Abstract
Helicobacter pylori strains frequently express Lewis X (Lex) and/or Ley on their cell surfaces as constituents of the O antigens of their lipopolysaccharide molecules. To assess the effect of Lex and Ley expression on the ability of H. pylori to colonize the mouse stomach and to adhere to epithelial cells, isogenic mutants were created in which fucT1 alone or fucT1 and fucT2, which encode the fucosyl transferases necessary for Lex and Ley expression, were deleted. C3H/HeJ mice were experimentally challenged with either wild-type 26695 H. pylori or its isogenic mutants. All strains, whether passaged in the laboratory or recovered after mouse passage, colonized the mice well and without consistent differences. During colonization by the mutants, there was no reversion to wild type. Similarly, adherence to AGS and KatoIII cells was unaffected by the mutations. Together, these findings indicate that Le expression is not necessary for mouse gastric colonization or for H. pylori adherence to epithelial cells.
Helicobacter pylori strains are persistent gastric colonizers of the human stomach whose presence increases the risk of peptic ulcer disease and gastric adenocarcinoma (5). These organisms are highly adapted for the human stomach, which presents formidable environmental challenges for any potential colonizer (5, 6). In addition to their conserved features, human gastric epithelial cells may express oligosaccharides corresponding to the Lewis A (Lea), Leb, Lex, and Ley histo-blood group antigens (27). H. pylori cells also are polymorphic in that they may express Lex or Ley (29, 36) and occasionally other related antigens as constituents of the O antigens of their lipopolysaccharide (LPS) molecules (22, 23). It has been suggested that expression of Le antigens may camouflage the bacterium or aid in bacterial adhesion (24). Additionally, H. pylori Lewis antigens undergo phase variation, that is, the random, reversible high-frequency switching of phenotype, a phenomenon which in some organisms, including Neisseria spp. and Haemophilus influenzae, contributes to virulence (1, 2, 4). Recently we have provided evidence that the host and bacterial Le phenotypes are related (7, 37), although this hypothesis has not been universally confirmed (16, 31).
Experimental challenge of mice with H. pylori leads to persistent infection which has many of the features of human colonization (19, 25). Mouse colonization assays have been used previously to assess the importance of various H. pylori genes in gastric colonization (10, 14). We sought to determine whether H. pylori strains deficient in Lewis antigen expression would be able to colonize mice. In previous studies, H. pylori strains with low levels of Lewis antigen expression were unable to colonize mice (3, 21, 25). However, the strains studied produced low overall levels of O antigens, and a galE mutant produced no O antigen. Thus, the lack of colonization could reflect lack of expression either of the entire O antigen or of its Le constituents specifically. We now report that mutants of H. pylori in which both 1,3-fucosyl transferases are mutated, leading to the absence of Lex and Ley expression, are able to colonize the mouse stomach in a manner essentially identical to that of the wild-type strain. The mutant and wild-type cells also bound equally to gastric epithelial cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Stock cultures were maintained at −70°C in brucella broth (BBL Becton Dickinson Microbiology Systems, Cockeysville, Md.) supplemented with 15% glycerol. The isolates were routinely subcultured on Trypticase soy agar plates with 5% sheep blood (TSB [BBL]) in a humid microaerobic atmosphere with 5% CO2 at 37°C for 48 h. The identity of H. pylori was confirmed by characteristic colony morphology and biochemical activities (35).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Reference |
|---|---|---|
| Strains | ||
| H. pylori | ||
| 26695 | Wild type | 33 |
| 98-950 | 26695/0651::aphA | This study |
| 98-1014 | 26695/0379::cat/0651::aphA | This study |
| E. coli DH5α | endA1 hsdR17 (rK− mK+) supE44 thi-I recA gyrA (Nalr) relAl Δ(argG-lacZYA) U169 deoR [Φ80 ΔlacΔ(lacZ)M15] | |
| Plasmids | ||
| pBlueScript | ColE1 Ampr; PCR cloning vector | |
| pILL600 | Ampr Kmr; source of kanamycin cassette | 18 |
| pSAT101::cm | pBlueScript, H. pylori recA (5′ end), Cmr; source of chloramphenicol cassette | 32 |
| pTT379 | pBlueScript, 0379 | This study |
| pTT651 | pBlueScript, 0651 | This study |
| pTT0379K | pBlueScript, 0379::aphA | This study |
| pTT0651K | pBlueScript, 0651::aphA | This study |
| pTT0379C | pBlueScript, 0379::cat | This study |
| pTT0651C | pBlueScript, 0651::cat | This study |
Construction of α-1,3-fucosyltransferase (FucT) gene mutants.
