Abstract
The important human pathogen Helicobacter pylori requires the abundant expression and activity of its urease enzyme for colonization of the gastric mucosa. The transcription, expression, and activity of H. pylori urease were previously demonstrated to be induced by nickel supplementation of growth media. Here it is demonstrated that the HP1338 protein, an ortholog of the Escherichia coli nickel regulatory protein NikR, mediates nickel-responsive induction of urease expression in H. pylori. Mutation of the HP1338 gene (nikR) of H. pylori strain 26695 resulted in significant growth inhibition of the nikR mutant in the presence of supplementation with NiCl2 at ≥100 μM, whereas the wild-type strain tolerated more than 10-fold-higher levels of NiCl2. Mutation of nikR did not affect urease subunit expression or urease enzyme activity in unsupplemented growth media. However, the nickel-induced increase in urease subunit expression and urease enzyme activity observed in wild-type H. pylori was absent in the H. pylori nikR mutant. A similar lack of nickel responsiveness was observed upon removal of a 19-bp palindromic sequence in the ureA promoter, as demonstrated by using a genomic ureA::lacZ reporter gene fusion. In conclusion, the H. pylori NikR protein and a 19-bp operator sequence in the ureA promoter are both essential for nickel-responsive induction of urease expression in H. pylori.
The gram-negative human pathogen Helicobacter pylori colonizes the mucus overlaying the gastric epithelium, leading to chronic gastritis that can subsequently develop into peptic ulcer disease and gastric cancer (12). Approximately half of the world's population is colonized by H. pylori, constituting a major public health problem (12).
One of the factors required for gastric colonization by H. pylori is its urease enzyme (13, 14, 37). Urea hydrolysis by urease yields ammonia and bicarbonate, and these products have important functions in H. pylori colonization and infection. Ammonia contributes to acid resistance by neutralizing the microenvironment of the bacterium (8, 27), serves as a nitrogen source (10), is essential for chemotactic behavior (25), and causes damage to the gastric epithelium (30). Bicarbonate also contributes to the virulence of H. pylori, as it protects against the bactericidal activity of peroxynitrite, a nitric oxide metabolite (20). Urease is also involved in the resistance of H. pylori to opsonization and phagocytosis and in the adherence of H. pylori to epithelials cells, although these functions do not depend on enzymatic activity or ammonia production via urease (21, 22, 26).
H. pylori produces large amounts of urease, and it has been estimated that up to 10% of the total protein content of H. pylori consists of urease (3). Active urease is a multimeric enzyme that consists of six UreA and six UreB subunits and 12 Ni2+ ions functioning as a cofactor (19, 23). The 27-kDa UreA and 62-kDa UreB urease subunits are encoded by the ureA and ureB genes, respectively, which are followed by a second operon encoding the UreIEFGH accessory proteins (23). The UreEFGH accessory proteins are involved in assembly and activation of urease, while the UreI protein probably functions as an acid-activated urea transporter (23, 27, 36).
Transcription of the H. pylori urease gene cluster occurs from two promoters, one upstream of the ureA gene and one in the intergenic region between ureB and ureI (1, 29). Transcription from these two promoters and subsequent pH-dependent differential mRNA decay lead to the formation of ureAB, ureABIE′, ureIE′, and ureF′GH mRNAs (1). Urease production in other ureolytic bacteria is known to be regulated by changes in environmental conditions, such as pH, urea availability, nitrogen availability, or growth phase (5). Uniquely, not only urease activity but also the expression of urease in H. pylori is regulated by the availability of the nickel cofactor (33). Nickel supplementation of brucella medium resulted in a 4-fold induction of urease expression at the protein level and a 12-fold induction of urease enzyme activity. The regulatory system mediating this nickel-responsive induction of urease expression has not yet been described (33).
