Abstract
Group B streptococci (GBS) are a major cause of pneumonia, sepsis, and meningitis in newborns and infants. GBS initiate infection of the lung by colonizing mucosal surfaces of the respiratory tract; adherence of the bacteria to host cells is presumed to be the initial step in and prerequisite for successful colonization (G. S. Tamura, J. M. Kuypers, S. Smith, H. Raff, and C. E. Rubens, Infect. Immun. 62:2450-2458, 1994). We have performed a genome-wide screen to identify novel genes of GBS that mediate adherence to fibronectin. A shotgun phage display library was constructed from chromosomal DNA of a serotype Ia GBS strain and affinity selected on immobilized fibronectin. DNA sequence analysis of different clones identified 19 genes with homology to known bacterial adhesin genes, virulence genes, genes involved in transport or metabolic processes, and genes with yet-unknown function. One of the isolated phagemid clones showed significant homology to the gene (scpB) for the GBS C5a peptidase, a surface-associated serine protease that specifically cleaves the complement component C5a, a chemotaxin for polymorphonuclear leukocytes. In this work we have demonstrated that affinity-purified recombinant ScpB and a peptide ScpB fragment (ScpB-PDF), similar to the peptide identified in the phagemid, bound fibronectin in a concentration-dependent manner. Adherence assays to fibronectin were performed, comparing an isogenic scpB mutant to the wild-type strain. Approximately 50% less binding was observed with the mutant than with the wild-type strain. The mutant phenotype could be fully restored by in trans complementation of the mutant with the cloned wild-type scpB gene, providing further evidence for the role of ScpB in fibronectin adherence. Our results suggest that C5a peptidase is a bifunctional protein, which enzymatically cleaves C5a and mediates adherence to fibronectin. Since binding of fibronectin has been implicated in attachment and invasion of eukaryotic cells by streptococci, our results may imply a second important role for this surface protein in the pathogenesis of GBS infections.
Group B streptococci (GBS) are important human pathogens which are responsible for a broad range of diseases in human newborns (2) and immunocompromised hosts (33). Infections in newborns occur in the first 6 days of life (early-onset disease) or in the first weeks (weeks 1 to 12) of life (late-onset disease). The pathogenesis of GBS infections is a multifactorial process that includes the ability to adhere, colonize, and invade epithelial and endothelial cells and then replicate and evade host immune defenses (29, 41). Adherence is thought to be the initiating step for entry into host cells, promoting invasion of deeper tissues and ultimate dissemination of the bacteria to the bloodstream and multiple organ systems (42).
The process of colonization involves microbial and host receptor-ligand interactions. Many streptococci express proteins that bind specifically to proteins of the extracellular matrix (ECM) and/or serum. The ECM surrounds cells in connective tissue such as skin, bones, and cartilage. It is heterogeneous, and various ECM proteins, including fibronectin (Fn), fibrinogen, laminin, collagen, and integrins are utilized as adhesin receptors by several different pathogens (47). The most widely described interaction is the binding to Fn, which is a large dimeric glycoprotein present in the ECM in a fibrillar form. Fn contains repeats of a characteristic amino acid triplet sequence, arginine-glycine-aspartic acid [RGD]), which can function as the binding site for some bacteria (39). Fn is also one of the major adherence targets for group A streptococci (GAS) (35). Adhesins include protein F/SfbI (13, 40), serum opacity factor (18), and Fn binding protein FBP54 (10). Fn binding proteins, such as SfbI from Streptococcus pyogenes, have also been demonstrated to act as invasins (26). Furthermore, SfbI induces a protective immune response against S. pyogenes in the sera and lungs of mice after intranasal vaccination (12), emphasizing the significance of Fn binding for pathogenesis of streptococcal infections.
Unlike GAS, Streptococcus dysgalactiae, and Staphylococcus aureus, GBS do not bind Fn in its soluble form (43), suggesting that Fn binding of GBS is a low-avidity interaction that requires multiple GBS adhesins (29). So far the GBS surface structure(s) that mediates binding to Fn has not been identified.