Construction of the isogenic FucT mutants of wild-type H. pylori strain 26695, in which we interrupted either one or both copies of the FucT genes, was performed as follows. Open reading frames (ORFs) HP0379 (fucT1) and HP0651 (fucT2) were first amplified by PCR using the primers indicated in Table 2. EcoRI and XhoI restriction sites were incorporated into the 5′ ends of the primers to ensure directional cloning of the genes. The digested and purified PCR products then were cloned into pBluescript and transformed into Escherichia coli DH5α, and recombinants (pTT379 and pTT651) were selected on Luria-Bertani plates containing 50 μg of ampicillin per ml. Each ORF was disrupted by insertion of an antibiotic resistance cassette. An aphA cassette (kanamycin resistance) was released from pILL600 by BamHI digestion and ligated into a BglII site at position 204 in both fucT ORFs. These constructs were used to transform E. coli DH5α with selection on Luria-Bertani plates containing 50 μg of ampicillin per ml and 30 μg of kanamycin per ml. The plasmids containing the mutated ORF HP0379 and HP0651 were designated pTT379K and pTT651K, respectively, and used to transform H. pylori strain 26695 by natural transformation. After we mutated each of the two ORFs alone, we then inactivated the second FucT ORF in several single mutants. To construct plasmids in which HP0379 or HP0651 was disrupted by a cat (chloramphenicol resistance) cassette, the cat cassette was amplified from pSAT101::cm (32) by PCR, purified, digested with BglII, and ligated with BglII-digested and purified ORFs of pTT379 and pTT651. H. pylori strains that had a single aphA insertion in either HP0379 (fucT1) or HP0651 (fucT2) were transformed using plasmids pTT379C and pTT651C containing the reciprocal cat insertion, and recombinants were selected on TSB plates containing both kanamycin and chloramphenicol. Correct insertion of the resistance cassettes into the intended target gene was confirmed by PCR. Silver staining and Western blotting of proteinase K-treated whole-cell lysates (22, 26) confirmed that the LPS of each of the mutants contained intact O antigens and that the Lewis antigen, when present, migrated on polyacrylamide gel electrophoresis as part of the O antigen.
TABLE 2.
Oligonucleotide primers used in this study
| Primer designation | H. pylori gene(s) | Nucleotide positiona | Orientationb | Primer sequence (5′→3′)c |
|---|---|---|---|---|
| F1-f | HP0379, HP0651 | 1 to 24 | F | GGCGGGAATTCATGTTCCAACCCCTATTAGACGCC |
| F1-r | HP0379 | 1251 to 1275 | R | GGCGGCTCGAGCAAACCCAATTTTTTAACCAACTTT |
| F2-r | HP0651 | 1404 to 1428 | R | GGCGGCTCGAGCTTTTTAACCCATCTCCTTATGGGTG |
| F1-FO | HP0379 | −200 to −179 | F | GCGTGCTAGGGTTTTATTCGG |
| F1-RO | HP0379 | +1334 to +135. | R | ATTAGGGGCCAATATCGCTGG |
| F2-FO | HP0651 | −112 to −91 | F | AGAGGTTTTAAAACGCAACGC |
| F2-RO | HP0651 | +1498 to +151 | R | ACATGCTCAAAAACCCCACGC |
For primers F1-1, F1-r, and F2-r, location refers to position within ORFs in strain 26695. For primers F1-FO, F1-RO, F2-FO, and F2-RO, − or + indicates position upstream or downstream from the nucleotide of the initiation codon.
F, forward; R, reverse.
Added restriction sites are underlined: GAATTC, Eco RI; CTCGAG, XhoI.
Animals and housing.
Female C3H/HeJ mice approximately 3 to 5 weeks old were purchased from Jackson Laboratory (Bar Harbor, Maine). Up to six animals per solid-bottom cage were housed in the Animal Care Facilities of Vanderbilt University in a room with a 12-h light-dark cycle at 21 to 22°C, and animals were fed a standard commercial rodent chow. Access to food and water was free throughout all experiments. No special pretreatment (such as acid inhibition or antibiotics) was used before orogastric H. pylori inoculation or before the animals were sacrificed. All experiments and procedures carried out with the animals had been approved by the Institutional Animal Care Committee of Vanderbilt University.
H. pylori inoculation and animal sacrifice.
C3H/HeJ mice were inoculated with cells of wild-type strain 26695 or its isogenic single (ΔfucT2), and double (ΔfucT1/2) mutants. The bacteria had been taken from the freezer immediately prior to mouse challenge. The mice received inocula of 109 to 1010 bacterial cells three times in a 3- to 4-day period. Mice were sacrificed at predetermined times up to 12 weeks after challenge. Animals were anesthetized in a CO2 chamber and sacrificed by cervical dislocation. At the time of sacrifice, blood was collected, and serum was obtained by centrifugation and frozen at −20°C for later determination of serum antibodies. The stomach was harvested, and one-third was macerated into a suspension for quantitative H. pylori culture, as described previously (38). The macerated specimens were inoculated onto Trypticase soy agar-5% sheep blood plates containing vancomycin, nalidixic acid, bacitracin, and amphotericin B in the concentrations described previously (38). In each case, colonies were identified as H. pylori based on their resistance to the antibiotics, characteristic morphology on plates, and the urease and oxidase activities of multiple colonies. Isolates from these studies (mouse passaged) were characterized and then, after three or fewer in vitro passages, were used to challenge fresh mice.
