Abstract
Recent data have shown that the respiratory pathogen Chlamydia pneumoniae expresses an altered gene transcription profile during gamma interferon (IFN-γ)-induced persistent infection in vitro. In the present study, we examined, by proteomics, expression of C. pneumoniae proteins labeled intracellularly with [35S]methionine/cysteine under normal conditions or IFN-γ-mediated persistence. The identity of differentially expressed proteins during persistent infection was determined by matching spots to those of proteins identified in C. pneumoniae elementary bodies by matrix-assisted laser desorption ionization mass spectrometry. Upon treatment with 50 U of IFN-γ per ml, a marked upregulation of major outer membrane protein (MOMP), heat shock protein 60 (Hsp-60/GroEL), and proteins with functions in DNA replication (GyrA), transcription (RpoA, PnP), translation (Rrf), glycolysis (PgK, GlgP), and type III secretion (SctN) was observed at 24 h of infection. In contrast, no significant decreases in bacterial protein expression were found in C. pneumoniae-infected cells due to IFN-γ treatment. Upregulation of C. pneumoniae proteins involved in diverse functions during persistent infection may allow the organism to resist the inhibitory effects of IFN-γ while retaining basic functions. Future studies should examine the differential expression of chlamydial proteins during the developmental cycle under IFN-γ pressure to obtain a finer representation of the gene products involved in establishing persistence.
Chlamydia pneumoniae is an obligate intracellular pathogen implicated in both acute and chronic respiratory diseases. Activation of a cell-mediated immune (CMI) response plays an important role in protection and pathogenesis of C. pneumoniae infection (6, 16). Stimulation of host cells by IFN-γ inhibits growth of C. pneumoniae primarily by induction of indoleamine 2,3-dioxygenase (IDO) activity, which deprives the organism of tryptophan (12, 14, 22). In a fashion similar to that used with Chlamydia trachomatis, our group (15) and others (3, 11) have shown that subinhibitory concentrations of IFN-γ can lead to the formation of a persistent state of C. pneumoniae in vitro, a phenomenon that is characterized by the formation of pleomorphic reticulate bodies (RBs) that are maintained in a viable but culture-negative state.
Features of C. trachomatis persistence induced by IFN-γ include downregulation of important antigens, such as the major outer membrane protein (MOMP), 60-kDa cysteine-rich outer membrane complex protein B (OmcB/Omp2), and chlamydial lipopolysaccharide (1). In contrast, levels of chlamydial heat shock protein 60 (GroEL) remain unaltered. A recent study by Shaw et al. reported an upregulation of additional proteins in C. trachomatis serovars A and L2 with molecular masses of 30 and 40 kDa in response to IFN-γ treatment, as examined by two-dimensional (2D) gel electrophoresis of infected HeLa cells (20).
Only limited amounts of data have characterized gene expression profiles of C. pneumoniae under persistent infection (3, 11). Using reverse transcriptase PCR, Byrne et al. (3) have shown that DNA replication genes (i.e., dnaA, polA, mutS, and minD) are expressed at normal levels in C. pneumoniae during IFN-γ-mediated persistent infection; however, expression of cell division-related genes (i.e., ftsK and ftsW) is downregulated. In a separate study, Mathews et al. (11) detected upregulation of C. pneumoniae genes coding for proteins with functions in cell wall structure (ompA and ompB), glycolysis (pyk), and peptidoglycan synthesis (nlpD) as a result of IFN-γ treatment. Taken together, these studies indicate that under IFN-γ-induced persistence, C. pneumoniae expresses an altered gene transcription profile. To our knowledge, the characterization of the coinciding protein expression patterns of persistent C. pneumoniae infection caused by IFN-γ treatment has not been determined.