Nickel-responsive regulation of gene expression has been observed in several bacteria, but the molecular mechanisms have been studied only for a few systems (15). One of the best-studied examples is the regulation of expression of nickel uptake in Escherichia coli, where the expression of the nickel transport operon nikABCDE is controlled by the NikR protein (6, 7, 9). NikR represses the transcription of the nickel uptake genes by binding to an operator sequence located in the target promoter region upon increased cytoplasmic nickel availability (6, 7, 9, 15), in a fashion similar to that of the family of Fur metalloregulatory proteins (16).
Urease activity is essential for gastric colonization by H. pylori (13, 14, 37), and its regulation is probably also necessary for successful colonization. H. pylori contains a gene (2, 32) which encodes a protein (HP1338 or JHP1257) homologous to the E. coli NikR protein (6, 7, 9). Here we report on the role of the H. pylori NikR ortholog in the nickel-responsive induction of urease expression and activity and identify a nickel-responsive operator sequence in the urease promoter.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The H. pylori strains and plasmids used in this study are listed in Table 1. H. pylori was routinely maintained on Columbia agar plates supplemented with 7% saponin-lysed horse blood, 0.004% triphenyltetrazolium chloride (Sigma), and Dent selective supplement (Oxoid) (4) at 37°C under microaerobic conditions (10% CO2, 5% O2, and 85% N2). Broth cultures were grown under the same conditions in brucella broth (Difco) supplemented with 3% newborn calf serum (Gibco) (BBN). Nickel chloride (Sigma) was filter sterilized and used at various concentrations. E. coli strains DH5α MCR (Gibco) and ER1793 (New England Biolabs) were grown aerobically at 37°C in Luria-Bertani medium (28). When appropriate, growth media were supplemented with ampicillin (100 μg/ml), kanamycin (20 μg/ml), or chloramphenicol (10 μg/ml).
TABLE 1.
H. pylori strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Reference or source |
|---|---|---|
| H. pylori strains | ||
| 26695 | Wild-type strain | 32 |
| 26695nikR | 26695 nikR::Kmr | This study |
| 1061 | Wild-type strain | 17 |
| AV433 | 1061 ureA::lacZ Kmr | 33 |
| 1061BJD3.8 | 1061 ureA(Δ−50/−112)::lacZ Kmra | This study |
| 1061BJD3.9 | 1061 ureA(Δ−50/−90)::lacZ Kmr | This study |
| 1061BJD3.10 | 1061 ureA(Δ−50/−70)::lacZ Kmr | This study |
| Plasmids | ||
| pAV348 | pBluescript derivative containing a 1.5-kb DNA fragment containing the H. pylori 1061 nikR gene | This study |
| pAV364 | pAV348 with the Kmr cassette of pJMK30 (34) inserted in the unique SphI restriction site | This study |
| pBW | H. pylori promoter-probe vector; Kmr | 11 |
| pBJD3.3 | pBW derivative containing the H. pylori 1061 ureA promoter fused to the lacZ gene of pBW; Kmr | 33 |
| pBJD3.8 | pBJD3.3 derivative [ureA(Δ−50/−112)::lacZ]; Kmr | This study |
| pBJD3.9 | pBJD3.3 derivative [ureA(Δ−50/−90)::lacZ]; Kmr | This study |
| pBJD3.10 | pBJD3.3 derivative [ureA(Δ−50/−70)::lacZ]; Kmr | This study |
ureA(Δ−50/−112), ureA lacking the sequences from positions −50 to −112.
Recombinant DNA techniques.
Restriction enzymes and DNA-modifying enzymes were used according to the manufacturer's instructions (New England Biolabs). Standard protocols were used for the manipulation of DNA and the transformation of E. coli (28) and H. pylori (4). Plasmid DNA was prepared by using Qiaprep spin columns (Qiagen), and PCR was carried out by using Taq polymerase (Promega).
Construction of an H. pylori nikR mutant.
