp of DNA immediately upstream from flp-1. The PCR product was digested with both EcoRV and SphI and ligated to pJN030 that had been previously digested with both enzymes (as described above). The resultant plasmid containing only flp-1 and flp-2 was designated pJN031. The kan cartridge was blunt-end ligated into the NruI site in pACYC 184 to produce pJN032, which was used as a vector equivalent control for pJN031.
Repair of the flp-1 flp-2 mutation.
Plasmid pJL301, containing the H. ducreyi cdtABC gene cluster with a kan cartridge inserted into the cdtA gene (32), was used to electroporate the flp-1 flp-2 double mutant 35000HP.402, and a kanamycin- and chloramphenicol-resistant transformant (35000HP.402.301) was selected on GC-heme agar containing kanamycin (30 μg/ml). The cdtA locus of this transformant was confirmed by Southern blot analysis to have been disrupted (data not shown). Primers p31 and p32 were used in PCR with 35000HP chromosomal DNA to amplify a 3.1-kb PCR product containing the wild-type flp-1 and flp-2 genes together with flanking DNA. Eight micrograms of this PCR product was used to electroporate the flp-1 flp-2 cdtA triple mutant 35000HP.402.301 as described previously (24). Following the 6-h incubation on CA plates after electroporation, the cells were suspended in 6 ml of HFF medium and dispensed into 12 wells of a 24-well microtiter plate (Falcon), each containing 1.5 ml of HFF medium. The plate was incubated for 24 h. Each well was then washed three times with HFF medium, 2 ml of fresh HFF medium was added to each well, and the plate was incubated again for 24 h. After two more consecutive enrichment cycles, one well was observed to contain microcolony-like structures. This well was treated with 1 ml of trypsin-EDTA for 5 min at room temperature, and 10-fold serial dilutions of the resultant cell suspension were spread on CA plates. Two hundred of the resultant colonies were plated on CA plates containing either kanamycin or chloramphenicol. A transformant (35000HP.402.301REP) found to be both kanamycin resistant and chloramphenicol sensitive was purified by single-colony isolation. PCR and Southern blot analyses indicated the absence of the cat cartridge in this transformant; nucleotide sequence analysis confirmed that it contained wild-type flp-1 and flp-2 ORFs.
Polyclonal antiserum to Flp1.
A synthetic peptide (KLKTKFSDLATGISSANGTTSLNSFK) derived from the last 25 amino acids of the Flp1 protein was obtained from BioSyn, Lewisville, Tex. The peptide was conjugated to keyhole limpet hemocyanin as previously described (29) and was used to immunize a rabbit to obtain polyclonal antiserum to Flp1. Because the first 14 amino acids in this peptide are also present in the predicted Flp2 protein from H. ducreyi, this antiserum contained antibodies that bound both Flp1 and Flp2 as determined by Western blot analysis (data not shown).
Western blot analysis for Flp expression.
Whole-cell lysates were prepared as previously described (31). Western blot analysis was accomplished as described previously (32) except that Immobilon-P polyvinylidene difluoride membranes (Millipore) were used in place of nitrocellulose and antibodies bound to target antigens were detected by using the Renaissance Enhanced Luminol chemiluminescent detection system (New England Nuclear, Boston, Mass.).
Adherence assays.
Adherence of bacteria to human cells and to plastic was measured quantitatively. Tissue culture plates (24-well) (Costar, Corning, N.Y.) were inoculated with 105 HFF/well, and the cells were grown to confluence (ca. 60 h). H. ducreyi strains were grown overnight in 35 ml of the Columbia broth-based medium. A 5-ml portion of each bacterial culture was transferred to individual 15-ml conical centrifuge tubes (Becton Dickinson), and the bacterial cells were harvested by centrifugation at 2,450 × g for 10 min at room temperature. The bacterial cells were suspended in HFF medium, and the OD600 was adjusted to 1.0. Portions (5 μl) were added in duplicate to individual wells in the 24-well tissue culture plate, and the bacterial cells were centrifuged onto the confluent monolayers for 5 min at 500 × g at room temperature, after which the plates were incubated for 2 h at 33°C. The wells were then washed three times with HFF medium to remove nonadherent bacteria. After nonadherent cells were removed by washing, 1 ml of trypsin-EDTA (GIBCO-BRL) solution was added to each well, and the plate was incubated for 5 min at room temperature. Then, the contents of each well was suspended by repeated pipetting; serial (10-fold) dilutions of the suspension were spread onto CA plates, which were incubated for 48 h. Plates containing between 30 and 300 colonies were counted to determine the number of adherent bacteria. To measure nonspecific binding to plastic, the same attachment assay was employed except that HFF monolayers were not used in the wells.
Microcolony formation assay.
Microcolony formation assays were performed in the same manner as the quantitative adherence assay except for two changes: the OD600 of the bacterial suspension was adjusted to 0.1, and the 24-well plates containing HFF monolayers were incubated at 33°C for 24 h after inoculation with bacteria. Each well was then washed three times with HFF medium and stained with crystal violet.
Autoagglutination assay.
Each H. ducreyi strain was inoculated into 35 ml of the Columbia broth-based medium and grown overnight at 33°C. A 5-ml portion of each culture was harvested as described above. The bacterial cells were suspended in phosphate-buffered saline by vigorous vortexing, and the OD600 of each suspension was adjusted to 1.0. The OD of this suspension was measured over time.
Analysis of outer membrane antigens.
LOS (8) and outer membrane proteins (28) were characterized as described previously.