To identify Fn adhesins of pathogenic GBS, we have employed a shotgun phage display approach to screen a GBS genomic library for sequences which encode peptides that bind to solid-phase Fn. Using this screen, we have identified gene fragments from GBS with homology to other gram-positive adhesins and virulence genes, confirming the feasibility of our approach. One of the isolated Fn binding clones revealed homology to the C5a peptidase (ScpB), a highly specific endopeptidase found in GAS (48), GBS (15), and group G streptococci (GGS) (8). It cleaves and inactivates C5a, a component of the human complement system, thereby contributing to the ability of GBS to evade phagocytosis (3).
Based on the identification of a C5a peptidase-related peptide, we hypothesized that this protein is bifunctional, serving as a protease and mediating binding to Fn. In this report we demonstrate that GBS C5a peptidase binds to immobilized Fn and serves as a bacterial adhesin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. A derivative of phagemid vector pHEN1 (16), pG3H6, which contains a six-histidine tag, a c-Myc tag, and a SmaI site for blunt-end cloning, was kindly provided by L. Frykberg (Swedish University of Agricultural Sciences, Uppsala, Sweden). Helper phage VCSM13 was purchased from Stratagene. Escherichia coli BL21(DE3) was used for expression and purification of recombinant ScpB (rScpB) and rScpB-PDF in expression vector pGEX-4T3. E. coli XL-1 Blue was used as a recipient for cloning the phagemid library. Allelic replacement experiments with the temperature-sensitive vector pVE6007 were performed with E. coli DH5α. E. coli MC1061 was used as the host strain for shuttle vector pDC123.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, phenotypea, or sequence | Reference or source |
|---|---|---|
| S. agalactiae strains | ||
| A909 | Wild type, serotype Ia | 23 |
| COH1 | Wild type, serotype III, Tetr | 25 |
| 78-471 | Wild type, serotype II | 7 |
| TOH97 | COH1, scpB::Ω-Kan2, Tetr, Kanr | This study |
| BEC971 | TOH97, pBEC101 (scpB+), Tetr, Kanr, Cmr | This study |
| E. coli strains | ||
| XLI-Blue | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) supE44 relA1 λ−lac [F−proAB lacIqZΔM15 Tn10(Tet)] | Stratagene, La Jolla, Calif. |
| BL21(DE3) | F−ompT gal (dcm) (lon) hsdSB(rB− mB−) endA1 hsdR17(rK− mK+) | Novagen |
| DH5α | supE44 thi-1 recA1 gyrA(Nalr) relA1 Δ(lacZYA-argF)169 [φ80dlacΔ(lacZ)M15] | Gibco BRL, Inc. |
| MC1061 | F−araD139 Δ(araABC-leu)7696 Δ(lac)X74 galU galK hsdR2(rK− mK+) mcrB1 rpsL(Strr) | 46 |
| Plasmids | ||
| pGEX-4T3 | Apr, expression vector, 4.9 kb | Amersham Pharmacia |
| pG3H6 | Apr, phagemid vector, 4.5 kb | 17 |
| pVE6007 | ori (Ts), Cmr, temperature-sensitive shuttle vector, 3.4 kb | 24 |
| pDC123 | phoZ, Cmr, shuttle-expression vector with blue-white | 5 |
| pT7Blue | Apr, 2.9 kb, T-overhang PCR cloning vector | Novagen |
| pCIV2 | Ω-kan2, Kanr, 5.9 kb | 30 |
| pBEC101 | pDC123 with scpB, Cmr | This study |
| Primers | ||
| CBP1_pG3H6 | 5′-GTT ATT ACT CGC GGC CCA-3′ | This study |
| CBP2_pG3H6 | 5′-GGC CCC ATT CAG ATC CTC-3′ | This study |
| CBP3_pG3H6 | 5′-CAG CTG CAG CAC CAC CAC-3′ | This study |
| CBP4_pG3H6 | 5′-GCC CGT TTG ATC TCG AGG T-3′ | This study |
| CBP6_C5a | 5′-AGC TAC TAA TCC CAA GAA G-3′ | This study |
| CBP17_C5a | 5′-ACC AGG AGA GAA TCG TTT G-3′ | This study |
| CBP19_C5a | 5′-CAT GAA AGG ATC CGA CAC ATT GCG-3′ | This study |
| CBP20_C5a | 5′-GAT CGT TTC TCT CGA GAG TAC GAG-3′ | This study |
| CBP25_C5a | 5′-GAT GGA TCC GTC AAA ACC CTG CAG-3′ | This study |
| CBP26_C5a | 5′-GTT CTC GAG ACA CGC ATC AAA AGC A-3′ | This study |
Kanr, kanamycin resistance; Tetr, tetracycline resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
Streptococcal strains were grown in Todd-Hewitt broth at 37°C with aeration, on Todd-Hewitt agar (THA) plates, or on blood agar plates at 37°C; mutant TOH97 was grown with the addition of 1 mg of kanamycin (KAN) per ml. E. coli strains were cultured in 2× YT medium supplemented with 100 μg of ampicillin (AMP) per ml (XL-1 Blue/pG3H6 and BL21/pGEX-4T3) or on Luria-Bertani (LB) agar (DH5α) with 10 μg of chloramphenicol (CHL) per ml (MC1061/pDC123) and 50 μg of XP (5-bromo-4-chloro-3-indolylphosphate) (Sigma) per ml for blue-white selection.