Characterization of H. pylori isolates.
For determination of Lex and Ley expression, H. pylori isolates grown for 48 h on TSB plates were harvested and washed twice with 3 ml of 0.15 M NaCl per plate. Lex and Ley antigen expression was determined by specific enzyme-linked immunosorbent assays (ELISAs) as described previously (35). To determine genotypes of H. pylori isolates recovered from mice, PCR was done using primers specific for fucT1 and fucT2 as shown in Table 2.
Serum antibodies to H. pylori.
For evaluation of the time course of the systemic immune response to H. pylori, sera from the sacrificed mice were studied. Levels of immunoglobulin G (IgG) antibody directed toward H. pylori whole-cell antigens in the serum specimens were measured by ELISA, as described previously (35). Seroconversion to H. pylori was defined as a serum IgG response to the H. pylori antigens that was greater than the mean plus three standard deviations for the unchallenged mice.
Binding of H. pylori to gastric epithelial cells.
KatoIII and AGS-CDM cells are gastric adenocarcinoma cell lines obtained from the American Type Culture Collection. The cells were grown in RPMI 1640 medium (Gibco BRL, Grand Island, N.Y.) containing 10% fetal bovine serum in polystyrene tissue culture flasks at 37°C. AGS-NY2 cells were kindly provided by Steven Moss and grown in Ham's F12 medium. In some experiments, epithelial cells were treated with 100U of gamma interferon (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) for 48 h before use and then washed twice in phosphate-buffered saline containing bovine serum albumin (1%) and sodium azide (0.02%). To evaluate binding of H. pylori, adherent cells from control or gamma interferon-treated cultures were collected by gentle scrapping with a rubber policeman and then washed by centrifugation at 200 × g for 5 min twice before being resuspended in medium at the appropriate concentration for use in the experiments. Cell viability was assessed by trypan blue exclusion. COS-1 and ID12 cells were cultured in Dulbecco’s modified Eagle’s medium with Geneticin (G418 sulfate; Gibco BRL) as described previously (8). Binding of H. pylori to the surface of epithelial cells was evaluated by flow cytometry using a modification of techniques described elsewhere (12; Y. Minohara and P. B. Ernst, unpublished data). Briefly, the test strains of H. pylori were labeled with PKH26 (Sigma, St. Louis, Mo.). Gastric epithelial cells were incubated with labeled H. pylori cells for 1 h at room temperature and washed three times with phosphate-buffered saline. Subsequently, the cells were resuspended in 400 ml of 1% paraformaldehyde and analyzed by flow cytometry (FACScan; Becton Dickinson, San Jose, Calif.) and the binding was expressed as the relative mean fluorescence intensity (12).
RESULTS
Lex and Ley expression of ΔfucT1/2 and ΔfucT2 mutants.
Using antigen-specific ELISAs for analysis of isolates before mouse challenge, the ΔfucT2 mutant was shown to have greatly reduced Lex expression and slightly reduced Ley expression compared to the wild-type strain (Table 3). The ΔfucT1/2 mutant showed values of less than 10 optical density units for both Lex and Ley, which confirms that inactivation of both α-1,3-fucosyltransferase genes blocks Lex and Ley expression (34). Since the test strains expressed O antigens but the ΔfucT1/2 mutant was deficient in Le expression, we now could specifically examine the role of Le expression in mouse gastric colonization.
TABLE 3.