Elucidation of the C. pneumoniae genome sequence has provided fixed information predicting the structure and function of bacterial proteins (9, 18, 21). Recently, proteomics has emerged as an approach to examine global protein synthesis as well as being an indicator of gene expression in bacteria. In addition, proteomic analysis can be an important tool for identifying virulence determinants, antigens, and vaccine candidates, since changes in the proteome depend on developmental stages, disease states, or environmental conditions (24). A recent study by Vandahl et al. (25) generated an electrophoretic map of the proteome of the infectious, extracellular form of C. pneumoniae, better known as the elementary body (EB). In this map, 263 C. pneumoniae proteins encoded from 167 genes were identified by matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS). In the present study, we used proteomics to examine the effects of IFN-γ-induced persistence on intracellular C. pneumoniae protein expression.
MATERIALS AND METHODS
Bacterial isolate.
C. pneumoniae A-03 was previously isolated in our laboratory from an atheroma of a patient with coronary artery disease (17). HEp-2 cells (ATCC CCL-23) maintained in RPMI 1640 medium (Cellgro, Herndon, Va.) with 10% fetal bovine serum, 2 mM l-glutamine, 1% (vol/vol) nonessential amino acids, 10 mM HEPES, 10 μg of gentamicin per ml, and 25 μg of vancomycin per ml were used for propagation of C. pneumoniae A-03 as described previously (13). EBs were harvested and purified by disruption of HEp-2 cell monolayers with sterile glass beads, sonication, and centrifugation over a Renografin density gradient as published elsewhere (4). EB suspensions were stored in sucrose-phosphate-glutamic acid buffer at −70°C at concentrations of 400 μg of total protein per ml, as determined by Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.).
Protein extraction.
Procedures for protein extraction were performed according to Vandahl et al. (25). EB preparations were pelleted by centrifugation in a tabletop microcentrifuge. Pellets were resuspended in 30 μl of 50 mM Tris HCl (pH 7.0) containing 1% sodium dodecyl sulfate (SDS), sonicated for 15 s, and boiled for 5 min. Samples were then treated with 2 μl of a mixture of 50 mM MgCl2, 476 mM Tris HCl, 24 mM Tris base, 1 mg of DNase I per ml, and 0.250 mg of RNase A per ml (pH 8.0) on ice for 10 min. Finally, samples were suspended in 90 μl of buffer containing 7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 40 mM Tris base, 65 mM dithiothreitol (DTT), and 2% Pharmalyte 3-10 (Amersham Pharmacia, Piscataway, N.J.) and frozen at −70°C. In addition to EBs, this procedure was used to extract proteins from C. pneumoniae-infected HEp-2 cells.
Infection protocol.
The conditions for inducing persistence of C. pneumoniae in vitro were identical to those in our previous study describing the formation of pleomorphic RBs that are maintained in a viable, but culture-negative state as a result of IFN-γ treatment (15). Briefly, HEp-2 cells were seeded in six-well plates at 106 cells per well and allowed to adhere overnight. Cells were subsequently inoculated with 2 × 107 inclusion-forming units (IFU) of C. pneumoniae A03 per well, centrifuged at 800 × g for 1 h at 4°C, and incubated at 37°C in 5% CO2 for 30 min. Infected cells were maintained in RPMI medium with or without 50 U of human recombinant IFN-γ per ml (Promega, Madison, Wis.) for 24 h. To detect chlamydial protein synthesis, infected HEp-2 cells were pulse-labeled for 2 h in methionine/cysteine-free RPMI medium containing 100 μCi of [35S]methionine/cysteine per ml (ICN Biomedicals, Irvine, Calif.) and 3 mg of cycloheximide per ml in the presence or absence of 50 U of IFN-γ per ml. Uninfected HEp-2 cells treated or untreated with IFN-γ were used as controls to determine potential incorporation of [35S]methionine/cysteine into host cell proteins. At the end of the 2-h labeling period, HEp-2 cells were washed in cold phosphate-buffered saline, scraped with a rubber policeman, and pelleted by centrifugation at 16,000 × g, and proteins were extracted as described above for EBs.
2D gel electrophoresis.