Construction of scpB mutant TOH97.
For allelic replacement mutagenesis of scpB, the broad-host-range vector pVE6007, which mediates CHL resistance and replicates at 30°C but not at 37°C, was used. A 2.5-kb fragment of scpB was amplified from chromosomal DNA of GBS strain 78-471 (7) and cloned directly into pT7Blue (Novagen). The resultant plasmid was digested with KpnI and HindIII, and the scpB-containing fragment was cloned into pVE6007 digested with the same enzymes. A unique BglII site in scpB was used to insert the 2.3-kb BamHI fragment of pCIV2 containing Ω-kan2 (30). The omega-KAN cassette terminates translation of ScpB at Glu-154. The resulting construct, pTH14, was transformed into DH5α and grown on selective LB agar plates containing CHL (5 μg/ml) and KAN (40 μg/ml) at 30°C. Plasmid DNA prepared from E. coli DH5α was then transformed into GBS strain COH1, and transformants were selected by plating on THA plus CHL (10 μg/ml) and growing at 30°C. Allelic exchange mutagenesis was carried out as described by Yim and Rubens (50).
Complementation of scpB mutant TOH97.
C5a peptidase was PCR amplified with the Long Template Expand PCR system (Boehringer Mannheim) by using the primers CBP6_C5a and CBP17_C5a. The PCR product contained the ribosomal binding site and the cell wall-anchoring LPTTND motif inserted into a T-vector derivative of pDC123 (5). The ligation was used to transform E. coli MC1061 and selected on solid medium containing 10 μg of CHL per ml and 50 μg of XP per ml to monitor alkaline phosphatase activity. The resultant complementation vector, pBEC101, was sequenced, and the integrity and orientation of the insert were confirmed. Three micrograms of pBEC101 was transformed into competent scpB mutant TOH97. Competent GBS were derived by the method of Framson et al. (11). Transformants, designated BEC97, were grown on THA containing 10 μg of CHL per ml and 1 mg of KAN per ml.
Phenotypic assays.
The quantification of type III capsule expression in COH1 and the mutant TOH97 was analyzed as described by Chaffin et al. (6). Beta-hemolysin and CAMP factor analysis was performed as described by Nizet et al. (28).
Construction of phagemid library.
Genomic DNA was isolated from Fn binding Streptococcus agalactiae strain A909 as described previously (19). The DNA was sonicated, and fragments of between 100 and 1,000 bp were isolated by preparative gel electrophoresis, treated with T4 DNA polymerase to generate blunt ends, and ligated into the SmaI site of the phagemid vector pG3H6. AMP-resistant transformants were harvested, grown in liquid culture (2× YT) at 37°C, and, in the logarithmic growth phase (optical density at 600 nm of 0.6), superinfected with 2.5 × 109 of PFU helper phage VCSM13. After 1 h of incubation at 37°C, the culture was pelleted and resuspended in 2× YT containing 100 μg of AMP per ml, 10 μg of tetracycline per ml, 50 μg of KAN per ml, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Induction of expression continued overnight at 30°C (phage rescue).
Phage particles were isolated from the supernatants of the expression cultures by polyethylene glycol precipitation. The phage titer was determined by reinfection of log-phase E. coli XL1-Blue cells and plating on LB agar containing 100 μg of AMP per ml as described by Scott and Smith (34).
Panning of the phagemid library.