Virulence testing.
The relative virulences of the H. ducreyi wild-type and mutant strains used were determined in a blinded manner using the temperature-dependent rabbit model for experimental chancroid (40). Lesion characteristics on days 2, 4, and 7 postinfection were scored using the following numeric values: 0, no change; 1, erythema; 2, induration; 3, nodule; and 4, pustule or necrosis. On day 7 postinfection, material excised from lesions caused by injection of 105 CFU was cultured on CA plates. Statistical analyses were performed as described previously (4, 47).
Nucleotide sequence accession number.
The nucleotide sequence of the H. ducreyi flp gene cluster has been deposited at GenBank and assigned accession number AY083157.
RESULTS
Identification of the flp gene cluster.
Random sequencing of a genomic library of H. ducreyi strain 35000 led to the discovery of an ORF that encoded a predicted protein with homology to the tadA (for tight adhesion) gene product in A. actinomycetemcomitans (26). In this periodontal pathogen, the tadA ORF is located amidst a cluster of 14 ORFs (Fig. 1A) that encode products involved in the ability of the organism to form surface fibrils and adhere tightly to glass (26). Additional nucleotide sequence analysis revealed the presence of several contiguous ORFs extending several kilobases downstream from the tadA ORF in H. ducreyi (data not shown). Eventually, a 12.8-kb region of H. ducreyi DNA was shown to contain 15 ORFs (Fig. 1B) that encoded predicted protein products that were most similar to those encoded by the gene cluster in A. actinomycetemcomitans that contained the tadA ORF.
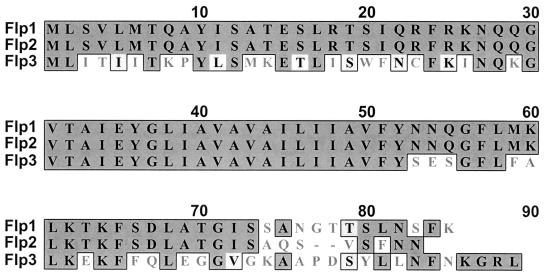
The first three ORFs in this gene cluster encoded predicted protein products that were all similar to that encoded by the flp-1 ORF in A. actinomycetemcomitans (25). The first ORF, designated flp-1, encoded a predicted protein that was 46% identical to the Flp1 protein of A. actinomycetemcomitans. The second and third ORFs encoded predicted proteins that had 88 and 50% identity, respectively, with the protein encoded by the H. ducreyi flp-1 ORF (Fig. 2); these two ORFs were designated flp-2 and flp-3. The predicted Flp2 protein had 50% identity with the Flp2 protein of A. actinomycetemcomitans. The other 12 ORFs were given designations consistent with those of their counterparts in A. actinomycetemcomitans (Fig. 1B). Because the flp-1, flp-2, and flp-3 ORFs are located at the beginning of this set of ORFs, the group of 15 ORFs will be referred to as the flp gene cluster hereafter.
FIG. 2.
Alignment of the amino acid sequences of Flp1, Flp2, and Flp3 from H. ducreyi. The dark shaded boxes indicate identity, and the light shaded boxes indicate conservative substitutions.
The flp gene cluster appears to constitute a polycistronic operon.
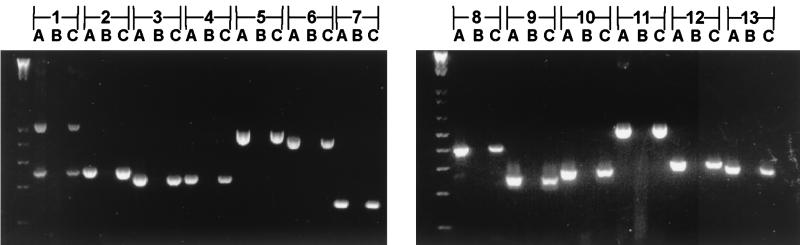
RT-PCR analysis was used to determine whether this gene cluster might comprise a single polycistronic operon. Oligonucleotide primers were designed for the 3′ region of each ORF and the 5′ region of the adjacent ORF for all ORFs in the gene cluster so that a product would result only if the two ORFs were cotranscribed. All primer sets were used in RT-PCR on total RNA isolated from H. ducreyi strain 35000 as described in Materials and Methods. This analysis revealed that all 15 of the ORFs in the flp gene cluster are apparently cotranscribed (Fig. 3).
FIG. 3.
RT-PCR analysis of the flp gene cluster (45). Refer to Fig. 1 for the annealing position for each primer. The reaction conditions were as follows: lanes A, RT-PCR using H. ducreyi chromosomal DNA; lanes B, RT-PCR using total RNA isolated from H. ducreyi without RT; lanes C, RT-PCR using total RNA from H. ducreyi with RT. Reaction numbers are indicated above the lanes. Reaction 1 contained primers p01 and p02. Due to the sequence identity between flp-1 and flp-2, p01 anneals to the middle of both of these ORFs, and this primer pair generated products of 557 and 255 bp that span from the middle of flp-1 to the beginning of flp-3 and from the middle of flp-2 to beginning of flp-3, respectively. Both products were subjected to nucleotide sequence analysis to confirm their identities. Reaction 2 contained primers p03 and p04, which amplify a 255-bp product that spans from flp-3 to orfB. Reaction 3 contained primers p05 and p06, which amplify a 215-bp product that spans from orfB to orfC. Reaction 4 contained primers p07 and p08, which amplify a 225-bp product that spans from orfC to rcpA. Reaction 5 contained primers p09 and p10, which amplify a 494-bp product that spans from rcpA to rcpB. Reaction 6 contained primers p11 and p12, which amplify a 448-bp product that spans from rcpB to orfD. Reaction 7 contained primers p13 and p14, which amplify a 157-bp product that spans from orfD to tadA. Reaction 8 contained primers p15 and p16, which amplify a 395-bp product that spans from tadA to tadB. Reaction 9 contained primers p17 and p18, which amplify a 238-bp product that spans from tadB to tadC. Reaction 10 contained primers p19 and p20, which amplify a 260-bp product that spans from tadC to tadD. Reaction 11 contained primers p21 and p22, which amplify a 539-bp product that spans from tadD to tadE. Reaction 12 contained primers p23 and p24, which amplify a 283-bp product that spans from tadE to tadF. Reaction 13 contained primers p25 and p26, which amplify a 253-bp product that spans from tadF to tadG. Size standards (in 100-bp increments) are present in the first lane in each panel.
