Abstract
The apical membrane antigen 1 (AMA1) has emerged as a promising vaccine candidate against malaria. Advanced evaluation of its protective efficacy in humans requires the production of highly purified and correctly folded protein. We describe here a process for the expression, fermentation, refolding, and purification of the recombinant ectodomain of AMA1 (amino acids 83Gly to 531Glu) of Plasmodium falciparum (3D7) produced in Escherichia coli. A synthetic gene containing an E. coli codon bias was cloned into a modified pET32 plasmid, and the recombinant protein was produced by using a redox-modified E. coli strain, Origami (DE3). A purification process was developed that included Sarkosyl extraction followed by affinity purification on a Ni-nitrilotriacetic acid column. The recombinant AMA1 was refolded in the presence of reduced and oxidized glutathione and further purified by using two ion-exchange chromatographic steps. The final product, designated AMA1/E, was homogeneous, monomeric, and >99% pure and had low endotoxin content and low host cell contamination. Analysis of AMA1/E showed that it had the predicted primary sequence, and tertiary structure analysis confirmed its compact disulfide-bonded nature. Rabbit antibodies made to the protein recognized the native parasite AMA1 and inhibited the growth of the P. falciparum homologous 3D7 clone in an in vitro assay. Reduction-sensitive epitopes on AMA1/E were shown to be necessary for the production of inhibitory anti-AMA1 antibodies. AMA1/E was recognized by a conformation-dependent, growth-inhibitory monoclonal antibody, 4G2dc1. The process described here was successfully scaled up to produce AMA1/E protein under GMP conditions, and the product was found to induce highly inhibitory antibodies in rabbits.
Plasmodium falciparum causes more than three million deaths each year, mostly among children below the age of five (30). The spread of multi-drug-resistant strains of the parasite has underlined an urgent need for a malaria vaccine. Evidence exists from both animal models and human studies that antibodies to erythrocytic and exoerythrocytic parasite antigens can induce protection. Apical membrane antigen 1 (AMA1) is one of the most promising erythrocytic-stage vaccine targets under investigation. Present on the extracellular merozoite stage of the parasite, AMA1 is amenable to host immune intervention during the process of invasion. Indeed, immunization in animal models with affinity-purified or recombinant forms of AMA1 along with adjuvants permissible for human use can induce a protective response against homologous parasite challenge in vivo (1, 5, 7, 23). Homologues of the AMA1 gene have been identified in all of the commonly studied species of Plasmodium (4, 8, 16, 18, 20, 24, 25, 29), and knockout studies have revealed that the expression of AMA1 protein is vital for parasite survival (28).
P. falciparum AMA1 is an integral membrane protein synthesized as a 72-kDa polypeptide (apparent molecular mass, 83 kDa) (24); it is localized in the apical rhoptries of the merozoites present within late-stage schizont (22). Around the time of schizont rupture and erythrocyte invasion, AMA1 of P. falciparum has been shown to be processed to a smaller 66-kDa protein, which is further proteolytically cleaved to 44- and 48-kDa soluble fragments (15, 17). Compared to several other blood stage antigens, AMA1 of P. falciparum shows limited interstrain polymorphism (11). During natural infection, AMA1 induces both B- and T-cell responses (19, 26), and antibodies to both recombinant P. falciparum AMA1 and affinity-purified, naturally induced anti-AMA1 inhibit the growth or invasion of the P. falciparum parasite in vitro (14). The ectodomain of AMA1 comprises a region constituting 16 interspecies conserved cysteine residues. These cysteine residues are cross-linked to form eight disulfide bridges, which in turn divide the ectodomain into three subdomains (13). Correct folding vis-à-vis the presence of these disulfide bonds has been shown, in the cases of the recombinant Plasmodium chabaudi and P. falciparum AMA1 proteins, to be critical for the induction of inhibitory anti-AMA1 antibodies (1, 14).
Although the function of AMA1 remains unclear, there is a growing need to focus resources on a human trial to evaluate the protective potential of AMA1 of P. falciparum in human volunteers. As a step in that direction, we have expressed a synthetic gene encoding 449 amino acids encompassing the three subdomains of the AMA1 ectodomain from P. falciparum in Escherichia coli. The protein, designated r-AMA1/E (“r” stands for recombinant, and “E” represents the E. coli codon bias of the synthetic gene), was refolded and purified, and the final protein product was designated AMA1/E. Biochemical characterization and evidence of correct folding of AMA1/E are presented. In addition, the in vitro parasite invasion data with antibodies raised against AMA1/E reaffirm the potential of AMA1 to be an important component of a future malaria vaccine.
MATERIALS AND METHODS
Cloning and expression.
A nucleotide construct encoding 449 amino acids of AMA1 of the P. falciparum 3D7 clone (residues 83Gly to 531Glu) was commercially synthesized with an E. coli codon bias (Retrogen, San Diego, Calif.). The synthetic gene insert was ligated to the NcoI and NotI sites of a modified pET32 plasmid called pWRMAL. The modifications in the plasmid included the replacement of the thioredoxin and other N-terminal tags with sequences that resulted in minimal non-AMA1 amino acids fused to the final recombinant protein and the addition of a gene for tetracycline resistance (E. Angov, unpublished data). To correct a reading frame error, the recombinant vector was cut with NcoI, end filled with Klenow fragment, and religated. The final construct contained 18 amino acids fused to the N terminus and 11 amino acids fused to the C terminus. The resulting protein construct was designated r-AMA1/E (MAHHHHHHPGGSGSGTMH-[AMA1 amino acids 83 to 531]-AAALEHHHHHH). The recombinant plasmid was transformed into E. coli Sure II cells, and the insert was sequenced on both strands. For protein expression, the recombinant plasmid was transformed into a redox-modified host E. coli strain [Origami (DE3); Novagen, Madison, Wis.]. Origami (DE3) cells are tetracycline and kanamycin resistant. The expression of r-AMA1/E protein was confirmed by IPTG (isopropyl-β-d-thiogalactopyranoside) induction in shake flask cultures, and glycerol stocks were prepared.
Fermentation (GMP production).
The expression of r-AMA1/E protein was carried out in a 10-liter bioreactor (New Brunswick Scientific, Edison, N.J.) at the lab scale and in a 300-liter bioreactor (New Brunswick Scientific) at the Walter Reed Army Institute of Research Pilot Bioproduction Facility. Medium consisting of Super Broth containing 0.8% glycerol and 12.5 μg of tetracycline ml−1 was inoculated with a 3-liter overnight culture started from production seed lot no. 0788. The bioreactor temperature was maintained at 27°C (pH 7.2), and agitation was maintained at 400 rpm. At a cell density for which the optical density at 600 nm (OD600) was 7.0, IPTG was added to a final concentration of 0.1 mM. One hour later, cells were harvested by centrifugation and frozen at −80°C. Aliquots (10 to 150 g) from this production were used to develop a purification and refolding process of r-AMA1/E protein at the lab prior to scale-up (1,500 g) in a GMP environment.
Plasmid stability.
The presence of recombinant plasmid in E. coli Origami (DE3) cells after fermentation was determined by plating an appropriate dilution of cells on Luria-Bertani agar plates containing either tetracycline (12.5 μg ml−1) or ampicillin (100 μg ml−1) (selective plates) and on Luria-Bertani agar plates containing no antibiotic (nonselective plates). The percent plasmid retention (the number of colonies on selective plates divided by the number of colonies on nonselective plates) was calculated by using colony counts on appropriate dilution plates containing between 30 and 300 colonies.
Metal affinity purification (lab scale).