Procedures for 2D electrophoresis were carried out with the Investigator 2-D Electrophoresis System (Genomic Solutions, Inc., Ann Arbor, Mich.). Protein extracts were mixed in IPG rehydration buffer (8 M urea, 4% CHAPS, 0.04 M Tris Base, 0.065 M DTT, 0.01% bromophenol blue) in a final volume of 400 μl as follows. (i) To generate an electrophoretic map of purified C. pneumoniae EBs, 100 μg of protein extracts was mixed in IPG buffer. (ii) To study intracellular C. pneumoniae protein expression, 300,000 cpm of 35S-labeled chlamydial proteins was mixed in IPG buffer. Diluted samples were loaded onto IPG strips and allowed to swell overnight. The following focusing parameters were applied: 5,000 V of maximum voltage, 124 V of holding voltage, 80 μA of maximum current per gel, 100,000 V·h, and a duration of 24 h. After focusing was completed, IPG strips were equilibrated in buffer containing 6 M urea, 2% DTT, 30% glycerol, and 1× Tris-acetate. 2D electrophoresis was run with SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) at 4°C for 4.5 h under a 500-V maximum voltage and 20,000 mW per gel, with 200 mM Tricine used as a cathode buffer, and 0.4% SDS plus 625 mM Tris-acetate (pH 8.3) used as an anode buffer. Following the electrophoretic run, gels were fixed in 45% methanol-7.5% acetic acid for 1 h. Gels containing purified EB proteins were stained with silver, whereas gels containing radiolabeled chlamydial proteins were treated for 30 min with Amplify fluorographic reagent (Amersham Pharmacia, Buckinghamshire, England), vacuum dried, and exposed to Kodak Biomax MR films.
Image analysis.
Silver-stained gels and X-ray films were scanned under a SenSys camera system (Photometrics, Tucson, Ariz.). Bioimage 2D Analyzer software (Genomic Solutions) was used to estimate Mr and pI coordinates of proteins and match spots of proteins localized in the EB with those of proteins expressed intracellularly. This software was also used to quantitate the volumes of radiolabeled protein spots to detect changes between normal and IFN-γ conditions. Statistical analysis of C. pneumoniae protein expression under normal or persistent infection conditions was done by collecting numerical data from a minimum of five experiments and performing a Mann-Whitney test with GraphPad software (http://graphpad.com/instatman/instat3.htm).
Identification of protein spots from purified EBs.
Protein spots from C. pneumoniae EBs were excised from silver-stained gels and treated with 20 μg of trypsin per ml in 50 mM ammonium bicarbonate at 37°C overnight (8). Two microliters of supernatant was mixed in an equal volume of saturated α-cyano-4-hydroxycinnamic acid, 50% (vol/vol) acetonitrile, 0.1% trifluoroacetic acid (TFA), and 0.8 μl of the resulting solution was applied to the MALDI-MS template. Masses of peptide fragments were determined by MALDI-time-of-flight (TOF) analysis with a Micromass mass spectrometer. Patterns of measured masses were matched against theoretical masses of proteins found in the annotated databases SWISS-PROT and TREMBL, accessible in the ExPASy Molecular Biology server (http://expasy.cbr.nrc.ca/). Searches were performed with the MS-FIT server (http://prospector.ucsf.edu/) with restrictions to proteins from 1 to 100 kDa and mass tolerances of 150 ppm. Partial enzymatic cleavages leaving one cleavage site, oxidation of methionine, and modification of cysteine with iodoacetamide were considered in these searches.
RESULTS
2D electrophoretic map of C. pneumoniae proteins expressed in infected HEp-2 cells.