Construction of mutants and complementation analysis.
Mutations in the tad gene cluster in A. actinomycetemcomitans have been correlated with decreased adherence to glass surfaces in vitro (26). To determine whether a similar mutant phenotype could be obtained in H. ducreyi, strains were constructed with specific mutations in three ORFs within the flp gene cluster. To construct a tadA mutant, a cat cartridge was inserted into the PmeI site located in the tadA ORF, and allelic exchange was used to construct the isogenic H. ducreyi tadA mutant 35000.400. To determine whether a more severe perturbation of this putative operon would result in a more drastic change in phenotype, the Ω-Cm cartridge, which has transcriptional terminators on both ends (18), was inserted, via allelic exchange, into the rcpA ORF located upstream from tadA in the flp gene cluster (Fig. 1). In A. actinomycetemcomitans, lack of expression of the RcpA protein is associated with little or no expression of fimbriae (surface fibrils) (21).
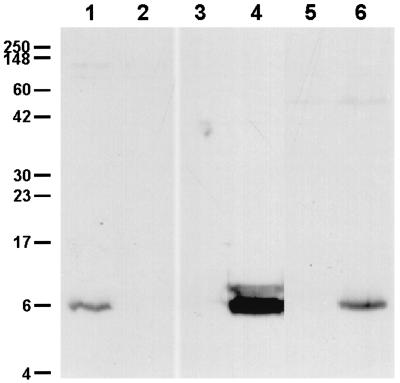
To permit complementation analysis, a nonpolar deletion mutation was constructed within the flp-1 and flp-2 genes located at the extreme 5′ end of this gene cluster. However, unlike the previous two mutants described above, this mutation was constructed in the human-passaged H. ducreyi strain 35000HP (6). A promoterless cat cartridge designed to permit transcription and translation of linked downstream ORFs (33) was inserted into a deletion that spanned part of both the flp-1 and flp-2 ORFs. Western blot analysis confirmed that this flp-1 flp-2 mutant lacked the ability to express the Flp1 and Flp2 proteins (Fig. 4, lane 2), whereas the wild-type strain expressed these macromolecules (Fig. 4, lane 1), as did the rcpA and tadA mutants (data not shown). When plasmid pJN031, which contained intact flp-1 and flp-2 ORFs, was introduced into the flp-1 flp-2 mutant, the complemented mutant expressed the Flp1 and Flp2 proteins as detected by Western blot analysis (Fig. 4, lane 4). Strain 35000HP.402(pJN032) containing the vector equivalent did not express any antigens reactive with this antiserum (Fig. 4, lane 3).
FIG. 4.
Western blot analysis of Flp1 and Flp2 expression by wild-type, mutant, and recombinant H. ducreyi strains. Whole-cell lysates were probed in Western blot analysis with polyclonal rabbit Flp1-keyhole limpet hemocyanin antiserum as the primary antibody. Lanes: 1, wild-type 35000HP; 2, flp-1 flp-2 mutant 35000HP.402; 3, 35000HP.402(pJN032) (vector equivalent control); 4, 35000HP.402(pJN031); 5, flp-1 flp-2 cdtA mutant 35000HP.402.301; 6, 35000HP.402.301REP. Molecular mass position markers in kilodaltons are on the left.
Effects of mutations on transcription of linked ORFs.
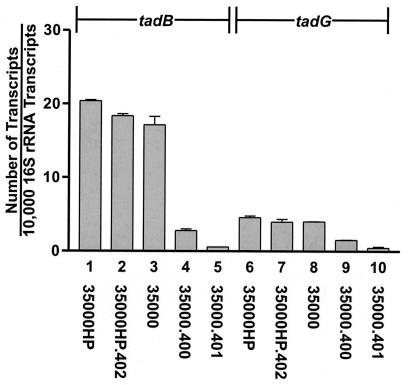
Real-time RT-PCR was used to determine whether the different mutations introduced into the flp gene cluster exerted polar effects on downstream ORFs. Transcripts derived from tadB and tadG were quantitated for all three mutants used in this study. As expected, the presence of the Ω-Cm cartridge (18) in the rcpA ORF in mutant 35000.401 resulted in much-decreased expression from both tadB and tadG (Fig. 5, bars 5 and 10, respectively). Similarly, insertion of the ΔE cat cartridge into the tadA ORF in mutant 35000.400 resulted in a decreased level of expression of these same two transcripts (Fig. 5, columns 4 and 9). In contrast, the presence of the promoterless cat cartridge (33) in the flp-1 flp-2 deletion mutant 35000HP.402 did not appreciably affect the levels of the tadB and tadG transcripts (Fig. 5, columns 2 and 7, respectively) in this mutant relative to those levels observed in the wild-type parent strain (Fig. 5, columns 1 and 6).