The region containing the H. pylori nikR ortholog and its upstream and downstream sequences was amplified from H. pylori strain 1061 by using primers F1337 (5′-TAGAAGAAATTGGCGCGTCA) and 1339R (5′-TCACGCCCATGTCATAGAA). The resulting nikR PCR fragment was cloned in pBluescript II SK(−) (Stratagene), resulting in pAV348 (Table 1). The nikR coding region in pAV348 was interrupted by insertion of the kanamycin resistance gene from pJMK30 (34) in the unique SphI site, resulting in plasmid pAV364 (Table 1). This plasmid was subsequently used for natural transformation of H. pylori 26695, and the kanamycin-resistant colonies isolated were designated 26695 nikR. Two colonies derived from independent transformations were tested and gave identical results in all experiments. Correct allelic replacement of the wild-type nikR gene with the interrupted version was confirmed by PCR-based analysis (data not shown).
Protein analysis.
H. pylori cultures were grown in unsupplemented or NiCl2-supplemented BBN for 20 to 24 h at 37°C with moderate shaking to an optical density at 600 nm (OD600) of 0.4 to 0.8, centrifuged at 4,000 × g for 10 min at 4°C, and resuspended in ice-cold phosphate-buffered saline to a final OD600 of 10. H. pylori cells were lysed by sonication for 15 s on ice with an MSE Soniprep 150 set at amplitude 10. Protein concentrations were determined by the bicinchoninic acid method (Pierce) with bovine serum albumin as the standard. Samples containing 15 μg of protein were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (28).
Urease activity.
The urease activity of fresh lysates was determined by measuring ammonia production from urea hydrolysis with the Berthelot reaction as described previously (33, 35). The amount of ammonia present in the samples was inferred from a standard NH4Cl concentration curve. Urease activity was expressed as micromoles of urea hydrolyzed per minute per milligram of protein.
ureA::lacZ transcriptional fusions.
Plasmids pBJD3.3, pBJD3.8, pBJD3.9, and pBJD3.10 were used to test the effect of the palindromic sequence at positions −49 to −67 on the nickel-responsive induction of the ureA promoter (Table 1). Plasmids pBJD3.8, pBJD3.9, and pBJD3.10 were all derived from pBJD3.3 (33), which contains the H. pylori 1061 ureA promoter cloned in front of a promoterless lacZ gene (11). The ureA promoter was modified by inverse PCR mutagenesis (38), resulting in deletions of sequences upstream of the H. pylori 1061 ureA transcriptional start site ranging from positions −50 to −70, from positions −50 to −90, and from positions −50 to −112. The ureA promoter regions of pBJD3.3, pBJD3.8, pBJD3.9, and pBJD3.10 were sequenced to verify correct removal of the desired sequences and the absence of other nucleotide substitutions or deletions. Transformation of H. pylori strain 1061 to kanamycin resistance by pBJD3.3 and its mutant derivatives resulted in integration of the plasmid by single homologous recombination, with the cloned promoter region preceding the lacZ reporter gene, whereas the wild-type promoter still preceded the intact urease operon (33). The insertion of the pBJD3.3 vector did not have a major effect on the expression, activity, or nickel induction of urease, as demonstrated previously (33). Transformation of H. pylori 1061 with pBJD3.3 and its mutant derivatives resulted in the kanamycin-resistant H. pylori strains AV433, 1061BJD3.8, 1061BJD3.9, and 1061BJD3.10 (Table 1). The β-galactosidase activities (in Miller units) (28) of these strains grown in either unsupplemented or nickel-supplemented BBN were determined with lysates from freshly sonicated cells as described previously (33).
Nucleotide sequence accession number.
The DNA sequence of the ureA promoter of H. pylori strain 1061 has been deposited in the GenBank sequence database under accession number AY078177.
RESULTS
Identification of the H. pylori NikR ortholog.
Analysis of the proteins encoded by the H. pylori strain 26695 genome (32) for orthologs of nickel regulatory proteins indicated that the HP1338 protein is homologous to the E. coli NikR protein, displaying 30% identity and 68% similarity. Orthologs of other known nickel regulatory proteins, such as the Ralstonia Cnr proteins (18, 31), were not apparent. The genetic organization of the genomic region containing the H. pylori HP1338 gene (subsequently referred to as nikR) is conserved between H. pylori strains 26695 and J99 (2, 32). Located downstream of the nikR gene is the HP1337 gene, which is annotated as a conserved hypothetical protein (2, 32); the upstream divergent operon encodes an ExbB-ExbD-TonB complex, which in other bacteria is involved in the transport of iron compounds across the outer membrane (24).