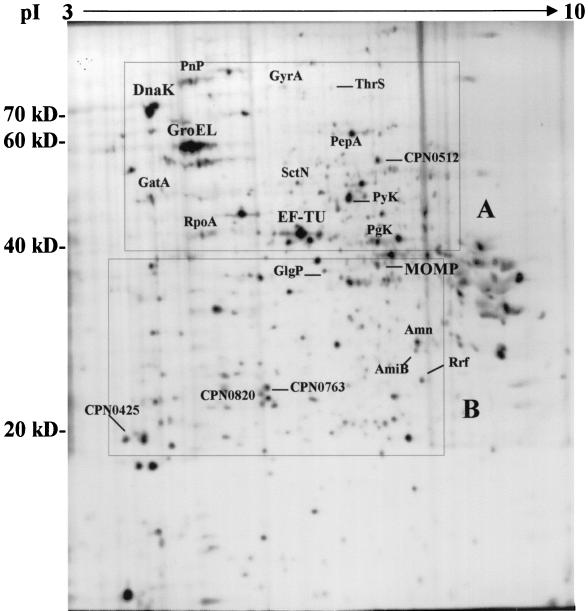
To determine intracellular expression of chlamydial proteins, C. pneumoniae-infected HEp-2 cells were pulse-labeled for 2 h with [35S]methionine/cysteine in the presence of cycloheximide at 24 h postinfection (p.i.). Figure 1 shows an electrophoretic map of C. pneumoniae proteins expressed in infected HEp-2 cells that were resolved in SDS-PAGE (10% polyacrylamide) for the second dimension. Approximately 600 protein spots could be detected by utilizing nonlinear pH 3 to 10 IPG strips. A 2D electrophoretic map of purified EBs containing proteins identified by MALDI-MS was matched to the electrophoretic map of infected HEp-2 cells by using Bioimage software to determine the identity of C. pneumoniae proteins expressed intracellularly. As shown in Fig. 1, we have determined by 24 h p.i., expression of heat shock protein 70 (Hsp-70/DnaK), heat shock protein 60 (Hsp-60/GroEL), MOMP, and DNA gyrase subunit A (GyrA), as well as proteins involved in transcription (RpoA and PnP), translation (EF-TU, ThrS, Rrf, PepA, and GatA), type III secretion (SctN), and glycolysis (PyK, PgK, and GlgP). In addition, expression of four hypothetical proteins with undetermined function was detected in C. pneumoniae-infected HEp-2 cells (CPN0425, CPN0512, CPN0763, and CPN0820). A summary of C. pneumoniae proteins identified in infected HEp-2 cells with experimental and theoretical Mr and pI values is shown in Table 1. Proteins have been grouped according to functional assignments provided by the Chlamydia Genome Project (http://chlamydia-www.berkeley.edu:4231/).
FIG. 1.
2D electrophoretic map of C. pneumoniae proteins expressed in infected HEp-2 cells. Chlamydial proteins were labeled for 2 h with [35S]methionine/cysteine at 24 h p.i., and 2D electrophoresis was performed as described in the text. Proteins were identified by computer-assisted matching to a 2D electrophoretic map of purified EBs containing proteins identified by MALDI-MS. Complete protein descriptions with Mr and pI coordinates are shown in Table 1. Boxed regions (A and B) represent sections of the 2D map that have been enlarged in Fig. 2.
TABLE 1.
C. pneumoniae proteins expressed in infected HEp-2 cells under normal conditions or IFN-γ-induced persistence
| Function | Designation | Mra | pIa | Effect of IFN-γb | Protein name |
|---|---|---|---|---|---|
| Outer membrane proteins | MOMP | 37.8 (36.0) | 6.8 (7.0) | + | Major outer membrane protein |
| Cell wall hydrolysis | AmiB | 25.2 (22.8) | 7.5 (8.7) | + | N-Acetylmuramoyl-l-Ala amidase |
| Chaperones | GroEL | 58.3 (58.2) | 5.5 (5.3) | + | Heat shock protein 60 |
| DnaK | 68.3 (71.3) | 5.1 (4.9) | NC | Heat shock protein 70 | |
| Glycolysis | PyK | 50.0 (54.6) | 6.5 (6.7) | NC | Pyruvate kinase |
| PgK | 41.9 (43.0) | 6.8 (6.1) | + | Phosphoglycerate kinase | |
| GlgP | 37.0 (94.5) | 6.1 (5.8) | + | Glycogen phosphorylase | |
| DNA replication | GyrA | 75.5 (93.8) | 5.7 (6.6) | + | DNA gyrase subunit A |
| Transcription | RpoA | 44.8 (41.8) | 5.2 (5.3) | + | DNA-directed RNA polymerase α chain |