All buffers were endotoxin free and were maintained chilled; all chemicals used during purification were American Chemical Society certified or the next best available grade. Purification was carried out at room temperature on a Waters-600 liquid chromatography system configured to run Amersham Pharmacia (Piscataway, N.J.) HR columns. Cell paste was thawed and suspended in 5× (wt/vol) buffer A (15 mM Na2HPO4, 5.1 mM KH2PO4, 450 mM NaCl; pH 7.4) and mixed until homogeneous. A solution of 20% sodium N-lauroylsarcosine (Sarkosyl) was added to a final concentration of 5% detergent. This suspension was mixed, and the E. coli cells were disrupted by high-pressure microfluidization (Model 1109; Microfluidic Corp., Newton, Mass.). The cell lysate was cleared by centrifugation at 22,000 × g, and the supernatant was diluted fourfold in buffer A before being loaded onto a Ni2+-nitrilotriacetic acid Superflow column (Qiagen, Valencia, Calif.) (0.5 ml of packed resin per g of paste). The Ni2+ column was pre-equilibrated with buffer B (buffer A containing 1.25% Sarkosyl; pH 7.4). After the lysate was loaded, the Ni2+ column was washed with 20 column volumes (CV) of buffer C (buffer A with 10 mM imidazole and 0.125% Sarkosyl; pH 7.4) followed by 20 CV of buffer D (20 mM sodium phosphate, 25 mM imidazole, 0.125% Sarkosyl; pH 8.0). Bound proteins were eluted from the column in buffer D containing 500 mM imidazole (pH 8.0).
Refolding.
The Ni2+ elution was diluted 40-fold (vol/vol) rapidly in degassed buffer E (20 mM sodium phosphate, 1 mM EDTA, 1 mM reduced glutathione [GSH], 0.25 mM oxidized glutathione [GSSG]; pH 8.0). The refolding buffer was prepared fresh, and refolding was carried out at room temperature (∼22°C) for a minimum of 15 h under nitrogen. The final protein product resulting from this refolding protocol was referred to as AMA1/E. Several other variations to the refolding protocol described above were also tested. One such variation included reduction of the Ni2+-eluted proteins with 5 mM dithiothreitol (DTT) for 1 h at 37°C before refolding. The protein that was yielded after reduction and refolding followed by ion-exchange purification was referred to as RR-AMA1/E.
Ion-exchange purification.
Ion-exchange column resins were sanitized with 0.2 N NaOH before use and then equilibrated to the initial binding conditions. After the refolding step, AMA1/E protein was concentrated on a DEAE Sepharose anion-exchange column (Amersham Pharmacia Biotech) (0.25 ml of packed resin per g of paste) and the column was pre-equilibrated with buffer E without the GSH or GSSG. After the protein was loaded, the column was washed with a minimum of 30 CV of the same equilibration buffer followed by 10 CV of buffer F (5 mM sodium phosphate, 50 mM NaCl, 1 mM EDTA; pH 8.0). AMA1/E was eluted in buffer F containing a final concentration of 100 mM NaCl (pH 8.0). AMA1/E eluted from the DEAE column was pH adjusted to 5.7 by the addition of 1 M NaH2PO4 · H20 and loaded on an SP Sepharose cation-exchange column (Amersham Pharmacia Biotech) (0.15 ml of packed resin per g of paste) pre-equilibrated with buffer G (50 mM sodium phosphate, 0.1 mM EDTA, 100 mM NaCl; pH 5.7). The column was washed with 20 CV of buffer G containing a final NaCl concentration of 275 mM (pH 5.7) followed by 10 CV of a pH exchange buffer (5 mM sodium phosphate, 0.1 mM EDTA; pH 7.1). AMA1/E was eluted from the column in formulation buffer (23.5 mM NaH2PO4 · H2O, 37.5 mM NaCl, 0.1 mM EDTA; pH 7.1).
Formulation, lyophilization, and storage.
Purified AMA1/E protein eluted from the SP column was quantified by a Bio-Rad (Richmond, Calif.) DC protein assay. AMA1/E was put in vials at 100 μg ml−1 (65 μg of protein per vial) in the final formulation buffer (23.5 mM NaH2PO4 · H2O, 30 mM NaCl, 0.1 mM EDTA, 3.15% sucrose; pH 7.1) and lyophilized.
Residual Sarkosyl and endotoxin content determination.
The residual Sarkosyl in purified AMA1/E protein preparations was measured by a reversed-phase high-performance liquid chromatography (HPLC) method (3). Endotoxin content was estimated by a chromogenic Limulus amebocyte lysate (LAL) endpoint assay (Associates of Cape Cod, Falmouth, Mass.). Dilutions of all protein samples and LAL standard were prepared in pyrogen-free vials. Positive control solutions prepared for the standard curves ranged from 1 to 0.06 endotoxin units (EU) ml−1 in twofold serial dilutions. A 96-well plate heater was used for incubation at 37°C for 20 min, and the assay was carried out per the manufacturer's instructions. The plates were read at 405 nm on a Vmax kinetic microplate reader (Molecular Devices Corp., Sunnyvale, Calif.).
Purity and stability analyses.
AMA1/E was evaluated for purity on precast polyacrylamide gels (4 to 12% gradient bis-Tris; Invitrogen, Carlsbad, Calif.) run under reduced and nonreduced conditions, with 5 to 10 μg of protein loaded per well. Gels were stained with Coomassie blue, destained, and scanned on a laser densitometer, and the acquired data were analyzed with ImageQuant 5.1 software (Molecular Dynamics, Sunnyvale, Calif.). Residual host cell protein (HCP) content was assessed by enzyme-linked immunosorbent assay (ELISA) and Western blotting with commercially available kits (Cygnus Technologies, Plainville, Mass.). The HCP standard recommended by the manufacturer was used. In addition to this control, a lysate of the host E. coli Origami (DE3) (expressing a Plasmodium vivax MSP1 protein construct) was also tested as a standard at protein concentrations between 15 and 1,000 ng ml−1 to determine whether the kit was capable of detecting proteins from this specific host E. coli. The total protein in the Origami (DE3) lysate was estimated by bicinchroninic acid protein assay (Pierce, Rockford, Ill.). HCP ELISA was performed twice with concentrations of AMA-1/E between 80 and 10,000 ng ml−1, per the standard procedure recommended by the manufacturer. Immunoblotting for HCP determination (Cygnus Technologies kit) was carried out with the HCP standard provided by the manufacturer and also with the Origami (DE3) E. coli lysate, with between 250 and 4,000 ng of protein per well being run on a reducing gel. The proteins were electrophoretically transferred to a nitrocellulose membrane, and the Western blot assay was performed per the manufacturer's instructions. The stability of AMA1/E was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting of protein samples drawn monthly from aliquots stored at −80, −30, 4, 22 (room temperature), and 37°C.
Primary structure analysis.
Purified AMA1/E protein was sequenced by using an automated Edman degradation method on an Applied Biosystems model 477A protein sequencer in line with HPLC (Applied Biosystems model 120A) for the detection of phenylthiohydantoin-derived amino acids. Protein samples were analyzed by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) [MALDI-TOF (MS)] (Voyager biospectrometry RP system; Applied Biosystems) with a sinapinic acid matrix. Lysozyme and cytochrome C were used as mass standards.
Reduction, alkylation, and free thiol analyses.
AMA1/E protein was reduced with a 100-fold molar excess of DTT over cysteines in the presence of either 4 M urea (for SDS-PAGE) or 4 M guanidine-HCl (GuHCl) (for reversed phase [RP]-HPLC and Ellman's test) at 50°C for 1 h. Alkylation was carried out in the presence of either 4 M urea or 4 M guanidine-HCl along with a 1,000-fold molar excess of iodoacetamide over cysteines for 1 h at room temperature in the dark. Free sulfhydryl groups were estimated in the presence and absence of 4 M guanidine-HCl by using Ellman's reagent (5,5′-dithio-bis-3-nitrobenzoic acid) (10). l-Cysteine was used to plot the standard curve.
GPC and RPC.