Inactivation of the nikR gene renders H. pylori nickel sensitive.
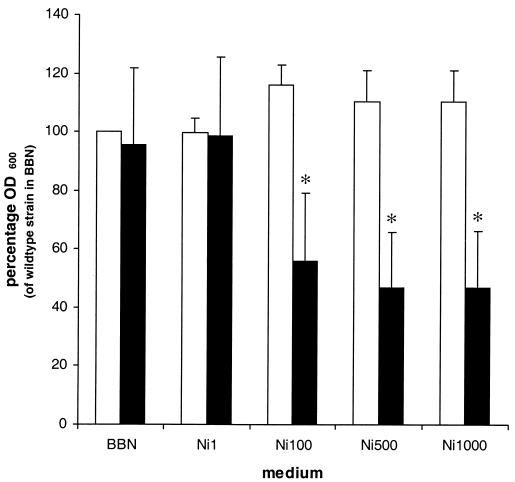
The nikR gene of H. pylori strain 26695 was interrupted by the insertion of a kanamycin resistance gene, and the effect of the nikR mutation on the nickel sensitivity of H. pylori was tested by comparing the growth of the wild-type strain and the nikR mutant in media with increasing NiCl2 concentrations. The growth of the nikR mutant in unsupplemented medium or in medium with 1 μM NiCl2 was not significantly different from that of the wild-type strain (Fig. 1). However, the growth of the nikR mutant was clearly affected by medium supplementation with 100 μM NiCl2 or higher (Fig. 1). This result indicates that NikR is required for the nickel resistance of H. pylori.
FIG. 1.
Insertional inactivation of the nikR gene renders H. pylori nickel sensitive. Wild-type H. pylori strain 26695 and its isogenic nikR mutant were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1), 100 μM (Ni100), 500 μM (Ni500), and 1,000 μM (Ni1000). Growth was monitored by measuring the OD600 24 h after inoculation. Growth is expressed as a percentage relative to the OD600 of the wild-type strain in unsupplemented medium (set at 100%; no error bar). White bars represent wild-type H. pylori 26695; blacks bars represent the nikR mutant. Results shown are the averages of four independent growth experiments; errors bars denote standard deviations. Asterisks indicate a significant difference in growth between the nikR mutant and wild-type H. pylori (the P value was <0.01, as determined by Student's t test).
Mutation of H. pylori nikR abolishes nickel-responsive induction of urease expression.
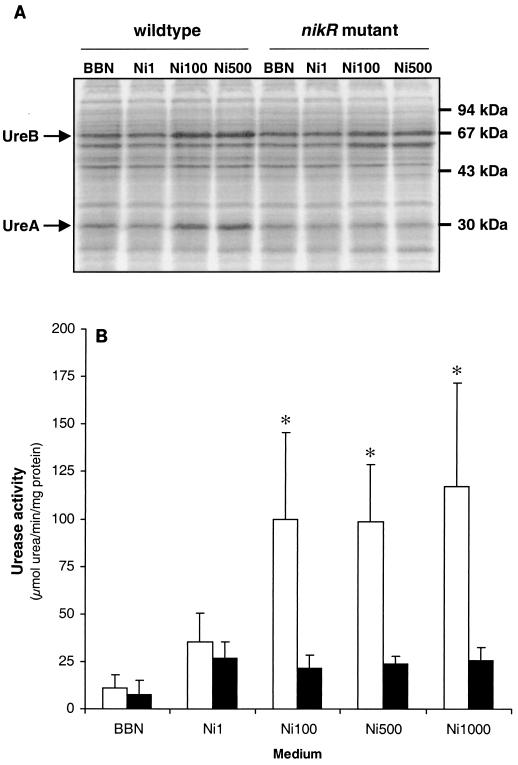
It was previously reported that medium supplementation with NiCl2 to 100 μM results in an approximately fourfold increase in the expression of the urease subunits UreA and UreB in H. pylori strain 26695 (33) (Fig. 2A). The expression of the urease subunits and urease enzyme activity were compared between the wild-type strain and the nikR mutant of H. pylori 26695 to determine the effect of the nikR mutation on the nickel-responsive induction of urease. The expression of the urease subunits UreA and UreB and urease enzyme activity did not differ significantly between the wild-type strain and the nikR mutant in unsupplemented BBN (Fig. 2). When BBN was supplemented with 1 μM NiCl2, the levels of urease subunit expression were unchanged (Fig. 2A), but urease enzyme activity was induced approximately threefold in both the wild-type strain and the nikR mutant (Fig. 2B). Differences between the wild-type and nikR mutant strains were, however, clearly apparent when the medium was supplemented with 100 μM NiCl2 or higher: the nikR mutant strain did not show any further induction of urease subunit expression (Fig. 2A) or urease enzyme activity (Fig. 2B), while the wild-type strain clearly showed a significant increase in the expression of the urease subunits UreA and UreB (Fig. 2A) as well as in urease enzyme activity (Fig. 2B).