| PnP | 79.8 (75.3) | 5.5 (5.4) | + | Polyribonucleotide nucleotidyltransferase | |
| Translation | ThrS | 70.2 (72.6) | 6.1 (5.8) | NC | Threonyl tRNA synthetase |
| PepA | 56.2 (54.7) | 6.2 (5.9) | NC | Leucine aminopeptidase | |
| GatA | 54.2 (53.6) | 5.1 (5.8) | NC | Glutamyl-tRNA amidotransferase A | |
| EF-TU | 44.0 (43.0) | 6.0 (5.4) | NC | Elongation factor Tu | |
| Rrf | 20.8 (20.1) | 8.0 (8.4) | + | Ribosome recycling factor | |
| Amino acid biosynthesis | Amn | 26.0 (32.6) | 7.5 (6.5) | + | AMP nucleosidase |
| Type III secretion | SctN | 52.1 (48.1) | 5.9 (5.8) | + | Type III secretion ATPase |
| Hypothetical proteins | 0512 | 56.1 (56.3) | 6.8 (6.2) | NC | Hypothetical protein CPN0512 |
| 0820 | 20.9 (21.6) | 5.7 (5.6) | NC | Hypothetical protein CPN0820 | |
| 0763 | 23.1 (26.7) | 5.9 (5.2) | NC | Hypothetical protein CPN0763 | |
| 0425 | 18.5 (21.3) | 4.6 (4.7) | NC | Hypothetical protein CPN0425 |
Theoretical values for Mr and pI of translated gene products are shown in parentheses and were obtained from the C. pneumoniae genome sequence database located in The Institute For Genomic Research server (http://www.tigr.org).
+, protein found to be significantly upregulated (P < 0.05) following IFN-γ treatment. NC, no significant change in expression.
Effects of IFN-γ-induced persistence on C. pneumoniae protein expression in infected HEp-2 cells.
To investigate whether differential expression of C. pneumoniae proteins occurred under IFN-γ-mediated persistence, infected HEp-2 cells were treated with 50 U of the cytokine per ml for 24 h (15), and intracellular chlamydial protein expression was examined by pulse-labeling for 2 h with [35S]methionine/cysteine in the presence of cycloheximide. Figure 2A represents an enlargement of boxed region A from Fig. 1. Upon treatment with IFN-γ, a significant increase in PnP, GyrA, GroEL, SctN, RpoA, and PgK expression was observed compared to that in cultures treated with medium alone. Moderate, but not statistically significant, increases (Fig. 3) in protein expression were observed for Pyk, EF-TU, DnaK, Thrs, PepA, GatA, and CPN0512, while expression of other unidentified proteins remained unchanged.
FIG. 2.
Effects of IFN-γ-induced persistence on C. pneumoniae protein expression. HEp-2 cells were infected with C. pneumoniae A-03 and incubated for 24 h in the absence or presence of 50 U of IFN-γ per ml. Chlamydial proteins were labeled for 2 h with [35S]methionine/cysteine, and 2D electrophoresis was performed as described in the text. Panels A and B coincide with the boxed regions shown in Fig. 1.
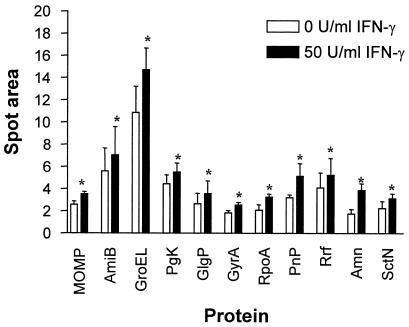
FIG. 3.
Statistical analysis of upregulated C. pneumoniae proteins in response to IFN-γ. Spot volumes were calculated from each protein under normal or IFN-γ conditions by using Bioimage. Numerical data were collected from a minimum of five experiments per experimental condition and subjected to a Mann-Whitney test. Bars indicate the means ± standard errors of the means. ∗, P < 0.05.