Seven wells of a 96-well microtiter plate were coated overnight at 4°C with highly purified human plasma Fn (Sigma) at a concentration of 5 μg/well in phosphate-buffered saline (PBS) (150 mM NaCl, 10 mM sodium phosphate, pH 7.2) or with 5% nonfat dry milk in PBS as a negative control. Each well was blocked with 200 μl of 5% nonfat dry milk per ml in PBS for 1 h at room temperature and washed with 200 μl of PBS plus 0.05% Tween. Fifty microliters of the streptococcal phage display library, containing 5 × 1010 phagemid particles, was added to each well and incubated for 2 to 3 h at room temperature. After intensive washing of the wells with PBS-0.05% Tween, the bound phage was used to directly infect cells of E. coli XL1-Blue in the microtiter plate. Therefore, 100 μl of log-phase E. coli XL1-Blue cells per well was added to the wells and incubated for 1 h at 37°C. Ten microliters from each well was used to determine the titer of the bound phage particles by 10-fold dilutions and plating on LB agar with 100 μg of AMP per ml. The remaining 90 μl/well was plated on LB agar plus AMP and used for an additional cycle of phage rescue and panning on Fn.
Sequencing of displayed inserts.
Individual clones were isolated after each panning cycle, and the insert DNA was amplified by standard PCR. Primers used for amplifying the insert hybridized on the plasmid pG3H6 to a region at the 5′ end of the insert (CBP1_pG3H6) and at the 3′ end of the insert (CBP2_pG3H6). The obtained PCR products were purified with the Qiagen PCR purification kit, and the DNA was sequenced from both ends by using the Big Dye terminator sequencing kit (PE Biosystems, Foster City, Calif.) with the primers CBP3_pG3H6 and CBP4_pG3H6. Analysis of the DNA and protein sequences was done with the BLAST (Basic Local Alignment Search Tool) programs of the National Center for Biotechnology Information, National Institutes of Health (1), to compare the GBS sequences with those in the GenBank database.
Construction and purification of fusion proteins.
The scpB gene and the scpB gene fragment (scpB-PDF) were amplified by PCR from chromosomal DNA of GBS strain COH1. Primers CBP19_C5a and CBP20_C5a contained BamHI and XhoI restriction sites for cloning of scpB into the expression vector pGEX-4T3. Primers CBP25_C5a and CBP26_C5a were used to amplify scpB-PDF and also contained BamHI and XhoI restriction sites. Amplified products were purified using the Qiagen PCR purification kit, digested with BamHI and XhoI, and ligated separately into the BamHI- and XhoI-digested expression vector pGEX-4T3. The ligated DNA was transformed into E. coli BL21(DE3). Recombinant clones were analyzed by restriction enzyme analysis. The rScpB used in this study contains amino acids (aa) 1 to 1090 of the protein and is missing the cell wall anchor. rScpB-PDF consists of the 112 aa (aa 116 to aa 227) found in the phage display screen. Production and purification of the two glutathione S-transferase (GST) fusion proteins from E. coli were performed according to the instructions of the manufacturer (Amersham Pharmacia), and they were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%) followed by Western blot as described by Laemmli (21). The recombinant fusion proteins were detected with a goat anti-GST antibody (Amersham Pharmacia) followed by a horseradish peroxidase (HRP)-conjugated rabbit anti-goat immunoglobulin G (heavy plus light chains) antibody (Pierce). GST protein alone was prepared as a control.
Binding of recombinant proteins to Fn.
The binding activities of purified rScpB and rScpB-PDF to immobilized Fn were measured by enzyme-linked immunosorbent assay (ELISA). Fn (2 μg/ml) was applied at 4°C overnight to 96-well U-bottom microtiter plates. After blocking with 5% BSA for 2 h, purified rScpB, rScpB-PDF, or rGST at various concentrations was added to the wells and incubated at 37°C for 2 h, and the wells were washed with PBS. A goat anti-GST antibody was added and allowed to bind for 1 h at room temperature. The wells were incubated with a secondary HRP-conjugated rabbit anti-goat immunoglobulin G antibody for 1 h at room temperature. The plate was washed three times with PBS and developed by adding substrate solution (ortho-phenyldiamine) (Sigma) according to the instructions of the supplier. After 15 min the reaction was stopped by addition of 10% sulfuric acid, and the color development was measured at 490 nm with an ELISA plate reader (Dynatech MR5000).