FIG. 2.
Insertional inactivation of the H. pylori nikR gene inhibits nickel-responsive induction of urease expression and activity. Wild-type H. pylori 26695 and its isogenic nikR mutant were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1), 100 μM (Ni100), 500 μM (Ni500), and 1,000 μM (Ni1000). (A) Changes in urease expression as monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining. H. pylori strains, UreA and UreB proteins, and relevant molecular mass markers are indicated. (B) Urease activity of the wild-type strain and the nikR mutant in unsupplemented and nickel-supplemented media. White bars represent wild-type H. pylori 26695; black bars represent the nikR mutant. Results shown are the averages of four independent growth experiments; error bars denote standard deviations. Asterisks indicate a significant difference in urease activity between wild-type H. pylori and the nikR mutant (the P value was <0.02, as determined by Student's t test).
Identification of a nickel-responsive operator sequence in the ureA promoter.
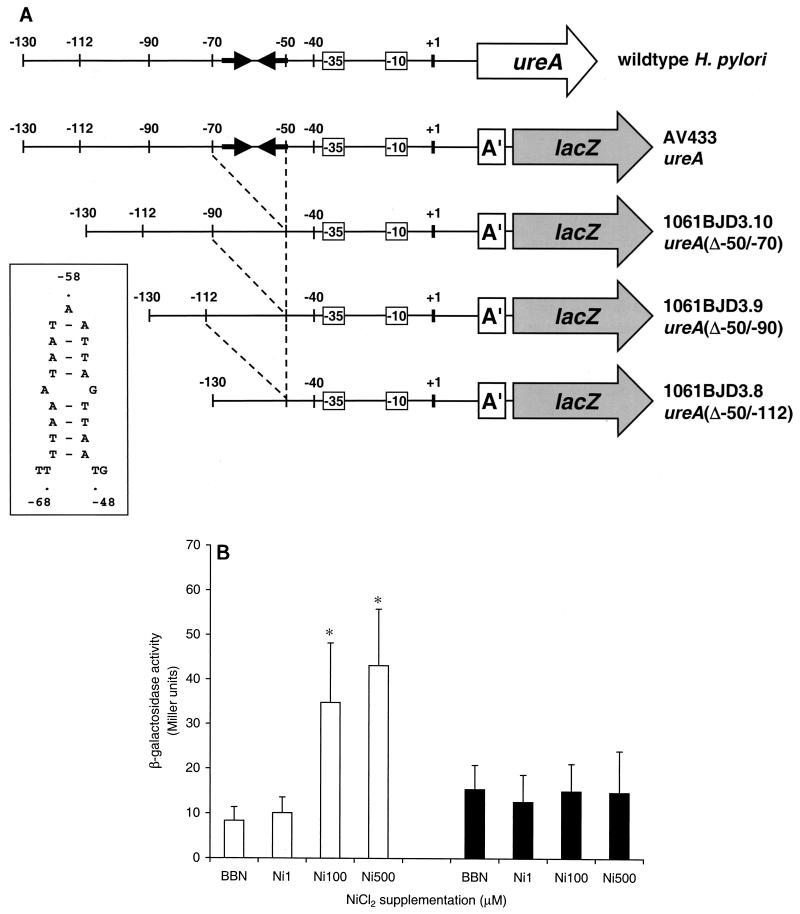
Nickel-responsive induction of urease expression in H. pylori is mediated at the transcriptional level via the ureA promoter (33). Metal-responsive regulatory proteins, such as NikR, often mediate their regulatory function by binding to palindromic operator sequences located in the promoter regions of the target genes (7, 16). The presence of a palindromic sequence in the ureA promoter was described previously, at positions −49 to −67 relative to the transcriptional start site of the ureA gene (Fig. 3A) (29). An H. pylori chromosomal ureA::lacZ reporter gene fusion in H. pylori strain 1061 (17) was used to assess the role of this palindrome in the nickel-responsive induction of urease expression (33). The wild-type promoter was compared with three mutated versions, which lacked the sequences from positions −50 to −70 (Δ−50/−70), from positions −50 to −90 (Δ−50/−90), or from positions −50 to −112 (Δ−50/−112), respectively. Effectively, all mutants lacked the palindromic sequence at positions −49 to −67 with no effect on the −35 and −10 regions (Fig. 3A). Removal of the palindromic sequence did not affect the transcription of the ureA promoter in unsupplemented growth medium (Fig. 3B); however, while the wild-type ureA::lacZ fusion was significantly