Figure 2B represents an enlargement of boxed region B from Fig. 1. Under IFN-γ pressure, significant increases in the expression of MOMP, GlgP, AmiB, Rrf, and Amn were observed, while levels of CPN0763, CPN0820, and CPN0425 were not altered compared to untreated controls. Overall, no significant decreases in bacterial protein expression were found in C. pneumoniae-infected cells due to IFN-γ treatment at 24 h p.i.
In order to measure differences in C. pneumoniae protein levels as a result of IFN-γ pressure, spot volumes were calculated from each identified protein by using Bioimage. The numerical values from this analysis were collected from five separate experiments and subjected to statistical analysis as described in Materials and Methods. Expression of 11 identified proteins was significantly increased following IFN-γ treatment compared to that in untreated controls (Fig. 3; P < 0.05). These increases above the protein expression levels measured from C. pneumoniae-infected HEp-2 cells without IFN-γ averaged 40% for PgK, AmiB, and Rrf; 50% for GyrA, GroEL, SctN, GlgP, and MOMP; and 70% for Pnp, RpoA, and Amn. These upregulated proteins are depicted in Table 1.
DISCUSSION
Infections caused by C. pneumoniae include sinusitis, bronchitis, and pneumonia (10). In certain patients, chronic respiratory disease may occur with the likelihood of developing asthmatic bronchitis (7). Over the last decade, a significant amount of data, including serological and direct detection studies, has suggested a potential role for chronic C. pneumoniae infection in human atherosclerosis (reviewed in references 5, 19, and 23). The mechanisms involved in establishing persistent infection with this organism remain unclear. However, in vitro studies have suggested that members of the genus Chlamydia may persist in tissue through an arrested state in the developmental cycle characterized by formation of enlarged atypical RB forms due to cytokine-induced amino acid deprivation (1). At the cellular level, activation of host cell IDO by IFN-γ is considered to be directly responsible for the development of persistent Chlamydia infection due to a depletion of intracellular tryptophan pools (1, 2, 14).
Recent completion of the C. pneumoniae genome (9, 18, 21) provides relevant information for the comprehensive investigation of proteins the organism can produce under various growth conditions. Vandahl et al. (25) recently reported a 2D reference map of the C. pneumoniae proteome containing 263 proteins identified in the EB by MALDI-MS. The present study used proteomics to examine the expression of C. pneumoniae proteins under normal conditions or the potential regulatory effects of IFN-γ. A 2D electrophoretic map of purified EBs containing proteins identified by MALDI-MS was compared to electrophoretic maps of infected HEp-2 cells to determine the identity of C. pneumoniae proteins expressed intracellularly. Ideally, a map of the metabolically active RBs would have represented a more accurate picture of C. pneumoniae proteins expressed during infection; however, purification of RBs in large amounts under standard growth conditions is problematic, and morphological effects caused by IFN-γ could further enhance the level of difficulty.
Comparison of electrophoretic maps of C. pneumoniae-infected HEp-2 cells grown with or without IFN-γ treatment showed significant upregulation of a variety of proteins with diverse metabolic functions. These included proteins involved in folding and stabilization (GroEL), glycolysis (PgK and GlgP), amino acid biosynthesis (Amn), type III secretion (SctN), transcription and RNA degradation (RpoA and PnP), DNA replication (GyrA), translation (Rrf), and structure of the cell wall (MOMP, AmiB). In contrast, we did not observe any significant downregulation of C. pneumoniae proteins in response to IFN-γ by 24 h p.i.
It is well known that bacteria overcome stressful conditions by qualitative and quantitative adjustments in protein synthesis. The synthesis or overexpression of a subset of proteins thus allows bacteria to face unfavorable environments and maintain essential cellular functions. In this respect, some of the proteins in C. pneumoniae showing upregulation in response to IFN-γ pressure are typically induced in other bacteria, such as Escherichia coli and Pseudomonas sp., by various stressful conditions, which include drastic changes in growth temperature (i.e., GroEL and PnP) (24, 26). Upregulation of GroEL in C. pneumoniae could be due to the presence of misfolded or aggregated proteins and is in agreement with C. trachomatis studies in which increased expression of this protein has been observed in IFN-γ-induced persistent cultures as examined by immunostaining (1). Persistence also causes a downregulation of C. trachomatis MOMP in HeLa cells, while our study found a significant upregulation of C. pneumoniae MOMP in response to IFN-γ, an event that has been confirmed at the transcriptional level by others (11). The differences in MOMP expression between C. trachomatis and C. pneumoniae caused by IFN-γ treatment may be associated with possible differences in susceptibilities to the cytokine between the two chlamydial species. However, differences in MOMP expression between C. pneumoniae and C. trachomatis as a result of IFN-γ pressure may also arise from the type of host cell utilized (i.e., HEp-2 versus HeLa cells).