HPLC analysis of purified protein was carried out with a Waters-510 HPLC pump connected to a Waters-712 WISP autosampler and controlled by Millenium Release 3.2 chromatographic software (Waters Corp., Milford, Mass.). A Waters-996 PDA detector was used to monitor the elution profiles. For gel permeation chromatographic (GPC) analysis, a Shodex protein KW-803 column (Waters Corp.) was used with a 10-μg protein injection. The buffer system consisted of 20 mM sodium phosphate and 100 mM K2SO4 (pH 7.15) at a flow rate of 0.5 ml min−1. The column was calibrated with molecular weight standards (Bio-Rad). Reversed-phase chromatographic (RPC) analysis was done with a C8 Aquapore RP-300 Å column (7 μm, 30 by 2.1 mm) (PE Brownlee, Norwalk, Conn.) at a flow rate of 0.5 ml min−1 and with 4 to 12 μg of protein per load. Solvent A contained 0.05% trifluoroacetic acid in H2O; solvent B contained 0.05% trifluoroacetic acid in acetonitrile. The solvent gradient consisted of 100% solvent A for 5 min, 100 to 30% solvent A over 15 min, 30 to 0% solvent A over 5 min, and back to 100% solvent A over 5 min.
Immune reagents.
Rat monoclonal antibody 4G2dc1 (used at 1.5 μg ml−1 on immunoblots and with ELISA), which recognizes a disulfide bond-dependent conformational epitope on P. falciparum AMA1 (17), was kindly provided by Alan W. Thomas, Biomedical Primate Research Center, Rijswijk, The Netherlands. A pool of immune human sera (used at a 1:1,000 dilution on immunoblots) was collected from an area of western Kenya where malaria is endemic; the same dilution of a pool of commercially obtained normal human sera (The Binding Site Limited, Birmingham, United Kingdom) was used as a negative control.
Immunoblotting.
Proteins were separated by SDS-PAGE and electrophoretically transferred to a nitrocellulose membrane (27). The blot was blocked with 0.5% casein and 0.3% Tween 20 containing phosphate-buffered saline (PBS). An appropriate dilution of primary antibody in PBST (PBS with 0.05% Tween 20) was incubated for 2 h. The blot was washed with PBST and then incubated with a 1:5,000 dilution of horseradish peroxidase (HRP)-conjugated secondary antibody (Southern Biotechnology Associates, Birmingham, Ala.) for 1 h. After being washed with PBST, the blot was developed with either Super Signal chemiluminescent substrate (Pierce) or BM Blue POD substrate (Roche, Indianapolis, Ind.) according to the manufacturer's recommendation.
IFA.
The recognition of P. falciparum 3D7 schizonts by anti-AMA1 antibodies was tested by indirect immunofluorescence assay (IFA). Thin blood smears were fixed with chilled methanol, and serial dilutions of sera in PBST were incubated for 2 h. Slides were washed three times with PBST and incubated with a 1:100 dilution of goat anti-rabbit immunoglobulin G (IgG) fluorescein isothiocyanate-labeled antibodies (Southern Biotechnology Associates) for 1 h. The slides were washed, and antifade solution (Molecular Probes, Inc., Eugene, Oreg.) was applied and read on a UV fluorescence microscope. IFA titers were determined as the last serum dilution with a positive recognition of the parasite compared to the negative adjuvant control rabbit serum diluted 1:20. The assay was done twice on separate days.
Rabbit immunization and total IgG purification.
Groups of three NZW rabbits were immunized three times with 100 μg of lab-grade refolded AMA1/E (animal codes R-1, R-2, and R-3), reduced and refolded protein RR-AMA1/E (R-4, R-5, and R-6), or its reduced and alkylated form, RA-AMA1/E (R-7, R-8, and R-10). Groups of three rabbits received 50 μg (V-2, V-3, and V-4) or 100 μg (V-9, V-10, and V-11) of AMA1/E protein produced under a GMP environment. A control group of 3 rabbits (R-9, V-45, and R-45) was given PBS along with the adjuvant. The formulation was prepared by adding 70% (vol/vol) Montanide ISA-720 (Seppic Inc., Paris, France) to 30% antigen to make a total of 1 ml of emulsion per dose. The immunization was given subcutaneously at multiple sites, with a 3-week interval between consecutive immunizations. Serum samples were collected 2 weeks after each immunization. The rabbits were bled out 2 weeks after the last immunization. Total IgG was purified from 9 ml of pooled rabbit sera (lab-grade AMA1/E or RA-AMA1/E group). The adjuvant control IgG was purified from a single animal (R9). IgG purification was done on a 5-ml protein-G Sepharose column (Amersham Pharmacia Biotech) with IgG binding and elution buffers (Pierce) according to the manufacturer's recommendation.
ELISA.
The antibody response was evaluated by ELISA. Ninety-six-well microtiter plates (Dynax, Chantilly, Va.) were coated with 100 ng of either RA-AMA1/E or AMA1/E per well, incubated overnight at 4°C, blocked for 1 h with PBST containing 5% casein (Sigma, St. Louis, Mo.), and washed with PBST. Consecutive dilutions of individual rabbit sera were incubated for 2 h at room temperature. The plates were washed, and 1:4,000 diluted HRP-conjugated secondary antibody was incubated for 1 h. The plates were washed and developed for 25 min with ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)]-peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). The OD405 was recorded, and comparative ELISA titers were calculated by using regression analysis on the titration curve. The ELISA was repeated three times for each individual serum in triplicate wells on separate days. Competitive ELISA was carried out with sera from three rabbits immunized with lab-grade AMA1/E and two rabbits in the RA-AMA1/E group. The sera were diluted 1:1,000 and preincubated in solution with 15 μg of either AMA1/E or RA-AMA1/E ml−1 or with bovine serum albumin (BSA) overnight at 4°C on a shaker. The tubes were centrifuged at 15,000 rpm for 15 min, and the supernatants were analyzed by ELISA (as described above) with AMA1/E coated on plates. The competition assay was done three times.
Parasite culture and GIA.
P. falciparum clone 3D7 cultures were prepared as described previously (12) in 48-well plates and kept in suspension cultures angled on a rotator platform or under static conditions. All cultures contained a final 10% heat-inactivated normal human serum in bicarbonate-containing RPMI 1640. The plates were gassed with 5% O2-5% CO2 and heat sealed in plastic bags. Synchronized cultures adjusted to 0.2% late ring stages were mixed with test or control serum or IgG (dialyzed into RPMI-NaOH) to a final hematocrit of 4%. In order to assess the antigen specificity of the antibody-mediated inhibitions, antigens (AMA1/E or RA-AMA1/E) were added to the IgG preparations before testing in the growth inhibition assay (GIA). The final concentration of antigens in the GIA was 5.3 μg ml−1 (limited by low solubility of RA-AMA1/E protein). Merozoites were released after approximately 34 h, and developing ring stages were harvested 14 h postinvasion, stained with Hoechst dye 33342, and analyzed by flow cytometry (12). The fluorescence signal was determined for a minimum of 40,000 erythrocytes gated on forward scatter. The fluorescent signal of ring-infected erythrocytes was about 20 times that of uninfected erythrocytes, and schizont-infected erythrocytes, if present, had another 20-fold increase in the signal. Almost all (>99%) of the parasites harvested from the assays were ring forms or early trophozoite stages, as confirmed by spot checks of Giemsa-stained thin smears. The percent inhibition was calculated from the mean parasitemia of triplicate test and control wells as 100% − (test/control). Sera from rabbits immunized with the adjuvant and PBS were used as controls in the GIA. Prebleeds from individual rabbits were also tested.
Statistical analysis.
Microsoft Excel was used to calculate the P values for two-tailed t tests and the correlation coefficients (r2).
RESULTS
Fermentation of E. coli Origami (DE3) expressing the r-AMA1/E protein at 10- and 300-liter scale.