induced in medium supplemented with 100 μM NiCl2 or higher (Fig. 3B), the deletion mutants did not show any induction at increasing NiCl2 concentrations [e.g., the ureA(Δ−50/−70) mutant] (Fig. 3B). The β-galactosidase activities of the ureA(Δ−50/−90) and ureA(Δ−50/−112) mutants did not differ significantly from that of the ureA(Δ−50/−70) mutant under all the tested medium conditions (data not shown).
FIG. 3.
Identification of a nickel-responsive operator sequence in the H. pylori ureA promoter. (A) Strategy used for identification of the operator sequence. The predicted palindromic structure is indicated by two converging arrows, and the palindrome structure and sequence are shown in the box next to the constructs. Promoter elements (−10 and −35) and the transcriptional start site (+1) are indicated. The boxed A′ indicates the truncated form of the ureA gene used for making the ureA::lacZ promoter fusions. (B) β-Galactosidase activities of the wild-type ureA::lacZ promoter fusion and a ureA(Δ−50/−70)::lacZ promoter fusion mutant in response to different nickel concentrations. White bars represent the wild-type ureA::lacZ promoter of H. pylori AV433; black bars represent the ureA(Δ−50/−70)::lacZ promoter of H. pylori 1061BJD3.10. Strains were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1), 100 μM (Ni100), and 500 μM (Ni500). Results shown are the averages of three independent experiments; error bars denote standard deviations. Asterisks indicate a significant increase in β-galactosidase activity compared to that of the wild-type promoter in unsupplemented medium (the P value was <0.05, as determined by Student's t test).
DISCUSSION
Urease enzyme activity plays an essential role in gastric colonization by H. pylori, since H. pylori mutants devoid of urease activity were unable to colonize the gastric mucosa in animal models (13, 14, 37) even when the gastric pH was neutralized by the administration of proton pump inhibitors (14). H. pylori produces very large amounts of urease, up to 10% of its total protein content (3). The expression and activity of H. pylori urease are upregulated by increased availability of the nickel cofactor, a novel type of transcriptional regulation for bacterial ureases (33). Here we have demonstrated that this nickel-responsive induction is mediated via the HP1338 protein, a NikR ortholog.