Lewis antigen expression by wild-type H. pylori strain 26695 and isogenic ΔfucT mutants after mouse challenge
| Passage | Bacterial genotype | Wk | No. of mice | Mean optical density units ± SD (range)a
|
|
|---|---|---|---|---|---|
| Lex | Ley | ||||
| Laboratory | Wild type | Prechallenge | NAb | 1,881 ± 179 (1,734-2,146) | 649 ± 67 (577-722) |
| 2 | 3 | 1,996 ± 108 (1,503-2,399) | 583 ± 58 (349-1,057) | ||
| 6 | 3 | 352 ± 58 (250-446) | 403 ± 139 (192-907) | ||
| 12 | NA | NA | NA | ||
| ΔfucT2 | Prechallenge | NA | 206 ± 24 (180-226) | 426 ± 34 (388-456) | |
| 2 | 5 | 237 ± 73 (36-494) | 348 ± 204 (126-419) | ||
| 6 | 4 | 355 ± 89 (45-509) | 426 ± 108 (224-993) | ||
| 12 | 4 | 613 ± 190 (3-1,111) | 838 ± 226 (6-1,474) | ||
| ΔfucT1/2 | Prechallenge | NA | 7 ± 2 (0-17) | −2 ± 1 (0-12) | |
| 2 | 4 | 3 ± 5 (0-16) | −3 ± 10 (−7-16) | ||
| 6 | 4 | 4 ± 4 (1-12) | 3 ± 5 (0-12) | ||
| 12 | 4 | 10 ± 4 (6-18) | 1 ± 2 (0-4) | ||
| Mousec | Wild type | Prechallenge | NA | 1,143 ± 197 (923-1,420) | 146 ± 33 (106-196) |
| 2 | 4 | 399 ± 130 (134-636) | 99 ± 58 (63-216) | ||
| 4 | 4 | 550 ± 134 (340-779) | 164 ± 67 (98-371) | ||
| 12 | 4 | 796 ± 247 (385-1,173) | 208 ± 83 (97-374) | ||
| ΔfucT2 | Prechallenge | NA | 314 ± 98 (106-665) | 90 ± 58 (52-193) | |
| 2 | 5 | 475 ± 90 (257-695) | 136 ± 46 (71-222) | ||
| 4 | 4 | 505 ± 109 (295-713) | 157 ± 54 (84-267) | ||
| 12 | 4 | 198 ± 218 (−14-608) | 108 ± 112 (−12-328) | ||
| ΔfucT1/2 | Prechallenge | NA | 6 ± 21 (−12-42) | 8 ± 19 (−3-42) | |
| 2 | 4 | 3 ± 5 (−3-16) | −3 ± 10 (−17-16) | ||
| 4 | 4 | 0 ± 14 (−14-53) | 0 ± 24 (−13-63) | ||
| 12 | 4 | −1 ± 4 (−11-3) | 14 ± 26 (−19-98) | ||
Values for one to five single colonies picked from preinoculation culture or from primary culture plate for each animal tested.
NA, not applicable.
Mouse-passaged strains were isolated from the animals used for laboratory passage and were used to challenge fresh mice after three or fewer in vitro passages.
Mouse challenge with laboratory-passaged H. pylori 26665 wild-type and mutant strains.
To examine the effect of elimination of the Lewis phenotype on gastric colonization in mice, laboratory-passaged wild-type 26695 and its isogenic ΔfucT2 and ΔfucT1/2 mutants were orally inoculated into C3H/HeJ mice. The results of these experimental challenges are summarized in Table 4. Compared with colonization by the wild-type 26695 strain (60%), challenge with the fucT mutants yielded higher colonization rates (87% for ΔfucT2 and 80% for ΔfucT1/2). The fucT mutants also showed higher levels of colonization at 2 and 6 weeks (between 103 and 105 CFU/stomach) than the wild-type strain (between 101 and 102 CFU/stomach) (Table 4). Thus, elimination of Lex and Ley expression did not interfere with the ability of laboratory strain 26695 to colonize the stomachs of mice.
TABLE 4.
Gastric colonization of C3H/HeJ mice with wild-type H. pylori strain 26695 or isogenic ΔfucT mutants detected by culture
| Passage | Bacterial genotype | No. of infected animals (mean log10H. pylori CFU ± SDa) at wk:
|
|||
|---|---|---|---|---|---|
| 2 (n = 5/strain) | 6 (n = 5/strain) | 12 (n = 5/strain) | Total (n = 15/strain) | ||
| Laboratory | Wild type | 3 (1.90 ± 0.10) | 3 (1.93 ± 0.21) | NEb | 6c (1.92 ± 0.15) |
| ΔfucT2 | 5 (3.58 ± 0.75) | 4 (3.35 ± 0.52) | 4 (3.40 ± 0.14) | 13 (3.45 ± 0.52) | |
| ΔfucT1/2 | 4 (4.05 ± 1.00) | 4 (3.60 ± 0.08) | 4 (3.73 ± 0.10) | 12 (3.79 ± 0.56) | |
| Mouse | Wild type | 4 (1.61 ± 0.50) | 4 (2.32 ± 1.03) | 4 (3.02 ± 0.36) | 12 (2.31 ± 0.87) |
| ΔfucT2 | 5 (3.18 ± 0.33) | 4 (2.57 ± 0.60) | 4 (1.83 ± 0.96) | 13 (2.58 ± 0.87) | |
| ΔfucT1/2 | 4 (1.34 ± 0.26) | 4 (1.91 ± 0.51) | 4 (2.76 ± 0.94) | 12 (2.02 ± 0.88) | |
The calculation includes only mice in which colonization was detected by culture.
NE, not examined.
Only 10 mice were challenged in total, since there was no 12-week challenge.
Mouse challenge with mouse-passaged H. pylori 26665 wild-type and mutant strains.