The synthetic gene cloned in the vector pWRMAL was sequenced, and the translation of this gene sequence revealed no amino acid changes from the published 3D7 clone sequence (GenBank accession no. U65407.1) Fermentation conditions were developed in a 10-liter bioreactor and later scaled up to a 300-liter GMP fermentation. The 10-liter fermentation routinely resulted in 150 g of cell paste, while the 300-liter fermentation resulted in 4.5 kg of cell paste. The final plasmid stability for the GMP fermentation was 36%. Although the use of Origami (DE3) increased the proportion of r-AMA1/E in the soluble fraction [compared to that observed with the conventional BL21(DE3) strain], protein fractionation experiments showed that the majority of r-AMA1/E was still localized in the insoluble fraction (data not shown).
Extraction of r-AMA1/E in Sarkosyl and its enrichment by Ni2+ affinity chromatography.
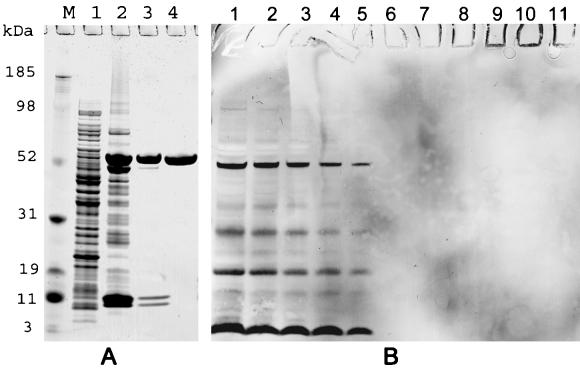
Aliquots were taken from the GMP cell paste lot, and a scalable refolding and purification process was developed. During cell lysis, soluble and insoluble forms of r-AMA1/E were extracted with a buffer containing 5% Sarkosyl. The r-AMA1/E constituted ∼1 to 2% of total cell protein, as estimated by laser densitometry of SDS-PAGE run under reduced conditions (Fig. 1A, lane 1). Following the first step of purification over a Ni2+ column, r-AMA1/E was enriched to ∼40% of total protein (Fig. 1A, lane 2). A large fraction of r-AMA/E present in the Ni2+ elution was aggregated, as seen by nonreduced SDS-PAGE (data not shown).
FIG. 1.
(A) SDS-PAGE analysis of AMA1/E during purification. The results shown are from a lab-grade purification starting with 20 g of cell paste. Protein analysis was done on a 4 to 12% gradient gel under reduced conditions, and the gel was stained with Coomassie blue. The elutions of DEAE and SP Sepharose columns were concentrated on a 3.5-kDa cutoff Centricon concentrator before being loaded. Lane M, molecular mass markers; lane 1, E. coli lysate loaded on the Ni2+ column; lane 2, Ni2+ column elution; lane 3, DEAE Sepharose column elution (5 μg of protein); lane 4, SP Sepharose column elution (5 μg of protein; lab-grade AMA1/E product). (B) Western blot analysis for E. coli protein detection (Cygnus HCP detection kit). Lanes 1 to 5, Origami (DE3) bacterial lysate with 4,000, 2,000, 1,000, 500, or 250 ng of protein, respectively; lanes 6 to 11, AMA-1/E product with 1, 10, 100, 500, 1,000, or 2,000 ng of protein per well, respectively.
Optimization of the refolding conditions.
In order to find the optimal refolding conditions, the Ni2+ elution was subjected to rapid dilution in refolding buffers of various GSH-to-GSSG ratios at pH 8.0 in phosphate buffer. Serial dilutions of these test refolding mixtures were coated on a microtiter plate, and ELISA reactivity against the conformation-specific inhibitory monoclonal antibody 4G2dc1 was used as a measure of folding efficiency, while the reactivity to a monoclonal antihexahistidine antibody was used to confirm equivalent coating efficiency. Ratios of GSH to GSSG tested for refolding included 1/0.1, 1/0.25, 1/1, and 0.1/1, while phosphate buffer containing EDTA (pH 8.0) alone was used as a control. The GSH-to-GSSG ratios of 1/0.1 and 1/0.25 were found to be equally efficient, both giving 4G2dc1 reactivity that was about five times higher than that of the phosphate buffer control. As the GSH-to-GSSG ratio of 1/0.25 had been previously reported for efficient refolding of P. chabaudi AMA1 and the same was more recently used to refold P. falciparum AMA1 expressed in E. coli (6, 14), we chose this ratio to refold r-AMA1/E. After being refolded, r-AMA1/E was designated AMA1/E. A minimum 40-fold dilution of the Ni2+ elution, to about 20 μg of protein ml−1, during refolding was found necessary to minimize aggregation. No significant increase in the monomer yield of AMA1/E was found when the Ni2+ elution was first reduced with DTT prior to refolding, and therefore, the GMP purification process was carried out with the Ni2+ elution without reduction. The presence of low concentrations of Sarkosyl (0.003%) in the refolding mix eliminated the need for a cosolvent during refolding.
Ion-exchange chromatography was used to purify AMA1/E to homogeneity.
After 15 h of incubation in the refolding buffer, AMA1/E was concentrated on a DEAE anion-exchange column and its monomeric form was eluted with 100 mM NaCl, while the impurities and AMA1/E aggregates remained bound to the column. The purity of AMA1/E after this step was ∼90% of the total protein eluted (Fig. 1A, lane 3). A pH adjustment step from 8.0 to 5.7 was needed to bind the majority of AMA1/E to the SP cation-exchange column. This pH change had no effect on the solubility of AMA1/E or its reactivity to immune reagents. AMA1/E bound to the cation exchanger was eluted with the final formulation buffer, eliminating the need for an additional buffer exchange step before formulation. The final yield of AMA1/E was about 0.75 to 1 mg liter of culture−1, with >99% purity estimated by laser densitometry of Coomassie blue-stained gels (Fig. 1A, lane 4). RR-AMA1/E also gave a similar yield and purity (data not shown).
Lyophilized formulation of AMA1/E along with sucrose and EDTA was stable.
A final 3.15% sucrose excipiant was added for stabilization and cake formation during lyophilization. AMA1/E, put in vials at 100 μg ml−1 in 0.65-ml aliquots, was found to be stable in its lyophilized form at 37, 22, and 4°C over a 24-week period, with no signs of breakdown or aggregation. Solution or lyophilized forms of AMA1/E stored at −30 or −70°C showed equivalent stability (data not shown).
Formulated AMA1/E product has very low residual endotoxin, host cell proteins, and Sarkosyl content.
The endotoxin content of purified AMA1/E under lab conditions was between 3 and 5 EU per 50 μg of protein but dropped to below 0.06 EU (lowest value detectable by LAL assay) per 50 μg of protein in GMP purification. No residual Sarkosyl was detected with an RP-HPLC-based assay (minimum detection limit, 0.0005%). The HCP content was determined by ELISA using an anti-E. coli antibody kit capable of quantitatively detecting 15 ng of HCP ml−1 (the lowest concentration of HCP tested) with the Origami (DE3) E. coli lysate. An AMA1/E sample at 10,000 ng ml−1 showed 54 and 44 ng HCP ml−1 (in two tests), giving a final purity of 99.4%. The purity of AMA1/E was also tested with a Western blot HCP determination kit (Cygnus). The Origami (DE3) lysate was used as a positive control at 4,000 to 250 ng of protein per well (Fig. 1B). All of the positive bands at 4,000 ng per well (Fig. 1B, lane 1) were also observed at 1,000 ng per well (Fig. 1B, lane 3). Below 1,000 ng per well, many E. coli protein bands were not detectable. No E. coli-specific bands were seen in AMA1/E lanes with up to 2 μg of AMA1/E loaded per well (Fig. 1B, lane 11).
AMA1/E had the predicted primary and tertiary structure with no free cysteines.