FIG. 5.
Real-time RT-PCR analysis of expression of the tadB and tadG ORFs in wild-type and mutant strains of H. ducreyi. Transcripts from the two ORFs were quantitated as described in Materials and Methods. Bars: 1 and 6, wild-type 35000HP; 2 and 7, flp-1 flp-2 mutant 35000HP.402; 3 and 8, wild-type 35000; 4 and 9, tadA mutant 35000.400; 5 and 10, rcpA mutant 35000.401. The error bars indicate standard deviations.
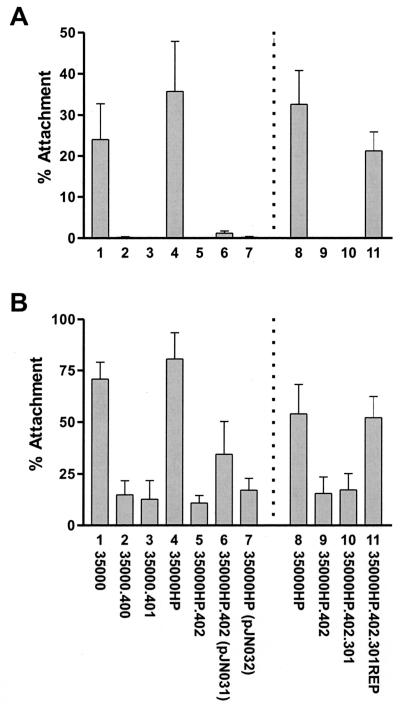
Mutations in the flp gene cluster affect attachment of H. ducreyi to HFF cells and plastic.
To determine whether perturbations in the flp gene cluster had an effect(s) similar to that observed with the tad locus of A. actinomycetemcomitans (26), all three H. ducreyi mutants were tested for the ability to bind plastic. The tadA, rcpA, and flp-1 flp-2 mutants (Fig. 6A, bars 2, 3, and 5) all exhibited a greatly reduced ability to attach to plastic. These mutants also exhibited a decreased ability to bind confluent HFF monolayers (Fig. 6B, bars 2, 3, and 5). Complementation of the flp-1 flp-2 mutant with the recombinant plasmid pJN031 resulted in partial restoration of attachment ability for HFF cells (Fig. 6B, bar 6), whereas the presence of only the vector equivalent pJN032 in this mutant had little or no effect on attachment to HFF cells (Fig. 6B, bar 7). Attachment of this complemented mutant to plastic (Fig. 6A, bar 6) was still much below that obtained with the wild-type parent strain (Fig. 6A, bar 1), although the level of attachment of this complemented mutant to plastic was sixfold higher than that obtained with the same mutant containing only the vector equivalent pJN032 (Fig. 6A, bar 7). When the flp-1 flp-2 mutant was tested for its ability to bind human keratinocytes (i.e., the HaCaT cell line) in vitro, it was found to attach at wild-type levels to these cells (data not shown).
FIG. 6.
Quantitative measurement of the binding of wild-type, mutant, and recombinant H. ducreyi strains to plastic and HFF cells. Assays were performed as described in Materials and Methods. (A) Twenty-four-well plastic tissue culture plates; (B) ATCC CRL-7014 HFF monolayers. The experiments represented by bars 1 through 6 are a composite of two separate experiments. Bars: 1, wild-type 35000; 2, tadA mutant 35000.400; 3, rcpA mutant 35000.401; 4, wild-type 35000HP; 5, flp-1 flp-2 mutant 35000HP.402; 6, 35000HP.402(pJN031); 7, 35000HP.402(pJN032). Two additional experiments were performed with four strains, and the composite results are depicted in bars 8 through 11. Bars: 8, wild-type 35000HP; 9, flp-1 flp-2 mutant 35000HP.402; 10, flp-1 flp-2 cdtA mutant 35000HP.402.301; 11, 35000HP.402.301REP. The error bars indicate standard deviations.
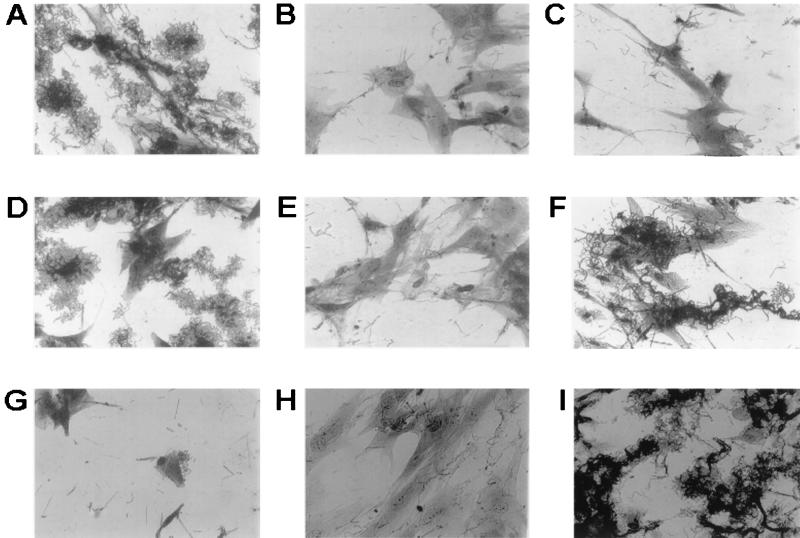
Mutations in the flp gene cluster affect microcolony formation.