To determine whether the results with the laboratory-passaged strains adequately reflected the situation for strains that had been passed in vivo, next we examined colonization by strains recovered from mice in the previous experiments. We found that the mouse-passaged ΔfucT mutants colonized the challenged mice as frequently as did the mouse-passaged wild-type parental strain (Table 4), and there were no significant differences in the frequency of colonization up to 12 weeks, when the experiment ended (80% for wild type, 87% for ΔfucT2, and 80% for ΔfucT1/2). Comparing sequential colonization rates, there was no evidence of spontaneous H. pylori clearance from the animals for up to 12 weeks, regardless of the challenge strain. The mean levels of colonization determined by quantitative culture were between 101 and 104 CFU/stomach, and there was no substantial difference between the wild type and the ΔfucT2 or ΔfucT1/2 mutant (Table 4). Since one possibility to explain the robust colonization by the mutant strains is reversion to wild type, we performed PCR analysis of the HP0379 and HP0651 genes of the ΔfucT2 and ΔfucT1/2 isolates after mouse passage. Both the laboratory- and mouse-passaged strains showed patterns identical to those of the preinoculation mutant strains, indicating the retention of the antibiotic resistance cassette (data not shown). These results indicated that there had been no reversion to wild type.
Lex and Ley expression by wild-type H. pylori and isogenic mutants after mouse passage.
To determine whether the expression of Lex and Ley of the individual H. pylori strains had changed during mouse passage, perhaps reflecting selection for particular phenotypes, Le typing was performed on five single colonies from each of the isolates obtained prechallenge and from each sacrifice time point (Table 3). In both experiments, involving the laboratory- or mouse-passaged wild-type strains, expression of Lex declined compared with that prechallenge, but the Ley expression changed little. The ΔfucT1/2 mutants showed no measurable Lex or Ley expression through the 12 weeks of colonization. The lack of Le expression in these mutants, consistent with the genotypic evidence of the stability of the insertion mutations, indicated no reversion to wild type.
Serum antibody response to H. pylori.
The serum antibody response to H. pylori whole-cell antigens was monitored from prechallenge to 12 weeks postchallenge in the 85 animals. The colonized animals showed progressive increases in the serum IgG response to H. pylori antigens through the 12-week experiments, as shown in the studies of the mice challenged with the mouse-passaged strains (Fig. 1). There were no significant differences in response based on the Lewis status of the challenge strain. For 20 mice that were H. pylori culture positive at 12 weeks postchallenge, there was no correlation between the antibody responses and bacterial CFU (data not shown). All 20 had seroconverted to the H. pylori antigens, which is consistent with their persistent infection. Since seroconversion was sensitive for detecting colonization, when we applied the same seroconversion criterion to the 17 culture-negative mice, we found that all 17 mice (100%) also met the criterion. In total, based on either the culture (80%) or seroconversion (96.5%) criterion, all (100%) of the 85 C3H/HeJ mice challenged with H. pylori in this study had become infected, regardless of the H. pylori genotype and Le expression (Tables 3 and 4).
FIG. 1.
Serum IgG responses to H. pylori whole-cell antigens in C3H/HeJ mice following challenge with wild-type (▪) and isogenic ΔfucT2 (•) and ΔfucT1/2 (▴) mutant mouse-passaged H. pylori strains. ×, negative control. Error bars indicate standard deviations. ODU, optical density units.
Effect of Le expression on H. pylori binding to gastric epithelial cells.
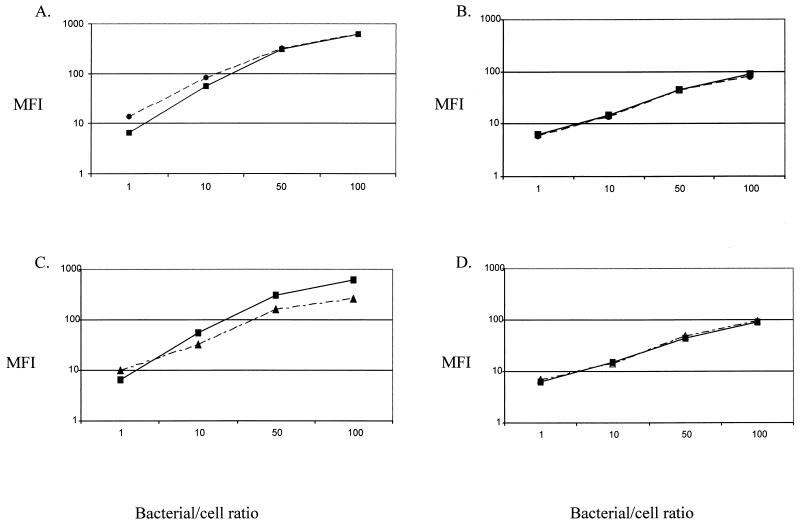
Since H. pylori naturally colonizes humans and not mice, we next performed in vitro experiments to assess the effect of Le expression on binding of H. pylori cells to human gastric epithelial cells. The assays entail labeling viable H. pylori cells with a chromophore, incubating with the epithelial cells at various bacteria/cell ratios, and, after washing, assaying the epithelial cells for fluorescence by flow cytometry. Studies using AGS-CDM, AGS-NY2, and KatoIII cells showed similar results; for bacteria/cell ratios of between 1:1 and 100:1, there was essentially no difference between the 26695 wild-type and Le knockout strains (Fig. 2). Treatment of KatoIII cells with gamma interferon increased the binding of both the wild-type and ΔfucT1/2 knockout strains, but in the same proportions (data not shown). Transfection of Cos cells ID12, which express class II major histocompatibility complex, slightly increased binding at the 1:1 ratio for both the wild type and the ΔfucT1/2 mutant but had no effect on binding at the 10:1 or 100:1 bacteria/cell ratio (data not shown). In total, Le expression had no significant effect on H. pylori binding in any of the cell binding assays studied.