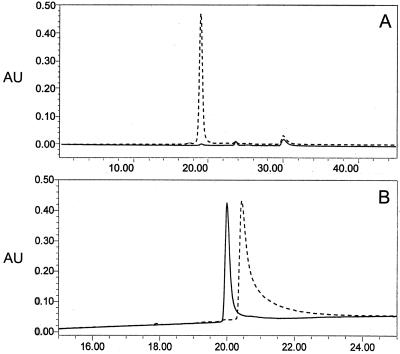
The primary sequence analysis of AMA1/E identified the first 24 N-terminal amino acids to be AHHHHHHPGGSGSGTMHGAEPAP (P. falciparumAMA1-specific residues are underlined). The methionine at the N terminus could not be identified. The MALDI-TOF (MS) analysis showed an average mass of 54,656 Da, while the predicted mass of AMA1/E was 54,633 Da. The final product was evaluated for homogeneity and the presence of multimers by analytical RPC and GPC. A single peak was seen on both GPC and RPC elution profiles, giving evidence of a homogeneous product (Fig. 2). The RPC elution profile of AMA1/E shifted towards higher hydrophobicity under reducing conditions (Fig. 2B, broken line). This indicates the exposure of the protein hydrophobic core upon DTT reduction, which otherwise remained buried due to its compact folded state, was stabilized by disulfide bond formation.
FIG. 2.
Analytical HPLC profile of the AMA1/E product. The detector output at 215 nm with absorbance units (AU) plotted against time (in minutes) is shown. (A) Gel permeation Shodex Protein KW-803 column elution profile with 10 μg of AMA1/E injected (broken line) and an equal volume of final formulation buffer with no protein injected (solid line); (B) reversed-phase C8 Aquapore RP-300 Å column (7 μm, 30 by 2.1 mm) elution profile with a 4-μg AMA1/E injection (solid line) or with 12 μg of AMA1/E reduced in the presence of 6 M GuHCl and 25 mM DTT and injected (broken line), for which a shift in retention time was observed under the same chromatographic conditions. See Materials and Methods for solvent and gradient information.
The primary structure of AMA1/E is expected to contain 16 cysteine residues, and the presence of any free cysteines in the final product would have indicated incorrect folding. The free cysteine content was determined by Ellman's test. Ellman's analysis was also carried out in the presence of 4 M GuHCl to unmask any sulfhydryl groups buried in the hydrophobic core of the protein. Ellman's test detected no free sulfhydryl groups in up to 5 μM AMA1/E, both in the presence and absence of 4 M GuHCl (minimum detection limit, 0.1 μM free sulfhydryl). The absence of free cysteines was further confirmed by treating AMA1/E with an alkylating agent before and after reduction. The mobility of AMA1/E on nonreduced SDS-PAGE gels showed no observable change after treatment with iodoacetamide (Fig. 3A, lanes 1 and 2), while its reductive alkylation caused a significant decrease in mobility (Fig. 3A, lane 3). A recombinant P. vivax MSP-1 p42 fragment (9), which was predicted to contain a single free cysteine, was used as a positive control in both the Ellman's and alkylation analysis, and this free cysteine was identified in both tests (data not shown). The tertiary structure analysis described above also suggests that, as in the case of P. chabaudi AMA1 (13), the majority of AMA1/E molecules also had all 16 cysteines cross-linked by disulfide bonds.
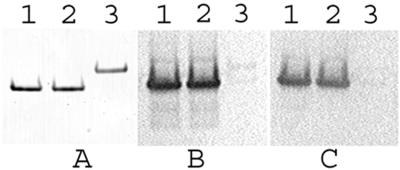
FIG. 3.
Relative SDS-PAGE mobility, alkylation analysis, and immune reactivity of AMA1/E. Lanes 1, AMA1/E protein (∼200 ng) in 4 M urea; lanes 2, AMA1/E in 4 M urea treated with iodoacetamide; lanes 3, AMA1/E in 4 M urea reduced with DTT followed by iodoacetamide treatment (see Materials and Methods for reaction details). (A) Proteins separated on a nonreducing 4 to 12% gradient gel and stained with Coomassie blue; (B) Western blot of the gel shown in panel A immunostained with monoclonal antibody 4G2dc1 and developed with HRP-POD substrate; (C) Western blot of the gel shown in panel A immunostained with an immune serum pool collected from western Kenya and developed with HRP-POD substrate.
AMA1/E reacts with conformation-dependent immune reagents.
AMA1/E reacted with the monoclonal antibody 4G2dc1 on immunoblots (Fig. 3B, lane 1). This monoclonal antibody recognizes a reduction-sensitive epitope on AMA1 of P. falciparum (17). Reactivity on immunoblots was also observed with a hyperimmune-serum pool from a malaria-endemic region of Kenya (Fig. 3C, lane 1). Alkylation of AMA1/E caused no change in its reactivity to the two immune reagents described above (Fig. 3B and C, lanes 2). However, a significant loss of reactivity to both immune reagents was observed upon reductive alkylation (Fig. 3B and C, lanes 3), further confirming the presence of critical reduction-sensitive epitopes on AMA1/E.
AMA1/E was found to be immunogenic in rabbits.
Immunization of rabbits with lab-grade AMA1/E and RR-AMA1/E at 100 μg per dose was done to determine if one form was immunologically superior to the other. A group of three rabbits was immunized with 100 μg of RA-AMA1/E per dose to determine whether disulfide bond-independent epitopes also contributed to the induction of inhibitory anti-AMA1 antibodies. The AMA1/E protein produced under GMP conditions was administered at 50 and 100 μg per dose to determine the immunogenicity at the two doses (50 μg is the expected human dose). No apparent signs of toxicity of the antigen-adjuvant combination were observed in the immunized animals. Table 1 shows the mean log ELISA titer of immunized groups with either AMA1/E or RA-AMA1/E coated on plates. Rabbits in the lab-grade AMA1/E group (R-1, R-2, and R-3) showed high titer antibodies against the AMA1/E protein. No significant difference in the titer was observed between the groups immunized with 50 and 100 μg of GMP protein (data not shown); hence, all six rabbits are represented by a single group in Table 1. The AMA1/E-specific titers observed in the group immunized with 50 and 100 μg of GMP-produced AMA1/E (V-2, V-3, V-4, V-9, V-10, and V-11) were higher than those of the group immunized with 100 μg of lab-grade AMA1/E (two-tailed t test, P = 2.5 × 10−4). The RR-AMA1/E group (R-4, R-5, and R-6) also had high ELISA titers against AMA1/E-coated wells. The mean titer of the RR-AMA1/E group was slightly lower than that of the lab-grade AMA1/E protein-immunized group, although the difference was not statistically significant (P = 8.2 × 10−1). The ELISA titers against the RA-AMA1/E protein-coated wells for the lab-grade, GMP-produced AMA1/E, and RR-AMA1/E groups were lower than those against the refolded-AMA1/E-coated wells (P = 2.5 × 10−2, 1.2 × 10−4, and 1.8 × 10−2, respectively). One of the three rabbits (R-8) immunized with RA-AMA1/E died during handling. Although the two remaining RA-AMA1/E-immunized rabbits (R-7 and R-10) had high titers of antibody against RA-AMA1/E coated wells, the titers were lower against the refolded-AMA1/E-coated wells. This difference was not statistically significant (P = 3.8 × 10−1).
TABLE 1.