Radiolabeling of bacteria and adherence assays.
GBS were grown to log phase and labeled with l-[4,5-3H]leucine (Amersham Pharmacia) as described previously (49).
The adherence assays with radiolabeled GBS were performed as described by Tamura and Rubens (43) with the following modifications: Fn (10 μg/ml) was applied to U-bottom microtiter plates (Falcon) overnight, and the bacteria remained on the plates for 2 h at 4°C after a centrifugation step. All determinations were performed in triplicate. To calculate the percent adherence of bacteria to Fn, the output counts per minute minus background (adherence to uncoated wells) were divided by the input counts per minute minus the background and multiplied by 100. CFU per milliliter were determined for each bacterium by plating dilutions of the radiolabeled bacteria, and the ratio of CFU to counts per minute was determined to verify comparable uptake of label for the wild-type and mutant strains in each experiment.
RESULTS
Derivation of a GBS genomic phage display library.
The expression of proteins as fusions with phage envelope proteins, called phage display, has been shown to be a powerful approach to identify and isolate peptides with specific binding properties from genomic libraries (17, 22, 27, 45, 51). To generate a GBS genomic phage display library, we isolated chromosomal DNA from the GBS type Ia strain A909 and then sonicated the DNA into 100- to 1,000-bp fragments. These fragments were cloned into the phagemid vector pG3H6 5′ to the gene encoding the phage coat protein g3p (gene 3 protein). Since sonication leads to random fragmentation of DNA, in contrast to restriction enzyme digestion, it ensures the generation of an almost complete representation of the genome within the library. DNA fragments which, after insertion, create a translational fusion with the g3p lead to the production of chimeric proteins that are presented on the capsid of the resulting phage.
To ensure that the library displays all potential reading frames, a particular GBS sequence has to be present multiple times in different lengths and fused in different reading frames. Our library contained 4 × 106 individual clones with inserts ranging from 100 to 1.000 bp. Given that GBS has an estimated genome size of 2.2 Mb, the library represents the GBS genome at least 200 times.
Affinity selection for clones with Fn binding activity.
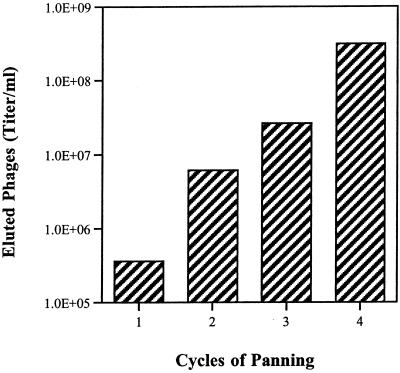
To identify phage expressing Fn binding peptides, we affinity screened the phage library by panning on Fn-coated plates. Bound phage were subsequently enriched by several cycles of binding and elution; in each cycle 3.5 × 1011 phagemid particles were used to ensure that several copies of each specific phage in the library were present. The phage titer increased with each cycle of binding, which is indicative of a specific enrichment. Four cycles of panning on Fn-coated plates resulted in an approximately 1,000-fold enrichment of eluted phage (Fig. 1). As a negative control, the phage library was also panned against 5% nonfat dry milk in PBS; 1,000-fold less binding was observed with this control experiment (data not shown), confirming that enrichment of the library for Fn binding phage was specific.
FIG. 1.
Enrichment of the phage display library as a consequence of subsequent cycles of Fn binding (panning). Enrichment is indicated by the amplification of the number of bound phage with each panning, which also indicates specific binding. Peptide-carrying phages have been enriched by a factor of 10 to 100 in each cycle.
Identification of Fn binding phage.
To identify the genes within specific phagemid clones encoding Fn binding activity, clones from each cycle of panning were isolated, and the genomic inserts were amplified by PCR and sequenced. A group of 23 clones did not display any homology to sequences in the public databases, including the database of unfinished microbial genomes. Homology to 19 known genes in the public databases was observed, and 13 of them had homology to putative or hypothetical genes (Table 2). Several genes were similar to those from other gram-positive bacteria, such as streptococci (mainly S. pyogenes) and staphylococci. Interestingly, a gene fragment with low homology to a region of a Fn binding protein (SFS) from Streptococcus equi (22) was identified, and it is listed in Table 2 because of its known Fn binding activity. DNA from clone CB 3-25 displayed high homology (98%) to a glutamine permease (GlnP) from GBS. glnP is transcribed in an operon with the glutamine transport gene glnQ, which is required for maximum Fn binding in GBS (unpublished data). Since studies with Campylobacter jejuni revealed that a GlnP homolog acts as an adhesin (31), these data provide additional evidence that the GBS GlnP binds Fn.