FIG. 2.
Binding of H. pylori strains to gastric epithelial cells (AGS-NY2 [A and C] and KatoIII [B and D]). Each panel compares binding of wild-type strain 26695 (solid lines) with that of its isogenic ΔfucT2 (A and B) or ΔfucT1/2 (C and D) mutant (dashed lines). Bacteria/cell ratios ranged from 1:1 to 1:100, and binding was estimated as the mean fluorescence intensity (MFI) by flow cytometry. Results are representative of those from two separate experiments.
DISCUSSION
To establish the role of Le expression in the ability of H. pylori strains to colonize the murine stomach, we created mutants with mutations in one or both of the 1,3-fucosyltransferase genes that are required for expression of both Lex and Ley (13, 20, 34). As expected, when both fucT1 and fucT2 both were inactivated, Le expression was blocked. In the present studies, we showed that this mutant genotype and its resulting phenotype persisted when the strains were passed in vivo for 12 weeks.
By using a variety of model systems, H. pylori virulence factors that affect the organism's ability to colonize the host have been identified (10). Such factors include motility, urease function, and superoxide dismutase activity (8, 9, 28). The results of this study clearly show that Le expression is not required for H. pylori colonization of C3H/HeJ mice, at least for up to 12 weeks. Since the initial studies were performed with laboratory-passaged strains that had had an unknown number of in vitro passages, we also recovered H. pylori cells from mice that had undergone challenge and used these mouse-passaged strains to assess whether in vivo passage led to any substantial differences. One limitation is that mouse passage may have selected for H. pylori cells better able to colonize the mouse stomach; however, the fucT genotypes and Le phenotypes were essentially unchanged, indicating that there had not been either reversion to wild type or upregulation of expression by a complementary mechanism. Recently, Suresh and colleagues (30) reported that a wild-type H. pylori strain lacking Lex and Ley expression was able to colonize both C57/BL6 and BALB/c mice. Although the genotype of that strain was not reported and the phenotype of the cells recovered from the mice was not determined, the present results both confirm and extend that work. Taken together, these results clearly indicate that H. pylori strains not expressing either Lex or Ley can colonize mice, at least for a period of 12 weeks. In studies reported by Martin et al. (21), a double fucT mutant lacking Lex or Ley expression colonized outbred HSD/ICR mice substantially more poorly than did its parental strain. Whether differences in the background H. pylori strains used or in the host mouse genotype (outbred versus C3H/HeJ in our studies) contributed to the differences observed remains to be determined. The C3H/HeJ LPS-nonresponder mice, with a mutation in Toll-like receptor 4, which recognizes bacterial LPS (17), may be more permissive for H. pylori colonization than most other mouse strains.
The ability of strains lacking Lex and Ley expression to colonize mice suggests several hypotheses. First, since some H. pylori strains recovered from humans do not express Lex or Ley and apparently are fully capable of colonizing humans (31), Le expression is not required for colonization of the mammalian stomach and has other functions for the bacterial cell. Alternatively, 12 weeks is not a sufficient indicator of long-term colonization, and eventually strains lacking Lex or Ley expression will be cleared from the host. However, up to 10% of H. pylori strains isolated from humans have no detectable Le expression; this is especially notable in strains lacking the cag island (35). Our previous studies (39) and that of Rasko et al. (26) show that humans can be colonized by genetically indistinguishable strains that vary in Lewis antigen expression; these clonal variants also illustrate that Le expression is not required for colonization of humans. However, since murine epithelial cells do not normally express Leb (15), this work is not fully analogous to the situation in primates, in which H. pylori expression of Lex and/or Ley generally mirrors that of the host (7, 37). An alternative hypothesis is that Le expression facilitates H. pylori binding to host epithelial cells (11). The results of our in vitro studies (Fig. 2) do not support that hypothesis. However, although several different cell lines were examined, with and without cytokine stimulation, these were transformed cells and may have lacked expression of critical ligands.
Together, these studies and observations in humans of H. pylori cells lacking Le expression indicate that H. pylori expression of Lex or Ley is not required for gastric colonization, at least over relatively short periods of time (weeks to months). These observations do not refute the hypothesis that Le expression is a bacterial phenotype subject to ongoing selection and that particular levels are adaptive among competing H. pylori strains for occupation of microniches. If the incremental fitness associated with expression of the phenotype most appropriate for a particular host or microniche was small (35), then the effects of this difference might not be evident except over long periods of observation.