Antibody responses and percent inhibition in GIA in vitro with rabbit sera
| Immunization group | No. of animals | Mean log ELISA titer (SD)a
|
Mean log IFA titer (SD)a
|
Mean GIA datab
|
||||
|---|---|---|---|---|---|---|---|---|
| AMA1/ E-coated wells | RA-AMA1/ E-coated wells | No. of expts | Positive schizonts | No. of expts | % Inhibition (SD) | No. of expts | ||
| Lab-grade AMA1/E, 100 μg | 3 | 5.60 (0.04) | 4.94 (0.20) | 3 | 4.41 (0.35) | 2 | 57 (17) | 3 |
| 68 (11)c | 3 | |||||||
| GMP AMA1/E, 50 and 100 μg | 6 | 6.01 (0.13) | 5.29 (0.22) | 3 | 4.71 (0.25) | 1 | 84 (4) | 2 |
| RR-AMA1/E, 100 μg | 3 | 5.57 (0.19) | 5.01 (0.17) | 3 | 4.06 (0.35) | 2 | 29 (22) | 2 |
| 41 (25)c | 2 | |||||||
| RA-AMA1/E, 100 μg | 2 | 4.83 (0.15) | 5.01 (0.17) | 3 | 2.43 (0.11) | 2 | 5 (1) | 1 |
| Adjuvant | 3 | 0.00 | 0.00 | 2 | 0.00 | 1 | −2 (2) | 2 |
Antibody titers were calculated for an OD414 of 0.5 by using regression analysis on titration curves. The log IFA titers were obtained by testing on methanol-fixed P. falciparum (3D7) schizonts.
The percent inhibition of parasite growth in a one-cycle assay under suspension conditions at a 1:5 serum dilution is shown. Percent inhibition was calculated from the mean parasitemia of triplicate test and control wells as 100% − [(test/control) × 100]. The inhibition in GIA was measured relative to the values obtained for control sera or culture media alone. Initial parasitemias were 0.2%, and the final control parasitemias ranged from 2.2 to 3.6%. Compared with growth in media alone (100%), there were no significant differences in the final parasitemias of 12 preimmune sera (mean ± standard deviation, 97% ± 4% [suspension] or 102% ± 1% [static]) or 3 adjuvant-only sera (99% ± 3% [suspension] or 106% ± 2% [static]).
Values under static GIA conditions.
Sera from all AMA1/E-immunized rabbits tested positive by IFA with late-stage schizonts of 3D7 parasites (Fig. 4A). Table 1 shows the mean log IFA titers of the groups. The IFA titer of the GMP-produced AMA1/E group was higher than that of the lab-grade AMA1/E group, although the difference was not significant (P = 2.7 × 10−1). The lab-grade AMA1/E group had slightly higher IFA titers than the RR-AMA1/E group, though this difference was also not significant (P = 2.8 × 10−1). The IFA titers of the lab-grade, GMP-produced AMA1/E, and RR-AMA1/E groups were significantly higher than the RA-AMA1/E group titers (P = 5.4 × 10−3, 1.5 × 10−5, and 8.6 × 10−3, respectively).
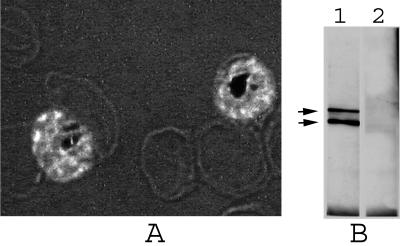
FIG. 4.
Recognition of the parasite AMA1 with anti-AMA1/E antibodies produced in rabbit. (A) Anti-AMA1/E antibodies (R-1) used for IFA on P. falciparum (3D7) parasites fixed with methanol. Merozoites contained within the late-stage schizonts are shown with bright fluorescence. Magnification, ×1,000. (B) Western blot of P. falciparum 3D7 parasite late schizont proteins extracted with SDS-PAGE loading buffer, separated on a nonreducing gel, Western blotted and immunostained with anti-AMA1/E antibodies, and developed with a chemiluminescent substrate. The top and bottom arrows represent ∼76 and 62 kDa, respectively. Lane 1, postimmune rabbit sera (R-1); lane 2, preimmune serum control.
Rabbit antibodies reacted with two bands, one at ∼76 kDa and the other at ∼62 kDa, on Western blots of an SDS extract of a schizont-rich preparation of P. falciparum 3D7 parasites (Fig. 4B, lane 1). These bands most likely correspond to the previously reported 83- and 66-kDa full-length and processed forms of AMA1 in P. falciparum (15, 22); the difference in apparent molecular mass observed here may be a result of a difference in PAGE conditions.
Anti-AMA1/E antibodies inhibit in vitro growth of the parasite.
The GIA of homologous P. falciparum 3D7 parasites was carried out with sera obtained from the immunized rabbits. Table 1 shows the mean percent inhibition under suspension conditions at a 1:5 dilution obtained for each of the immunized groups. The lab-grade and GMP-produced AMA1/E group sera showed significant inhibition of parasite growth compared to that of the adjuvant controls (P = 2.4 × 10−2 and 1.1 × 10−9, respectively). The lab-grade AMA1/E group sera analyzed under static GIA conditions gave even higher inhibition compared to that of the suspension culture, although the difference between static and suspension culture values was not significant (P = 8.4 × 10−2) (Table 1). Rabbits immunized with 50- or 100-μg doses showed no significant difference in the percent inhibition (data not shown). The RR-AMA1/E-immunized group sera showed a lower level of inhibition than the lab-grade AMA1/E group under suspension (P = 1.5 × 10−1) and static (P = 1.9 × 10−2) conditions; the difference was not statistically significant under suspension conditions. There was a positive correlation between log ELISA and log IFA titers (r2 = 0.84). There was also a positive correlation between log ELISA against the AMA1/E protein-coated wells and the percent GIA (r2 = 0.81). A positive correlation was also observed between the log IFA titer and percent inhibition (r2 = 0.77). The r2 values cited above were calculated with data from all of the immunized rabbits in all of the groups. No inhibition was seen in the RA-AMA1/E group compared to the lab-grade AMA1/E group (P = 3.1 × 10−2). In comparison to the growth in media alone (100%), there were no significant differences in the final parasitemia of 12 preimmune sera (mean ± standard deviation = 97% ± 4%) or the three adjuvant-alone sera (99% ± 3%). Whole serum from one of the rabbits (R-3), which showed 44% inhibition in the one-cycle suspension GIA, was used in a two-cycle suspension GIA at the same dilution. Inhibition of 87% was seen, which was indicative of cumulative inhibition over two cycles.
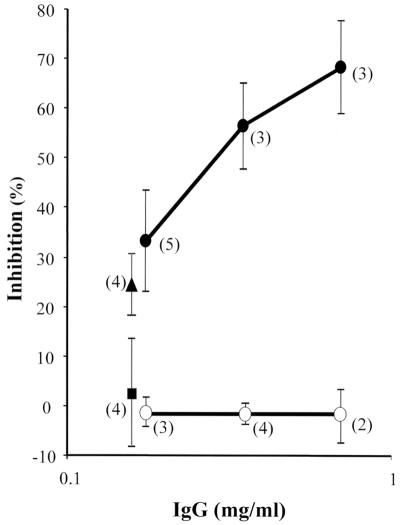
Inhibition of parasite growth was also observed with IgG purified from pooled sera of the rabbits within the lab-grade AMA1/E, RA-AMA1/E, and adjuvant control groups (Fig. 5). The percent inhibition of the AMA1/E group was significantly higher than that of the adjuvant control group at 0.18, 0.35, and 0.7 mg of IgG ml−1 (P = 7 × 10−4, 8 × 10−3, and 6 × 10−3, respectively). No inhibition was observed when the RA-AMA1/E group IgG was compared to the equivalent IgG concentration from the adjuvant control group. In order to determine if the inhibition caused by the IgG could be reversed, an identical concentration of AMA1/E or RA-AMA1/E proteins was added to the culture during the GIA. The addition of 5.3 μg of AMA1/Ε protein ml−1 to 0.18 mg of anti-AMA1/E IgG ml−1 significantly reversed inhibition compared to the addition of the same amount of RA-AMA1/E (P = 6 × 10−3) (Fig. 5). These data indicate the critical role of disulfide bonds in the formation of epitopes that can induce inhibitory anti-AMA1 antibodies.
FIG. 5.