TABLE 2.
Sequence analysis of GBS DNA isolated from phagemids encoding Fn binding fusion proteins
| Homology | Clonea | Homologous proteinb | % Amino acid identity (homology)/spanc |
|---|---|---|---|
| Genes with known function | CB 5-8 | DNA-directed RNA polymerase subunit (S. pyogenes M1) | 100 (33)/33 |
| CB 3-14 | C5a peptidase (S. agalactiae) | 98 (110)/112 | |
| CB 3-25 | GlnP (S. agalactiae) | 98 (91)/92 | |
| CB 2-12 | Phosphoglycerate kinase (S. pyogenes M1) | 96 (54)/56 | |
| CB 4-12 | ABC transporter (S. pyogenes M1) | 87 (64)/73 | |
| CB 5-6 | Fn binding protein (S. equi) | 48 (13)/27 | |
| Genes with hypothetical or putative function | CB 3-7 | Putative phosphoglucomutase (S. pyogenes M1) | 100 (49)/49 |
| CB 3-23 | Putative pyruvate formate lyase 2 (S. pyogenes M1) | 89 (42)/47 | |
| CB 2-36 | Putative CTP-synthetase (S. pyogenes M1) | 79 (142)/178 | |
| CB 2-14 | Conserved hypothetical protein (S. pyogenes M1) | 76 (130)/170 | |
| CB 3-5 | Putative aminodeoxychorismate lyase (S. pyogenes M1) | 76 (38)/50 | |
| CB 5-3 | Putative transcription factor (S. pyogenes M1) | 59 (42)/71 | |
| CB 3-2 | Putative phosphotransferase system, enzyme II, A component (S. pyogenes M1) | 56 (75)/132 | |
| CB 5-9 | Conserved hypothetical protein (S. pyogenes M1) | 52 (46)/87 | |
| CB 2-30 | ORFID (SA2098 hypothetical protein similar to glycerate dehydrogenase) (S. aureus) | 50 (56)/111 | |
| CB 2-38 | Putative integral membrane protein (S. pyogenes M1) | 49 (39)/79 | |
| CB 3-21 | Probable tail protein (Streptococcus phage φ01205) | 46 (62)/132 | |
| CB 2-34 | Putative C3-degrading proteinase (S. pyogenes M1) | 41 (55)/132 | |
| CB 3-16 | Hypothetical protein (S. pyogenes M1) | 40 (44)/109 |
The clone number indicates the number of phage rescued and the number of the clone.
Homology scores with a sum probability of <e−10 were considered significant. Homology scores of >e−10 were classified as having no significant database match. Only one individual mutant of particular interest with low homology is listed (CB 5-6).
The span is the length of the region, in amino acids, to which percent similarity and identity refer. Twenty-three clones showed no homology to any sequence present in the public database or to any unfinished genome sequence and are not listed.
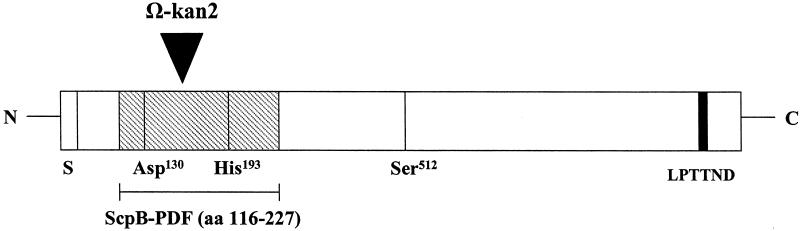
One of the more interesting clones contained DNA with significant homology (98% identity over 112 aa) to the GBS C5a peptidase gene, scpB. The phagemid insert encoding the putative Fn binding fragment (ScpB-PDF) (Fig. 2) lies in the N terminus of the protein (aa 116 to 227) and includes two of the active residues of this serine protease catalytic site, aspartic acid 130 and histidine 193.