The vast majority of H. ducreyi wild-type strains have been shown to form microcolonies when cultured overnight in the presence of HFF cells (4). To determine whether the tadA, rcpA, and flp-1 flp-2 mutants could form microcolonies in vitro, wild-type and mutant strains were cocultured with these human epithelial cells as described in Materials and Methods. After 24 h of growth, strains 35000 and 35000HP readily formed characteristic microcolonies when cultured in the presence of HFF (Fig. 7A and D, respectively). However, with all three mutants (Fig. 7B, C, and E), microcolonies either were not present or were very small in comparison to those produced by the wild-type parent strains. Complementation of the flp-1 flp-2 deletion mutation resulted in restoration of the ability to form microcolonies (Fig. 7F). As expected, the presence of the vector equivalent pJN032 in the flp-1 flp-2 mutant did not confer the ability to form microcolonies on this strain (Fig. 7G).
FIG. 7.
Microcolony formation by wild-type, mutant, and recombinant H. ducreyi strains. Assays were performed as described in Materials and Methods. (A) 35000 (wild type); (B) tadA mutant 35000.400; (C) rcpA mutant 35000.401; (D) 35000HP (wild type); (E) flp-1 flp-2 mutant 35000HP.402; (F) complemented flp-1 flp-2 mutant 35000HP.402(pJN031); (G) 35000HP.402(pJN032); (H) flp-1 flp-2 cdtA mutant 35000HP.402.301; (I) 35000HP.402.301REP. Magnification, ×400.
Repair of the flp-1 flp-2 mutant.
Because the complemented mutant's ability to attach to plastic and HFF cells was much lower than that of the wild-type strain, we used two approaches to minimize the possibility that the flp-1 flp-2 mutant 35000HP.402 contained an undetected secondary mutation in the flp gene cluster or elsewhere in its genome that affected its ability to attach to the two aforementioned targets. First, we reconstructed the flp-1 flp-2 mutant from strain 35000HP and showed that the phenotype of this new mutant in the attachment assays was the same as that of the original flp-1 flp-2 mutant and that complementation with pJN032 did not increase attachment levels beyond those obtained with the original complemented mutant (data not shown). Second, we repaired the mutation in the original flp-1 flp-2 mutant 35000HP.402. To accomplish this, we first introduced a cdtA mutation (32) into this mutant to mark it and eliminate any possibility of isolating a wild-type strain as a result of contamination of the experimental system. Next, this new flp-1 flp-2 cdtA triple mutant 35000HP.402.301 was electroporated with a 3.1-kb PCR product containing the wild-type flp-1 and flp-2 ORFs together with flanking DNA. A transformant with the ability to attach to plastic was selected as described in Materials and Methods and designated 35000HP.402.301REP. PCR and Southern blot analyses confirmed that this transformant lacked the promoterless cat cartridge, and Western blot analysis indicated that it expressed Flp1 and Flp2 (Fig. 4, lane 6). When this transformant (Fig. 6A and B, bars 11) was tested for its ability to attach to plastic and HFF cells, it attached to both these targets at levels very similar to those obtained with the wild-type parent strain (Fig. 6A and B, bars 8). Similarly, the repaired mutant (Fig. 7I) readily formed microcolonies.
Effects of mutations in the flp gene cluster on other factors potentially involved in adherence or microcolony formation.
Alterations in the tad locus genes of A. actinomycetemcomitans appeared to affect not only nonspecific adherence by this organism but also its ability to autoagglutinate in liquid suspension (26). Therefore, it was possible that the defects observed with the H. ducreyi mutants tested in the present study were artifactual and merely reflected the inability of extremely large clumps of autoagglutinated bacteria to adhere to HFF cells in tissue culture. To explore this possibility, we tested both wild-type and mutant strains in an autoagglutination assay as described in Materials and Methods and found no significant differences (data not shown). Similarly, all of these strains grew at similar if not identical rates in Columbia broth in vitro and possessed LOS and outer membrane protein profiles that were indistinguishable (data not shown).
Effects on virulence of mutations in the flp gene cluster.
Because the mutants described above exhibited reduced abilities to form microcolonies in vitro, it was conceivable that this might result in attenuated virulence in vivo, especially in view of the fact that a previous study indicated a possible association between lack of ability to form microcolonies and reduced virulence in the temperature-dependent rabbit model (4). Initially, the tadA mutant was tested in two independent experiments in this animal model. Data presented in Table 3 indicate that there was a very slight but significant reduction in lesion scores obtained with the tadA mutant 35000.400 compared to those obtained with the wild-type H. ducreyi 35000. When the purulent lesions resulting from injection of 105 CFU were excised and macerated, viable H. ducreyi organisms were recovered from every lesion tested for both the wild-type and tadA mutant strains (Table 3). In addition, there was no predominance of wild-type bacteria over mutant bacteria in terms of total numbers of H. ducreyi organisms recovered from these different lesions. When the flp-1 flp-2 mutant 35000HP.402 was tested in this rabbit model, it did not appear to be any less virulent than the wild-type parent strain 35000HP with regard to lesion production (Table 3).
TABLE 3.