NikR was originally identified in E. coli as the nickel-responsive repressor of the nickel uptake operon nikABCDE (9) and is homologous to members of the family of Fur regulatory proteins, which are involved mostly in metal-responsive repression of metal acquisition and oxidative stress defense (16). Biochemical characterization of the E. coli NikR protein indicated that it is a member of the ribbon-helix-helix group of regulatory proteins (6) and that, when complexed with nickel, it binds to a 5′-GTATGA-N16-TCATAC-3′ inverted repeat sequence in the E. coli nikA promoter (6, 7). Both H. pylori genome sequences (2, 32) contain only one sequence resembling this E. coli NikR-binding sequence, which is located directly upstream of the H. pylori nikR gene, indicating possible autoregulation of nikR expression (7).
The urease operon contains two promoters, which are located upstream of the ureA and ureI genes (1, 33), leading to the differential expression of the subunit and accessory proteins. Nickel supplementation induces transcription only from the ureA promoter (33), and this induction is dependent both on the NikR protein (Fig. 2) and on the presence of a 19-bp palindromic sequence in the ureA promoter (Fig. 3). This palindrome closely resembles the iron-responsive regulator Fur in terms of the structure and binding sequence (16). It would be of interest to test the ureA::lacZ promoter variants used in this study in a nikR mutant of H. pylori strain 1061; unfortunately, however, we have been unsuccessful in mutating nikR in this strain (data not shown). It is also unfortunate that the use of the pBW-based lacZ reporter gene system is currently successful only with H. pylori strain 1061, despite attempts to use it with H. pylori strain 26695 (4, 11).
The role of H. pylori NikR in urease regulation is opposite the role of E. coli NikR in nickel uptake, since H. pylori NikR induces the transcription of urease genes while E. coli NikR represses the transcription of nickel uptake genes. This difference may be connected to the location of the palindrome, which is located upstream of the −35 and −10 promoter sequences in the ureA gene. We hypothesize that in unsupplemented medium, the ureA promoter is not completely accessible to RNA polymerase, leading to normal levels of expression of urease. Supplementation of growth medium with higher concentrations of NiCl2 may lead to increased cytoplasmic availability of nickel, allowing the formation of a NikR-nickel complex which can subsequently bind to the palindrome in the ureA upstream region. Binding of NikR to the palindrome may make the ureA promoter more accessible to RNA polymerase, leading to increased levels of urease gene transcription.
The nikR mutant strain showed some increase in urease activity when the medium was supplemented with 1 μM NiCl2, to levels similar to those seen in the wild-type strain (Fig. 2B). This increase, however, was not accompanied by a significant increase in the expression of the urease subunits UreA and UreB (Fig. 2A) (33) and is probably due to more efficient nickel activation of an inactive urease apoenzyme already present. As suggested previously (33), these findings indicate that rather than the amount of urease protein, the amount of the cofactor nickel is a limiting factor for the urease activity of H. pylori. Increased availability of nickel at a low pH, combined with increased influx of urea via the acid-activated UreI protein (27, 36), would enable quick activation of inactive apourease and the associated increase in resistance to acid shock.
In conclusion, the NikR protein of H. pylori induces the transcription, expression, and activity of the essential virulence factor urease in response to nickel. Future studies should focus on the specificity of binding of NikR to the urease promoter as well as the possible roles of NikR in the regulation of nickel uptake, storage, or efflux. The nickel-responsive induction of urease via NikR may also play a role in other urease-producing bacteria and may allow the development of new or improved strategies to prevent or control infection with urease-positive bacteria.
Acknowledgments
This study was financially supported by grants 901-14-206 and DN93-340 from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek to A.H.M.V.V. and J.G.K., a CASE studentship from the Biotechnology and Biological Sciences Research Council and the National Institute for Biological Standards and Control to B.J.D., and grant Ki201/9-1 from the Deutsche Forschungsgemeinschaft to M.K.