Purified anti-AMA1/E IgG inhibited growth by P. falciparum 3D7 parasite in a GIA, and the inhibition was reversed by the addition of the antigen. Shown are the results of a one-cycle GIA with 0.18, 0.35, or 0.7 mg of purified rabbit IgG ml−1 in suspension culture GIA in 48-well plates. •, anti-AMA1/E IgG; ○, anti-RA-AMA1/E IgG. Antigens (5.3 μg ml−1) were added to anti-AMA1/E IgG concentrations of 0.18 mg ml−1. ▴, RA-AMA1/E antigen added; ▪, AMA1/E antigen added (symbols are offset for clarity). The means ± standard deviations are shown, with the number of experiments for each data point represented within parentheses.
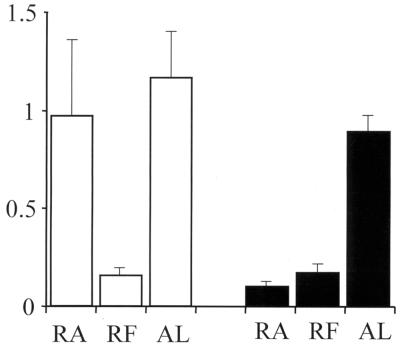
ELISA, IFA, and GIA data for antibodies to recombinant AMA1/E suggested that the conformational epitopes present on the refolded protein (in addition to the linear epitopes) were indeed highly immunogenic. A competition ELISA with sera from three rabbits in the lab-scale AMA1/E group (R-1, R-2, and R-3) and two rabbits in the RA-AMA1/E group (R-7 and R-10) was done by preincubation with 15 μg of AMA1/E or RA-AMA1/E ml−1 or with BSA as a control. Figure 6 shows the mean OD405 at a 1:16,000 serum dilution for the two groups along with the standard deviations for three experiments. Preincubation with the refolded AMA1/E protein resulted in, on average, 86 and 81% reduction in the OD405 in the AMA1/E and RA-AMA1/E groups, respectively, in comparison to the BSA control values for the same group (P = 1.99 × 10−13 and 1.55 × 10−10, respectively). Although the RA-AMA1/E protein preincubation resulted in an average 89% drop in the OD405 in the RA-AMA1/E-immunized group (P = 5.63 × 10−9), it resulted in an insignificant 16% drop in the refolded AMA1/E group (P = 0.071). These data further suggest that a large proportion of the antibodies against refolded AMA1/E were against disulfide bond-dependent epitopes.
FIG. 6.
Competitive ELISA with individual sera. Serum samples at a 1:1,000 dilution were preincubated with either RA-AMA1/E (RA), AMA1/E (RF), or BSA protein (AL). ELISA was done to assay for anti-AMA1/E-specific antibodies. Shown here are the average OD405s at a 1:16,000 dilution for AMA1/E-immunized rabbits (R-1, R-2, and R-3) (clear bars) and RA-AMA1/E-immunized rabbits (R-7 and R-10) (black bars). The standard deviations are represented by lines on top of the bars.
DISCUSSION
The availability of a significant quantity of a stable recombinant protein with pharmaceutical levels of purity is an essential step on the path of testing adjuvant combinations capable of inducing long-lasting and high-titer responses in humans. We describe here a process that was successfully scaled up to produce AMA1/E, a recombinant protein based on the ectodomain of P. falciparum AMA1. A 300-liter fermentation generated 4.5 kg of cell paste. Starting from 1.5 kg of cell paste, 70 mg of AMA1/E, enough material for over 800 doses, was purified, put in vials, and lyophilized under a GMP environment. While the final yield of AMA1/E was relatively low, the AMA1/E product satisfied all of the major criteria for proceeding to a phase I clinical trial. These include purity (>99%, as measured by SDS-PAGE and GPC), endotoxin content (0.06 EU per 50 μg of protein, LAL test), free thiol content (<0.1 μM free-SH groups per μM protein, Ellman's test), positive reactivity to immune reagents (monoclonal antibody 4G2dc1 and malaria immune sera, Western blotting), mass analysis [54,648 Da, MALDI-TOF (MS)], correct N-terminal sequence (first 21 residues, Edman method), host cell protein content (<0.5%, ELISA), Western blotting (no E. coli-specific band with up to 2 μg of AMA1/E loaded per well), residual Sarkosyl content (below detectable limits by RP-HPLC), and product stability (stable at 37°C for more than 6 months) in its lyophilized form (data not shown). The GMP-produced AMA1/E product was immunogenic in rabbits and raised high-titer inhibitory antibodies.
The correct folding of AMA1, as in the cases of several other Plasmodium antigens, has been shown to be critical for its immunological activity (1, 6, 14). Full-length P. falciparum AMA1 was first expressed in the eukaryotic insect cell system (21), and although the baculovirus product was soluble, the purification strategy was not designed for scale-up production. Prokaryotic expression of AMA1 from various species has been problematic, primarily due to the formation of insoluble aggregates, which is presumably due to incorrect folding of the protein. Previous work on P. chabaudi AMA1 expression in E. coli showed that it was necessary to include an in vitro refolding step in the process in order to obtain correctly folded protein (1). A similar approach was successful for obtaining correctly folded AMA1 from P. falciparum, and the antibodies made against it inhibited parasite growth in vitro (14). A scalable process for the production of recombinant AMA1 has not yet been described. Following the success with another malarial antigen (9), we attempted to express r-AMA1/E as a soluble protein in E. coli. A redox-modified strain of E. coli, Origami (DE3), with mutations in the glutathione and thioredoxin reductase pathways (2) was used for expression, with induction carried out at a low temperature and with a minimal IPTG concentration. Despite attempts to optimize the fermentation conditions to obtain soluble r-AMA1/E, a large fraction was still located in the insoluble pellet. Hence, a downstream purification process was developed to extract r-AMA1/E from both soluble and insoluble fractions and to refold it in vitro.
The use of Origami (DE3) cells was, in fact, a factor that attributed to low protein yield due to plasmid loss during scale-up fermentation. Although the vector pWRMAL contained both ampicillin and tetracycline resistance genes for plasmid maintenance, penicillin derivatives like ampicillin cannot be used during the production of a human-use vaccine. The E. coli Origami (DE3) strain used to enhance soluble protein expression carried a tetracycline resistance gene, resulting in low plasmid maintenance in the presence of tetracycline over the long growth period required for scale-up fermentation. Future studies will be directed to the selection of other bacterial hosts that allow the use of a selectable marker to increase plasmid maintenance.
The increase in reactivity to immune reagents observed after the refolding step and the homogeneity of the final product justified the need for the inclusion of this refolding step in the process, although this was a limiting factor during scale-up production. A minimum 40-fold dilution was necessary to gain optimal immune reactivity. An anion-exchange step was used after refolding to separate the monomeric AMA1/E from its aggregated forms, which eluted at a higher NaCl concentration. This monomer selection step resulted in some loss of product during purification. After anion exchange, a doublet at ∼10 kDa was found to coelute with AMA1/E, and although GPC was an option, we avoided it due to problems associated with scale-up. Instead, a cation-exchange step with SP-Sepharose was used to purify AMA1/E to homogeneity. Assays based on the immunodetection of HCP, in combination with laser densitometry of stained polyacrylamide gels and analytical GPC, were used to determine that the final product was >99% pure.
N-terminal sequencing and mass-spectrometric analysis confirmed the correct primary structure of AMA1/E. Ellman's test and alkylation analysis confirmed the absence of any free cysteines in the final product. The shifts in the RPC elution profile under reduced conditions and the immunoblot reactivity to a conformation-dependent inhibitory rat monoclonal antibody, 4G2dc1, under nonreduced conditions further confirm the disulfide-bonded nature of the antigen.