FIG. 2.
Schematic presentation of ScpB. Shaded area, Fn binding fragment (ScpB-PDF, aa 116 to 227); black area, cell wall anchor motif LPTTND at the C terminus; S, signal sequence; Asp-130, His-193, and Ser-512, amino acids in the serine protease active site. The insertion site of the Ω-kan2 cassette for construction of the mutant TOH97 is indicated as a large arrowhead above the protein.
Binding of recombinant C5a peptidase and peptides to Fn.
C5a peptidase (Scp) is a surface-associated peptidase produced by GAS, GBS, and GGS, which cleaves complement chemotaxin C5a and is thought to play an important role in immune evasion by attenuating recruitment of polymorphonuclear leukocytes to the site of infection (15). scpB from GBS is expressed by representative strains from all nine serotypes (15). The identification of a peptide from ScpB which bound Fn, as expressed in the GBS phage display library, led to our hypothesis that ScpB may also serve as a bacterial adhesin.
We further characterized the role of ScpB in Fn binding by using the type III strain COH1 rather than A909 (type Ia), from which the phage display library was created, since the adherence of COH1 to Fn had been fully characterized previously (43) and GBS strain COH1 was also shown to express ScpB (4). The DNA sequence of the COH1 scpB gene fragment is identical to the one from A909 (data not shown), and an insertion mutation in COH1 scpB had already been constructed (see below).
To generate rScpB and rScpB-PDF encoded by the DNA insert of the isolated phagemid clone, chromosomal COH1 DNA was amplified by PCR and cloned into the pGEX-4T3 expression vector. These plasmid constructs created GST fusion proteins allowing expression and affinity purification of each protein with GST-Sepharose. The expected molecular weight and purity of each recombinant protein were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the nature of the fusion proteins was confirmed by Western blotting using an anti-GST antibody (data not shown).
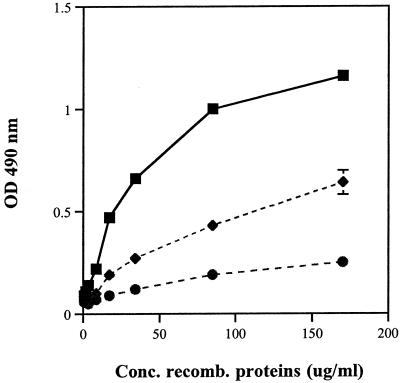
Increasing concentrations of the purified fusion proteins were incubated on ELISA plates containing 2 μg of immobilized Fn per ml, and the binding characteristics were quantified using anti-GST antibody (Fig. 3). Purified rGST proteins served as a negative control and bound poorly in comparison to the fusion proteins rScpB and rScpB-PDF. The recombinant proteins bound to Fn in a concentration-dependent manner, although adherence of rScpB-PDF was 50% lower than that of rScpB. These results confirmed the observations from the phage display library experiments, specifically, that the peptide spanning the region of ScpB from aa 116 to 227 contains an Fn binding domain. We next sought to confirm that ScpB could mediate adherence of GBS bacterial cells to immobilized Fn.
FIG. 3.
Binding of purified rScpB (▪) and rScpB-PDF (⧫) fusion proteins and the control rGST protein (•) to immobilized Fn. Fn-coated microtiter plates were blocked with 5% BSA in PBS, and various concentrations of recombinant proteins were allowed to bind for 2 h. Detection of bound proteins was performed with an anti-GST antibody and a secondary HRP-conjugated anti-goat IgG antibody (see Materials and Methods). The plates were developed with ortho-phenyldiamine, and color development was analyzed by optical density (OD) at 490 nm. Points represent means of triplicates, and standard deviations are indicated. Recombinant proteins bound in a concentration-dependent manner.
Generation and characterization of an scpB isogenic mutant.
We hypothesized that ScpB would mediate adherence of GBS to immobilized Fn. To address this hypothesis, we utilized an isogenic mutant of COH1, designated TOH97. This mutant was made by allelic replacement mutagenesis utilizing the Ω-kan2 cassette, which was inserted within the scpB gene at Glu-154. This insertion truncates the amino-terminal end and terminates transcription distal to the Ω-kan2 insertion site, including the region encoding the Fn binding peptide described above. Southern blot analysis with COH1 genomic DNA and PCR confirmed that the wild-type gene indeed had been replaced in TOH97 by scpB::Ω-kan2 (data not shown).