Recent 35S pulse-labeling and 2D electrophoresis work by Shaw et al. examined the effects of IFN-γ on protein expression patterns of C. trachomatis serovars A and L2 (20). Interestingly, downregulation of MOMP and numerous other proteins was only observed in serovar A, implying a lower sensitivity toward IFN-γ for serovar L2. Among the proteins found to be upregulated in both serovars, two were presumably identified as β-tryptophan synthase (serovar A) and α-tryptophan synthase (serovar L2) based on their Mr and pI coordinates, which matched those for the translated gene products in the C. trachomatis genome (20).
A molecular profile of C. pneumoniae persistence is beginning to emerge based on two current reports examining mRNA levels (3, 11) and the protein patterns obtained in this study. During IFN-γ-mediated persistent infection, C. pneumoniae remains metabolically active as determined by the ability of the organism to accumulate small quantities of DNA (3). This event correlates with the transcription of genes involved in replication (i.e., dnaA, polA, mutS, and minD) as described by Byrne et al. (3), as well as the detection of GyrA expression in our study, which is involved in negative supercoiling of circular DNA. Detection of upregulated genes coding for enzymes with functions in glycolysis, such as pyruvate kinase examined by Mathews et al. (11), as well as phosphoglycerate kinase (PgK) and glycogen phosphorylase (GlgP) examined by us, suggests that release of stored energy may be essential during persistence. Of particular interest, the expression of genes with possible roles in peptidoglycan synthesis is increased under IFN-γ treatment. These include nlpD, also described by Mathews et al. (11), and amiB in our study, which are two peptidases, the roles of which during persistent C. pneumoniae infection remain unclear at present.
In summary, C. pneumoniae upregulates the expression of proteins involved in diverse functions in response to IFN-γ. Regulation of these proteins may allow C. pneumoniae to resist the effects of IFN-γ while retaining essential metabolic activities. Continuing proteomic analysis of persistent C. pneumoniae infection may prove useful in the identification of potential virulence factors, biomarkers in patients with chronic infection, and therapeutic targets. Future studies should examine the differential expression of bacterial proteins at different periods of the developmental cycle to define the genes and gene products involved in establishing persistence. In addition, subsequent experiments will determine the roles of IDO and tryptophan depletion as possible mechanisms of IFN-γ-mediated shift in protein expression patterns of C. pneumoniae when induced into a persistent state.
Acknowledgments
We thank Ned Smith and Jian Cai, Mass Spectrometry Laboratory, University of Louisville, for assistance in peptide mass fingerprint analysis.
Robert Molestina was supported in part by a postdoctoral fellowship from Glaxo SmithKline.