Editor: R. N. Moore
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and, T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Bauerfeind, P., R. Garner, B. E. Dunn, and H. L. Mobley. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J. E., C. M. J. E. Vandenbroucke-Grauls, S. H. Phadnis, and J. G. Kusters. 1999. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect. Immun. 67:2433-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burne, R. A., and Y. M. Chen. 2000. Bacterial ureases in infectious diseases. Microbes Infect. 2:533-542. [DOI] [PubMed] [Google Scholar]
- 6.Chivers, P. T., and R. T. Sauer. 1999. NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 8:2494-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high-affinity nickel uptake in bacteria: Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 8.Clyne, M., A. Labigne, and B. Drumm. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Pina, K., V. Desjardin, M. A. Mandrand-Berthelot, G. Giordano, and L. F. Wu. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J. Bacteriol. 181:670-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Reuse, H., and S. Skouloubris. 2001. Nitrogen metabolism, p. 125-133. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 11.de Vries, N., E. J. Kuipers, N. E. Kramer, A. H. M. van Vliet, J. J. E. Bijlsma, M. Kist, S. Bereswill, C. M. J. E. Vandenbroucke-Grauls, and, J. G. Kusters. 2001. Identification of environmental stress-regulated genes in Helicobacter pylori by a lacZ reporter gene fusion system. Helicobacter 6:300-309. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eitinger, T., and M. A. Mandrand-Berthelot. 2000. Nickel transport systems in microorganisms. Arch. Microbiol. 173:1-9. [DOI] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 18.Grass, G., C. Grosse, and, D. Nies. 2000. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia sp. strain CH34. J. Bacteriol. 182:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawtin, P. R., H. T. Delves, and D. G. Newell. 1991. The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol. Lett. 61:51-54. [DOI] [PubMed] [Google Scholar]
- 20.Kuwahara, H., Y. Miyamoto, T. Akaike, T. Kubota, T. Sawa, S. Okamoto, and H. Madea. 2000. Helicobacter pylori urease suppresses bactericidal activity of peroxynitrite via carbon dioxide production. Infect. Immun. 68:4378-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makristathis, A., E. Rokita, A. Labigne, B. Willinger, M. L. Rotter, and A. M. Hirschl. 1998. Highly significant role of Helicobacter pylori urease in phagocytosis and production of oxygen metabolites by human granulocytes. J. Infect. Dis. 177:803-806. [DOI] [PubMed] [Google Scholar]
- 22.Makristathis, A., E. Rokita, E. Pasching, P. Apfalter, B. Willinger, M. L. Rotter, and, A. M. Hirschl. 2001. Urease prevents adherence of Helicobacter pylori to Kato III gastric epithelial cells. J. Infect. Dis. 184:439-445. [DOI] [PubMed] [Google Scholar]
- 23.Mobley, H. L. T. 2001. Urease, p. 179-191. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: Physiology and Genetics. ASM Press, Washington, D.C. [PubMed]
- 24.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokita, E., A. Makristathis, E. Presterl, M. L. Rotter, and A. M. Hirschl. 1998. Helicobacter pylori urease significantly reduces opsonization by human complement. J. Infect. Dis. 178:1521-1525. [DOI] [PubMed] [Google Scholar]
- 27.Sachs, G., D. R. Scott, D. L. Weeks, M. Rektorscheck, and K. Melchers. 2001. Regulation of urease for acid habitation, p. 277-283. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Shirai, M., R. Fujinaga, J. K. Akada, and T. Nakazawa. 1999. Activation of Helicobacter pylori ureA promoter by a hybrid Escherichia coli-H. pylori rpoD gene in E. coli. Gene 239:351-359. [DOI] [PubMed] [Google Scholar]
- 30.Smoot, D. T., H. L. Mobley, G. R. Chippendale, J. F. Lewison, and J. H. Resau. 1990. Helicobacter pylori urease activity is toxic to human gastric epithelial cells. Infect. Immun. 58:1992-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibazarwa, C., S. Wuertz, M. Mergeay, L. Wyns, and D. van Der Lelie. 2000. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J. Bacteriol. 182:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karpk, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vliet, A. H. M., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for the determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]
- 36.Weeks, D. L., S. Eskandari, D. R. Scott, and, G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 37.Wirth, H. P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16:994-996. [PubMed] [Google Scholar]