Lesion formation by wild-type and mutant strains of H. ducreyi in the temperature-dependent rabbit modela
| Strain | Inoculum size | Mean lesion score (SD)
|
P valueb | ||
|---|---|---|---|---|---|
| Day 2 | Day 4 | Day 7 | |||
| Expt A | |||||
| 35000 | 105 | 3.63 (0.52) | 4.00 (0) | 4.00 (0) | |
| 35000.400 | 105 | 3.63 (0.52) | 4.00 (0) | 4.00 (0) | 0.0156c |
| 35000 | 104 | 3.25 (0.46) | 3.38 (0.52) | 3.50 (0.53) | |
| 35000.400 | 104 | 3.00 (0) | 2.88 (0.35) | 2.38 (0.74) | |
| Expt B | |||||
| 35000 | 105 | 4.00 (0) | 4.00 (0) | 4.00 (0) | |
| 35000.400 | 105 | 3.75 (0.46) | 3.88 (0.35) | 3.88 (0.35) | 0.0125c |
| 35000 | 104 | 3.88 (0.35) | 3.88 (0.35) | 4.00 (0) | |
| 35000.400 | 104 | 3.25 (0.46) | 3.38 (0.52) | 3.38 (0.74) | |
| Expt C | |||||
| 35000HP | 105 | 4.00 (0) | 4.00 (0) | 4.00 (0) | |
| 35000HP.402 | 105 | 3.75 (0.46) | 3.88 (0.35) | 3.88 (0.35) | 0.2928d |
| 35000HP | 104 | 3.38 (0.52) | 3.00 (0.53) | 3.13 (0.35) | |
| 35000HP.402 | 104 | 3.38 (0.52) | 2.63 (0.91) | 2.75 (1.04) | |
| Expt D | |||||
| 35000HP | 105 | 3.75 (0.46) | 3.88 (0.35) | 3.88 (0.35) | |
| 35000HP.402 | 105 | 3.50 (0.53) | 3.63 (0.51) | 3.50 (0.53) | 0.9393d |
| 35000HP | 104 | 3.00 (0) | 3.00 (0.53) | 2.88 (0.64) | |
| 35000HP.402 | 104 | 3.50 (0.53) | 3.00 (1.07) | 3.00 (1.20) | |
Eight rabbits were used in each experiment.
P value calculated for the difference between wild-type and test strain lesion scores. P values were calculated by using the lesion scores from both inoculum sizes and from all 3 days. A P value of 0.0167 was needed for significance.
The tadA mutant was less virulent than the wild-type parent strain in both experiments.
The flp1 flp2 mutant was no less virulent than the wild-type parent strain in both experiments.
DISCUSSION
In this study, we identified a 15-gene cluster in H. ducreyi that was shown to encode products with homology to proteins involved in the formation of surface appendages in other bacteria, including A. actinomycetemcomitans (26) and C. crescentus (43). In A. actinomycetemcomitans, these surface structures (i.e., fibrils) are associated with the ability of the organism to bind nonspecifically to inert surfaces (21, 26). In H. ducreyi, inactivation of any one of several genes in the flp gene cluster resulted in a much-reduced ability to attach to plastic and to HFF cells in vitro (Fig. 6), as well as an inability to form microcolonies when cocultured with HFF cells (Fig. 7).
Inactivation of the flp-1 and flp-2 genes in mutant strain 35000HP.402 eliminated the ability of H. ducreyi to express the Flp1 and Flp2 proteins (Fig. 4); these two proteins are very similar to the proteins reported to constitute the majority of the pili (fimbriae) of both A. actinomycetemcomitans (25) and C. crescentus (43). Examination of the predicted protein (i.e., Flp3) encoded by the ORF immediately following the flp-2 ORF in H. ducreyi indicated that it has similarity to both Flp1 and Flp2. Why H. ducreyi has the genetic information to produce three very similar Flp proteins is not clear.
Regardless of the precise mechanism by which perturbations in the flp gene cluster caused defects in the attachment of H. ducreyi to plastic and HFF cells, these same mutants exhibited a striking decrease in the ability to form microcolonies in vitro (Fig. 7). In a previous study of a large number of H. ducreyi isolates, several strains were found to be unable to form microcolonies when cocultured with HFF cells in vitro (4). Several of these microcolony-negative strains proved to have reduced virulence when tested in the temperature-dependent rabbit model. However, all of these strains except one were also deficient in either the ability to attach to HFF cells in vitro or the ability to kill these human cells (4). The results of the present study indicate, for the first time, that microcolony formation is not essential for expression of virulence by H. ducreyi in this rabbit model, because the flp-1 flp-2 mutant, which did not form microcolonies, was still fully virulent in this model.
It is not clear why complementation of the flp-1 flp-2 mutant with the wild-type flp-1 flp-2 genes in trans did not allow this mutant to attach to plastic or HFF cells at levels approximating those obtained with the wild-type parent strain (Fig. 6). The construction and use of a second flp-1 flp-2 double mutant in complementation analysis yielded the same type of results, suggesting that perhaps the overexpression of the Flp1 and Flp2 proteins in the complemented mutant (Fig. 4, lane 4) inhibited proper assembly of the putative adhesive unit. When this double mutation was repaired, however, attachment levels were restored to levels similar or identical to those obtained with the wild-type parent strain (Fig. 6). This result indicated that the original flp-1 flp-2 mutant was unlikely to have possessed a second, undetected mutation that had altered its ability to bind plastic or HFF cells.