Editor: R. N. Moore
REFERENCES
- 1.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne, G. I., L. K. Lehmann, and G. J. Landry. 1996. Induction of tryptophan catabolism is the mechanism for gamma interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne, G. I., S. P. Ouellette, Z. Wang, J. P. Rao, L. Lu, W. L. Beatty, and A. P. Hudson. 2001. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 69:5423-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, L. A., C.-C. Kuo, and J. T. Grayston. 1998. Chlamydia pneumoniae and cardiovascular disease. Emerg. Infect. Dis. 4:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halme, S., J. Latvala, R. Karttunen, I. Palatsi, P. Saikku, and H.-M. Surcel. 2000. Cell-mediated immune response during primary Chlamydia pneumoniae infection. Infect. Immun. 68:7156-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammerschlag, M. R., K. Chirgwin, P. M. Roblin, M. Gelling, W. Dumornay, L. Mandel, P. Smith, and J. Schachter. 1992. Persistent infection with Chlamydia pneumoniae following acute respiratory illness. Clin. Infect. Dis. 14:178-182. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, O. N., M. Wilm, A. Shevchenko, and M. Mann. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112:513-530. [DOI] [PubMed] [Google Scholar]
- 9.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1995. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 10.Kuo, C.-C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathews, S., C. George, C. Flegg, D. Stenzel, and P. Timms. 2001. Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-γ treated cultures of Chlamydia pneumoniae. Microb. Pathog. 30:337-345. [DOI] [PubMed] [Google Scholar]
- 12.Mehta, S. J., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-γ: role of tryptophan catabolism. J. Infect. Dis. 177:1326-1331. [DOI] [PubMed] [Google Scholar]
- 13.Molestina, R. E., D. Dean, R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Characterization of a strain of Chlamydia pneumoniae isolated from a coronary atheroma by analysis of the omp1 gene and biological activity in human endothelial cells. Infect. Immun. 66:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pantoja, L. G., R. D. Miller, J. A. Ramirez, R. E. Molestina, and J. T. Summersgill. 2000. Inhibition of Chlamydia pneumoniae replication in human aortic smooth muscle cells by gamma interferon-induced indoleamine 2,3-dioxygenase activity. Infect. Immun. 68:6478-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantoja, L. G., R. D. Miller, J. A. Ramirez, R. E. Molestina, and J. T. Summersgill. 2001. Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect. Immun. 69:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penttila, J. M., M. Anttila, M. Puolakkainen, A. Laurila, K. Varkila, M. Sarvas, P. H. Makela, and N. Rautonen. 1998. Local immune responses to Chlamydia pneumoniae in the lungs of BALB/c mice during primary infection and reinfection. Infect. Immun. 66:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez, J. A., S. Ahkee, J. T. Summersgill, B. L. Ganzel, L. L. Ogden, T. C. Quinn, C. A. Gaydos, L. L. Bobo, M. R. Hammerschlag, P. M. Roblin, W. LeBar, J. T. Grayston, C.-C. Kuo, L. A. Campbell, D. L. Patton, D. Dean, and J. Schachter. 1996. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. Ann. Intern. Med. 125:979-982. [DOI] [PubMed] [Google Scholar]
- 18.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saikku, P. 1997. Chlamydia pneumoniae and atherosclerosis—an update. Scand. J. Infect. Dis. Suppl. 104:53-56. [PubMed] [Google Scholar]
- 20.Shaw, A. C., G. Christiansen, and S. Birkelund. 1999. Effects of interferon gamma on Chlamydia trachomatis serovar A and L2 protein expression investigated by two-dimensional gel electrophoresis. Electrophoresis 20:775-780. [DOI] [PubMed] [Google Scholar]
- 21.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 12:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summersgill, J. T., N. N. Sahney, C. A. Gaydos, T. C. Quinn, and J. A. Ramirez. 1995. Inhibition of Chlamydia pneumoniae growth in HEp-2 cells pretreated with gamma interferon and tumor necrosis factor alpha. Infect. Immun. 63:2801-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor-Robinson, D., and B. J. Thomas. 1998. Chlamydia pneumoniae in arteries: the facts, their interpretation, and future studies. J. Clin. Pathol. 51:793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VanBogelen, R. A., E. E. Schiller, J. D. Thomas, and F. C. Neidhart. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20:2149-2159. [DOI] [PubMed] [Google Scholar]
- 25.Vandahl, B. B., S. Birkelund, H. Demol, B. Hoorelbeke, G. Christiansen, J. Vandekerckohove, and K. Gevaert. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22:1204-1223. [DOI] [PubMed] [Google Scholar]
- 26.Vasseur, C., J. Labadie, and M. Hébraud. 1999. Differential protein expression by Pseudomonas fragi submitted to various stresses. Electrophoresis 20:2204-2213. [DOI] [PubMed] [Google Scholar]