AMA1/E was found to be highly immunogenic in rabbits in combination with Montanide ISA-720 adjuvant. Antibodies raised against the recombinant protein recognized the native parasite AMA1 protein by both IFA and Western blotting. Whole serum from the immunized rabbits showed growth inhibition of the homologous P. falciparum (3D7) parasites in vitro both under suspension and static GIA conditions. During this process development, we refolded the r-AMA1/E protein either directly (AMA1/E) or after DTT reduction of the Ni2+ column elution (RR-AMA1/E). The RR-AMA1/E product, based on its overall lower immunogenicity, lower percent GIA values of its antisera, and the observation that DTT reduction gave no significant gain in the monomeric protein yield, was not pursued further in the scale-up GMP process.
It has previously been shown that AMA1-based protective immunity can be passively transferred by IgG transfusion into naive animals (1, 23). Antibodies to recombinant AMA1 from P. falciparum have recently been reported to inhibit parasite invasion in vitro (14). These data suggest that AMA1-based protection is probably antibody mediated. The in vitro growth inhibition observed with whole sera and with purified IgG, in addition to the positive correlations observed between the ELISA titer (against AMA1/E coated wells) and the IFA titer, suggests that these measures of antibody response might serve as good correlates of AMA1-based protection in vivo. When comparing the same sera in parallel experiments, we have observed higher percent GIA values in static culture than in the suspension GIA (paired t test, P = 5 × 10−6). Fluid movement in suspension culture may better mimic the blood flow conditions encountered in vivo by the parasite than does static culture GIA. Some antibodies are known to show better inhibition in either static or suspension culture (12), and it remains to be seen whether suspension or static culture GIA better predicts protection following vaccination. However, it is encouraging to report that anti-AMA1/E antibodies are inhibitory under both conditions.
Previous data on mice vaccination with recombinant P. chabaudi AMA1 suggested that the presence of intact disulfide bonds in the vaccinating AMA1 antigen are necessary to induce protection (1, 6). The data presented here also suggest that disulfide bond-dependent motifs play a critical role in the induction of inhibitory anti-AMA1 antibodies. The higher ELISA titers obtained with refolded AMA1/E-coated wells than with RA-AMA1/E-coated wells in the AMA1/E-immunized group, the lower IFA titers in the RA-AMA1/E group, the inability of the anti-RA-AMA1/E antibodies to block parasite invasion, the ability of AMA1/E and not RA-AMA1/E protein to significantly reverse the in vitro growth inhibition, and the ability of AMA1/E and not RA-AMA1/E to out-compete binding of most of the anti-AMA1/E antibody to AMA1/E protein in ELISA all indicate that a majority of the immunologically significant epitopes of AMA-1 are sensitive to reduction.
In conclusion, this study details the process development for the production of a disulfide cross-linked AMA1 ectodomain recombinant protein that could be evaluated as a malaria vaccine candidate. In anticipation of clinical trials, the final AMA1/E product is undergoing safety, stability, and potency tests in animals.
Acknowledgments
We thank A. W. Thomas, Biomedical Primate Research Institute, Rijswijk, The Netherlands, for the monoclonal antibody 4G2dc1; E. Angov at WRAIR for the plasmid; and Greg E. Garcia and Deborah R. Doctor for N-terminal sequencing. Special thanks are due to Robin F. Anders, Carter Diggs, and Lorraine Soisson for invaluable advice on protein product analysis and purification. We thank Douglas Smoot for flow cytometry of parasites in the GIA.
This work was performed while S.D. and P.V.L. held National Research Council Research Associate awards at the WRAIR.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunization with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Bessette, P. H., F. Aslund, J. Beckwith, and G. Georgiou. 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA 96:13703-13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess, R. R. 1996. Purification of over produced E. coli RNA polymerase σ factors by solubilizing inclusion bodies and refolding from sarkosyl. Methods Enzymol. 273:145-149. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., and A. Saul. 1994. Sequence analysis of the apical membrane antigen 1 (AMA-1) of Plasmodium vivax. Mol. Biochem. Parasitol. 65:183-187. [DOI] [PubMed] [Google Scholar]
- 5.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, et al. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51:711-719. [DOI] [PubMed] [Google Scholar]
- 6.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 8.Dutta, S., P. Malhotra, and V. S. Chauhan. 1995. Sequence analysis of the apical membrane antigen-1 of Plasmodium cynomolgi bastianelli. Mol. Biochem. Parasitol. 73:267-270. [DOI] [PubMed] [Google Scholar]
- 9.Dutta, S., L. A. Ware, A. Barbosa, C. F. Ockenhouse, and D. E. Lanar. 2001. Purification, characterization, and immunogenicity of a disulfide cross-linked Plasmodium vivax vaccine candidate antigen, merozoite surface protein 1, expressed in Escherichia coli. Infect. Immun. 69:5464-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellman, G. L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:50-77. [DOI] [PubMed] [Google Scholar]
- 11.Escalante, A. A., H. M. Grebert, S. C. Chaiyaroj, M. Magris, S. Biswas, B. L. Nahlen, and A. A. Lal. 2001. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 113:279-287. [DOI] [PubMed] [Google Scholar]
- 12.Haynes, J. D., J. K. Moch, and D. S. Smoot. Erythrocytic malaria growth or invasion inhibition assays (GIA) with emphasis on suspension culture GIA. In Denise L. Doolan (ed.), Malaria methods and protocols, in press. The Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 13.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 14.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 16.Kappe, S. H. I., and J. H. Adams. 1996. Sequence analysis of apical membrane antigen-1 genes (AMA-1) of Plasmodium yoelii yoelii and Plasmodium berghei. Mol. Biochem. Parasitol. 78:279-283. [DOI] [PubMed] [Google Scholar]
- 17.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119-15124. [DOI] [PubMed] [Google Scholar]
- 18.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterization of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109:147-156. [DOI] [PubMed] [Google Scholar]
- 19.Lal, A. A., M. A. Hughes, D. A. Oliveira, C. Nelson, P. B. Bloland, A. J. Oloo, W. E. Hawley, A. W. Hightower, B. L. Nahlen, and V. Udhayakumar. 1996. Identification of T-cell determinants in natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in an adult population exposed to malaria. Infect. Immun. 64:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall, V. M., M. G. Peterson, A. M. Lew, and D. J. Kemp. 1989. Structure of the apical membrane antigen 1 (AMA-1) of Plasmodium chabaudi. Mol. Biochem. Parasitol. 37:281-284. [DOI] [PubMed] [Google Scholar]
- 21.Narum, D. L., G. W. Welling, and A. W. Thomas. 1993. Ion-exchange-immunoaffinity purification of a recombinant baculovirus Plasmodium falciparum apical membrane antigen, PF83/AMA-1. J. Chromatogr. A 657:357-363. [DOI] [PubMed] [Google Scholar]
- 22.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full length and processed form of PF83/AMA-1, an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 23.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson, M. G., P. Nguyen-Dinh, V. M. Marshall, J. F. Elliott, W. E. Collins, R. F. Anders, and D. J. Kemp. 1990. Apical membrane antigen of Plasmodium fragile. Mol. Biochem. Parasitol. 39:279-284. [DOI] [PubMed] [Google Scholar]
- 26.Thomas, A. W., J. F. Trape, C. Rogier, A. Goncalves, V. E. Rosario, and D. L. Narum. 1994. High prevalence of natural antibodies against P. falciparum 83-kilodalton apical membrane antigen Pf83/AMA-1. Am. J. Trop. Med. Hyg. 51:730-740. [DOI] [PubMed] [Google Scholar]
- 27.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Bio/Technology 24:145-149. [PubMed] [Google Scholar]
- 28.Triglia, T. J., S. Healer, R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 29.Waters, A. P., A. W. Thomas, J. A. Deans, G. H. Mitchell, D. E. Hudson, L. H. Miller, T. F. McCutchan, and S. Cohen. 1990. A merozoite receptor protein from P. knowlesi is highly conserved and distributed throughout Plasmodium. J. Biol. Chem. 265:17974-17979. [PubMed] [Google Scholar]
- 30.World Health Organization Tropical Disease Research. 1997. TDR twelfth program report, p. 57-76. World Health Organization, Geneva, Switzerland.