While both A. actinomycetemcomitans and C. crescentus use their respective tad (flp) loci to synthesize proteins which form fibrils, we could not detect such structures on the surface of H. ducreyi 35000 or 35000HP (S. Kachlany, J. Nika, and E. J. Hansen, unpublished observations). However, despite the apparent lack of detectable surface structures, a phenotype similar to that of an A. actinomycetemcomitans tadA mutant was obtained when the flp gene cluster was perturbed in H. ducreyi; that is, the organism became less adherent to an inert surface. There are several possibilities that could explain our inability to detect fibrils on H. ducreyi. First, a fibrillar structure encoded by the flp gene cluster in H. ducreyi may be significantly smaller than those described for A. actinomycetemcomitans and C. crescentus so that it was not detectable by the transmission electron microscopy methods utilized in the present study. Second, the number of fibrils formed by H. ducreyi may be very small, thus making detection of these structures more difficult. Third, any flp-encoded fibrils formed by H. ducreyi may be so fragile that they were destroyed during sample preparation. It is also possible that this putative H. ducreyi adhesin may not form a fibril at all but instead is assembled into an adhesive patch on the bacterial cell surface.
The recent description of the widespread existence of the tadA subfamily of secretion NTPases (39) suggests there exists strong selective pressure for the maintenance of this large gene cluster across a diverse spectrum of bacterial species. Moreover, it was recently proposed that tadA of A. actinomycetemcomitans is the first representative of a distinct subfamily of potential type IV secretion NTPase genes and that the flp-1-tadG gene cluster of this periodontal pathogen may encode a secretion system that is distinct from type II and type IV secretion systems (10). As it is found in both pathogenic and nonpathogenic species of bacteria, it is possible that this gene cluster encodes products that are not directly involved in pathogenesis. However, it was recently reported that the Flp1 protein of A. actinomycetemcomitans appears to share conserved sequence features with known type IV prepilin genes, and it was suggested that Flp proteins may be important for the colonization properties of diverse bacterial species (27). At the very least, it would appear that the protein products encoded by this gene cluster are important for some aspect of the normal life cycle of the organisms which contain these genes. The exact role of the H. ducreyi flp gene products in the pathogenesis of chancroid, if any, remains to be determined.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI32011 to E.J.H. and National Research Service Award F32-AI09845 to C.K.W. The H. ducreyi genome-sequencing project was funded by U.S. Public Health Service grant AI45091 to Robert. S. Munson, Jr.
We are grateful to Bruce A. Green and Slawomir Lukomski for providing plasmids containing antibiotic resistance cartridges, to Michelle Alfa for providing one of the HFF cell lines, and to N. E. Fusenig for providing the HaCaT keratinocyte cell line. We also thank Alexandros Kiupakis for advice on RNA isolation and Donald Sodora for guidance on the use of real-time RT-PCR. Finally, we are grateful to the members of the Hansen laboratory for their comments and suggestions concerning the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Alfa, M. J., and P. Degagne. 1997. Attachment of Haemophilus ducreyi to human foreskin fibrobalasts involves LOS and fibronectin. Microb. Pathog. 22:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, M. J., P. Degagne, and T. Hollyer. 1993. Haemophilus ducreyi adheres to but does not invade cultured human foreskin cells. Infect. Immun. 61:1735-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa, M. J., P. Degagne, and P. A. Totten. 1996. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect. Immun. 64:2349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa, M. J., M. K. Stevens, P. Degagne, J. Klesney-Tait, J. D. Radolf, and E. J. Hansen. 1995. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect. Immun. 63:1754-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Tawfiq, J. A., M. E. Bauer, K. R. Fortney, B. P. Katz, A. F. Hood, M. Ketterer, M. A. Apicella, and S. M. Spinola. 2000. A pilus-deficient mutant of Haemophilus ducreyi is virulent in the human model of experimental infection. J. Infect. Dis. 181:1176-1179. [DOI] [PubMed] [Google Scholar]
- 6.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684-1687. [DOI] [PubMed] [Google Scholar]
- 7.Al Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 8.Bauer, B. A., M. K. Stevens, and E. J. Hansen. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect. Immun. 66:4290-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer, M. E., and S. M. Spinola. 1999. Binding of Haemophilus ducreyi to extracellular matrix proteins. Infect. Immun. 67:2649-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brentjens, R. J., M. Ketterer, M. A. Apicella, and S. M. Spinola. 1996. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J. Bacteriol. 178:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brentjens, R. J., S. M. Spinola, and A. A. Campagnari. 1994. Haemophilus ducreyi adheres to human keratinocytes. Microb. Pathog. 16:243-247. [DOI] [PubMed] [Google Scholar]
- 15.Campagnari, A. A., L. M. Wild, G. E. Griffiths, R. J. Karalus, M. A. Wirth, and S. M. Spinola. 1991. Role of lipopolysaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect. Immun. 59:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope, L. D., S. R. Lumbley, J. L. Latimer, J. Klesney-Tait, M. K. Stevens, L. S. Johnson, M. Purven, R. S. Munson, Jr., T. Lagergard, J. D. Radolf, and E. J. Hansen. 1997. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 94:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellay, R., J. Frey, and H. M. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram negative bacteria. Gene 52:147-152. [DOI] [PubMed] [Google Scholar]