Acknowledgments
This work was supported in part by grants R01DK53707, R01GM63270, R01DK51677, and R21AI48173 from the National Institutes of Health; the Medical Research Service of the Department of Veterans Affairs; a European H. pylori Study Group Research Fellowship from the Digestive Disorders Foundation, United Kingdom (to E.E.-O.); and the Japan Clinical Pathology Foundation for International Exchange and Yoshida Scholarship Foundation (to T.T.).
Editor: V. J. DiRita
REFERENCES
- 1.Appelmelk, B. J., B. Shiberu, C. Trinks, N. Taposi, P. Y. Zheng, T. Verboom, J. Maaskant, C. H. Hokke, W. E. C. M. Schipohost, D. Blanchard, I. M. Simoons-Smit, D. H. van den Eijnden, and C. M. J. E. Vandenbroucke-Grauls. 1998. Phase variation in Helicobacter pylori lipopolysaccharide. Infect. Immun. 66:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelmelk, B. J., S. L Martin, M. A. Monteiro, C. A. Clayton, A. A. McColm, P. Zheng, T. Verboom, J. J. Maaskant, D. H. Van Den Eijnden, C. H. Hokke, M. B. Perry, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 1999. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in α3-fucosyltranserase genes. Infect. Immun. 67:5361-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appelmelk, B. J., M. A. Monteiro, S. L. Martin, A. P. Moran, and C. M. J. E. Vandenbroucke-Grauls. 2000. Why Helicobacter pylori has Lewis antigen. Trends Microbiol. 8:565-570. [DOI] [PubMed] [Google Scholar]
- 4.Appelmelk, B. J., M. C. Martino, E. Veenhof, M. A. Monteiro, J. J. Maaskant, R. Negrini, F. Lindh, M. Perry, G. Del Giudice, and C. M. Vandenbroucke-Grauls. 2000. Phase variation in H type I and Lewis a epitopes of Helicobacter pylori lipopolysaccharide. Infect. Immun. 68:5928-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser, M. J. 1999. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J. Infect. Dis. 179:1523-1530. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J., and D. Kirschner. 1999. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc. Natl. Acad. Sci. USA 96:8359-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubois, A., D. E. Berg, E. T. Incecik, N. Fiala, L. M. Heman-Ackah, J. Del Valle, M. Yang, H. P. Wirth, G. I. Perez-Perez, and M. J. Blaser. 1999. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology 116:90-96. [DOI] [PubMed] [Google Scholar]
- 8.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, K. A. 1999. Animal models of Helicobacter gastritis. Curr. Top. Microbiol. Immunol. 241:123-154. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, N. J., M. A. Monteiro, G. Faller, E. J. Walsh, A. P. Moran, I. S. Roberts, and N. J. High. 2000. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol. Microbiol. 35:1530-1539. [DOI] [PubMed] [Google Scholar]
- 12.Fan, X., S. E. Crowe, S. Behar, H. Gunasena, G. Ye, H. Haeberle, N. Van Houten, W. K. Gourley, P. B. Ernst, and V. E. Reyes. 1998. The effect of class II MHC expression on adherence of Helicobacter pylori and induction and apoptosis in gastric epithelial cells: a mechanism for Th1 cell-mediated damage. J. Exp. Med. 187:1659-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, Z., N. W. C. Chan, M. M. Palcic, and D. E. Taylor. 1997. Cloning and heterologous expression of and α 1,3 fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J. Biol. Chem. 272:21357-21363. [DOI] [PubMed] [Google Scholar]
- 14.Ghiara, P., M. Marchetti, M. J. Blaser, M. K. R. Tummuru, T. L. Cover, E. D. Segal, L. S. Tompkins, and R. Rappuoli. 1995. Role of Helicobacter pylori virulence factors vacuolating cytotoxin, CagA, and urease in a mouse model of disease. Infect. Immun. 63:4154-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guruge, J. L., P. G. Falk, R. G. Lorenz, M. Dans, H. P. Wirth, M. J. Blaser, D. E. Berg, and J. I. Gordon. 1998. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 95:3925-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heneghan, M. A., C. F. McCarthy, and A. P. Moran. 2000. Relationship of blood group determinants on Helicobacter pylori lipopolysaccharide with host Lewis phenotype and inflammatory response. Infect. Immun 68:937-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawahara, T., S. Teshima, A. Oka, T. Sugiyama, K. Kishi, and K. Rokutan. 2001. Type I Helicobacter pylori lipopolysaccharide stimulates toll-like receptor 4 and activates mitogen oxidase 1 in gastric pit cells. Infect. Immun 69:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labigne-Roussel, A., P. Courcoux, and L. Tompkins. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchetti, M., B. Arico, D. Burroni, N. Figura, R. Rappuoli, and P. Ghiara. 1995. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science 267:1655-1658. [DOI] [PubMed] [Google Scholar]
- 20.Martin, S. L., M. R. Edbrooke, T. C. Hodgman, D. H. van den Eijnden, and M. I. Bird. 1997. Lewis X biosynthesis in Helicobacter pylori. Molecular cloning of and α (1,3) fucosyltransferase gene. J. Biol. Chem. 272:21349-21356. [DOI] [PubMed] [Google Scholar]
- 21.Martin, S. L., A. A. McColm, and B. J. Appelmelk. 2000. H. pylori adhesin and Lewis X. Gastroenterology 119:1414-1415. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro, M. A., K. H. Chan, D. A. Rasko, D. E. Taylor, P. Y. Zheng, B. J. Appelmelk, H. P. Wirth, M. Yang, M. J. Blaser, S. O. Hynes, A. P. Moran, and M. B. Perry. 1998. Simultaneous expression of type 1 and type 2 Lewis blood group antigens Helicobacter pylori lipopolysaccharides. J. Biol. Chem. 273:11533-11543. [DOI] [PubMed] [Google Scholar]
- 23.Monteiro, M. A., B. J. Appelmelk, D. A. Rasko, A. P. Moran, S. O. Hynes, L. L. MacLean, K. H. Chan, F. S. Michael, S. M. Logan, J. O'Rourke, A. Lee, D. E. Taylor, and M. B. Perry. 2000. Lipopolysaccharide structures of Helicobacter pylori genomic strains 26695, J99, mouse model H. pylori Sydney strain, H. pylori P4666 carrying sialyl LewisX and H. pylori UA915 expressing Lewis B: classification of H. pylori lipopolysaccharides into glycotype families. Eur. J. Biochem. 267:305-320. [DOI] [PubMed] [Google Scholar]
- 24.Moran, A. P., M. M. Prendergast, and B. J. Appelmelk. 1996. Molecular mimicry of host structures by bacterial lipopolysaccharides and contribution to disease. FEMS. Immunol. Med. Microbiol. 16:105-115. [DOI] [PubMed] [Google Scholar]
- 25.Moran, A. P., E. Sturegard, H. Sjunnesson, T. Wadstrom, and S. O. Hynes. 2000. The relationship between O-chain expression and colonisation ability of Helicobacter pylori in a mouse model. FEMS Immunol. Med. Microbiol. 29:263-270. [DOI] [PubMed] [Google Scholar]
- 26.Rasko, D. A., T. J. Wilson, D. Zopf, and D. E. Taylor. 2000. Lewis antigen expression and stability in Helicobacter pylori isolates from serial gastric specimens. J. Infect. Dis. 181:1089-1095. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto, J., T. Watanabe, H. Tokumaru, H. Takagi, H. Nakazato, and K. O. Lloyd. 1989. Expression of Lewisa, Lewisb, Lewisx, sialyl-Lewisa, and sialyl-Lewisx blood group antigens in human gastric carcinoma and in normal gastric tissue. Cancer Res. 49:745-752. [PubMed] [Google Scholar]
- 28.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoons-Smit, I. M., B. J. Appelmelk, T. Verboom, R. Negrini, J. L. Penner, G. O. Aspinall, A. P. Moran, S. F. Fei, B. S. Shi, W. Rudnica, A. Savio, and J. de Graaff. 1996. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. J. Clin. Microbiol. 34:2196-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suresh, M. R., M. B. Fanta, J. Kriangkum, Q. Jiang, and D. E. Taylor. 2000. Colonization and immune responses in mice to Helicobacter pylori expressing different Lewis antigens. J. Pharm. Pharm. Sci. 3:259-266. [PubMed] [Google Scholar]
- 31.Taylor, D. E., D. A. Rasko, R. Sherburne, C. Ho, and L. D. Jewell. 1998. Lack of correlation between Lewis antigen expression by Helicobacter pylori and gastric epithelial cells in infected patients. Gastroenterology 115:1113-1122. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance in low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G., Z. Ge, D. A. Rasko, and D. E. Taylor. 2000. Lewis antigen in Helicobacter pylori: biosynthesis and phase variation. Mol. Microbiol. 36:1187-1196. [DOI] [PubMed] [Google Scholar]
- 35.Webb, G. F., and M. J. Blaser. 2002. Dynamics of bacterial phenotype selection in a colonized host. Proc. Natl. Acad. Sci. USA 99:3135-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth, H. P., M. Yang, M. Karita, and M. J. Blaser. 1996. Expression of the human cell surface glycoconjugates Lewis x and Lewis y by Helicobacter pylori isolates is related to cagA status. Infect. Immun. 64:4598-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth, H. P., M. Yang, R. M. Peek, Jr., K. M. Tham, and M. J. Blaser. 1997. Helicobacter pylori Lewis expression is related to the host Lewis phenotype. Gastroenterology 113:1091-1098. [DOI] [PubMed] [Google Scholar]
- 38.Wirth, H. P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirth, H. P., M. Yang, R. M. Peek, Jr., J. Hook-Nikanne, M. Fried, and M. J. Blaser. 1999. Phenotypic diversity in Lewis expression of Helicobacter pylori isolates from the same host. J. Lab. Clin. Med. 133:488-500. [DOI] [PubMed] [Google Scholar]