- 19.Fortney, K. R., R. S. Young, M. E. Bauer, B. P. Katz, A. F. Hood, R. S. Munson, Jr., and S. M. Spinola. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, B. W., A. A. Campagnari, W. Melaugh, N. J. Phillips, M. A. Apicella, S. Grass, J. Wang, K. L. Palmer, and R. S. Munson, Jr. 1997. Characterization of a transposon Tn 916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179:5062-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond, G. W., C. J. Lian, J. C. Wilt, W. L. Albritton, and A. R. Ronald. 1978. Determination of the hemin requirement of Haemophilus ducreyi: evaluation of the porphyrin test and media used in the satellite growth test. J. Clin. Microbiol. 7:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond, G. W., C. J. Lian, J. C. Wilt, and A. R. Ronald. 1978. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob. Agents Chemother. 13:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, E. J., J. L. Latimer, S. E. Thomas, M. Helminen, W. L. Albritton, and J. D. Radolf. 1992. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J. Bacteriol. 174:5442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue, T., I. Tanimoto, H. Ohta, K. Kato, Y. Murayama, and K. Fukui. 1998. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42:253-258. [DOI] [PubMed] [Google Scholar]
- 26.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 28.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammel, C. J., N. P. Dekker, J. Palefsky, and G. F. Brooks. 1993. In vitro model of Haemophilus ducreyi adherence to and entry into eukaryotic cells of genital origin. J. Infect. Dis. 167:642-650. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, D. A., J. Klesney-Tait, S. R. Lumbley, C. K. Ward, J. L. Latimer, C. A. Ison, and E. J. Hansen. 1999. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 67:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, D. A., M. K. Stevens, J. L. Latimer, C. K. Ward, K. Deng, R. Blick, S. R. Lumbley, C. A. Ison, and E. J. Hansen. 2001. Characterization of Haemophilus ducreyi cdtA, cdtB, and cdtC mutants in in vitro and in vivo systems. Infect. Immun. 69:5626-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukomski, S., R. A. Hull, and S. I. Hull. 1996. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J. Bacteriol. 178:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marrs, C. F. 1994. Type 4 pili in the families Moraxellaceae and Neisseriaceae, p. 127-143. In V. L. Miller, J. B. Kaper, D. A. Portnoy, and R. R. Isberg (ed.), Molecular genetics of bacterial pathogenesis. ASM Press, Washington, D.C.
- 35.Orndorff, P. E., and C. A. Bloch. 1990. The role of type 1 pili in the pathogenesis of Escherichi coli infections: a short review and some new ideas. Microb. Pathog. 9:75-79. [DOI] [PubMed] [Google Scholar]
- 36.Palmer, K. L., W. E. Goldman, and R. S. Munson, Jr. 1996. An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol. Microbiol. 21:13-19. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, K. L., and R. S. Munson, Jr. 1995. Cloning and characterization of the genes encoding the haemolysin of Haemophilus ducreyi. Mol. Microbiol. 18:821-830. [DOI] [PubMed] [Google Scholar]
- 38.Paranchych, W. 1990. Molecular studies on N-methylphenylalanine pili, p. 61-78. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis. Academic Press, London, United Kingdom.
- 39.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell, B. K., J. A. Richardson, J. D. Radolf, and E. J. Hansen. 1991. A temperature-dependent rabbit model for production of dermal lesions by Haemophilus ducreyi. J. Infect. Dis. 164:359-367. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.San Mateo, L. R., K. L. Toffer, P. E. Orndorff, and T. H. Kawula. 1999. Neutropenia restores virulence to an attenuated Cu,Zn superoxide dismutase-deficient Haemophilus ducreyi strain in the swine model of chancroid. Infect. Immun. 67:5345-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smyth, C. J., M. B. Marron, J. M. Twohig, and S. G. Smith. 1996. Fimbrial adhesins: similarities and variations in structure and biogenesis. FEMS Immunol. Med. Microbiol. 16:127-139. [DOI] [PubMed] [Google Scholar]
- 45.Spinola, S. M., T. J. Hiltke, K. R. Fortney, and K. L. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 64:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinola, S. M., L. M. Wild, M. A. Apicella, A. A. Gaspari, and A. A. Campagnari. 1994. Experimental human infection with Haemophilus ducreyi. J. Infect. Dis. 169:1146-1150. [DOI] [PubMed] [Google Scholar]
- 47.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. R. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, S., B. Schilling, L. Tarantino, M. V. Tullius, B. W. Gibson, and R. S. Munson, Jr. 2000. Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J. Bacteriol. 182:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Telzak, E. E., M. A. Chiasson, P. J. Bevier, R. L. Stoneburner, K. G. Castro, and H. W. Jaffe. 1993. HIV-1 seroconversion in patients with and without genital ulcer disease. Ann. Intern. Med. 119:1181-1186. [DOI] [PubMed] [Google Scholar]
- 50.Tennent, J. M., and J. S. Mattick. 1994. Type 4 fimbriae, p. 127-146. In P. Klemm (ed.), Fimbriae: adhesion, biogenesis, and vaccines. CRC Press, Boca Raton, Fla.
- 51.Totten, P. A., J. C. Lara, D. V. Norn, and W. E. Stamm. 1994. Haemophilus ducreyi attaches to and invades human epithelial cells in vitro. Infect. Immun. 62:5632-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyndall, M. W., A. R. Ronald, E. Agoki, W. Malisa, J. J. Bwayo, J. O. Ndinya-Achola, S. Moses, and F. A. Plummer. 1996. Increased risk of infection with human immunodeficiency virus type 1 among uncircumcised men presenting with genital ulcer disease in Kenya. Clin. Infect. Dis. 23:449-453. [DOI] [PubMed] [Google Scholar]
- 54.Ward, C. K., S. R. Lumbley, J. L. Latimer, L. D. Cope, and E. J. Hansen. 1998. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol. 180:6013-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]