The absence of C5a peptidase expression by TOH97 was verified by Western blot analysis of mutanolysin surface extracts with C5a peptidase antiserum (monoclonal antibody F1) (kindly provided by J. F. Bohnsack, University of Utah), confirming the interruption of the C5a peptidase reading frame (data not shown). The Ω-kan2 cassette contains a transcriptional terminator preventing transcription of downstream genes. Sequence analysis using the Genetics Computer Group software package revealed a putative terminator at the 3′ end of scpB. The lmb gene, which codes for the laminin binding protein, is positioned downstream of scpB and is under the control of its own promoter (36). Therefore, it was unlikely that insertion of the Ω-kan2 cassette affects Lmb expression. Furthermore, serotype III strain COH1 has been shown not to bind to laminin (43).
Further phenotypic analysis of the mutation showed no effect on bacterial growth in Todd-Hewitt broth, RPMI plus 5% Casamino Acids, or plasma. Colony morphology and expression of β-hemolysin, CAMP factor, hippuricase, hyaluronidase, and caseinase were indistinguishable from those of the wild type, as were the amounts of type III capsule (6).
Analysis of Fn binding of the mutant.
To address whether C5a peptidase mediates Fn binding of GBS, we compared the binding of COH1 and TOH97 as described previously (43). All experiments were performed at 4°C to prevent growth of the bacteria and to reduce metabolic activity, since bacterial metabolic enzymes such as proteases can influence adherence.
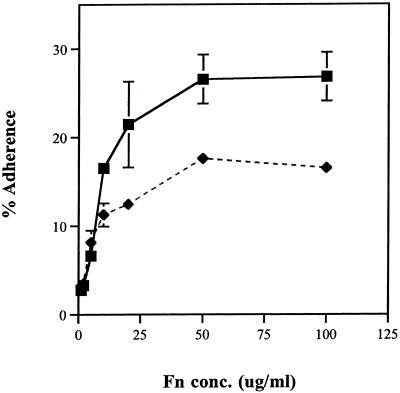
In agreement with our previous results (43), the wild-type strain COH1 adheres to Fn at 30 to 50% of the input inoculum for Fn concentrations of 20 to 100 μg/ml (Fig. 4). Coating wells with Fn concentrations of >20 μg/ml did not increase the binding significantly, whereas adherence dropped to 5 to 10% at a concentration of 2 μg/ml. The ScpB− mutant TOH97 showed a 50% decrease in adherence compared to COH1. Both COH1 and TOH97 showed less than 0.01% adherence to either uncoated wells or wells coated with BSA, indicating that the adherence to immobilized Fn was specific. These data suggest that C5a peptidase contributed significantly to the adherence of GBS. However, since binding was not completely abolished, the bacteria may produce other Fn binding adhesins.
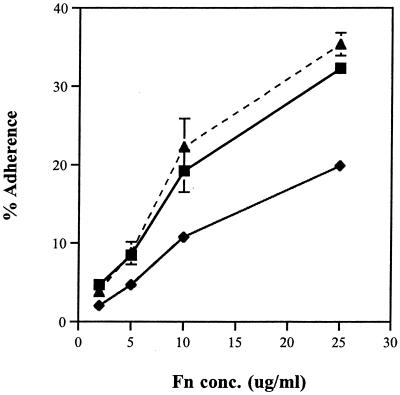
FIG. 4.
Adherence of radiolabeled COH1 and TOH97 to immobilized Fn. Microtiter plates were coated with various concentrations of Fn, and [3H]leucine-labeled COH1 (▪) and TOH97 (⧫) were added. Shown is the percentage of adherent bacteria relative to the input. Points represent means of triplicates, and bars represent the standard deviation. TOH97 adhered to Fn approximately 50% less than wild-type COH1.
Complementation of TOH97 with scpB in trans restores Fn binding.
We hypothesized that cloning of the wild-type scpB gene into the scpB mutant, TOH97, would restore Fn binding. The recombinant plasmid pBEC101 containing the scpB gene, including its ribosomal binding site and the carboxy-terminal cell wall-anchoring LPTTND motif, was transformed into the mutant TOH97. Clones growing on THA plates with CHL and KAN were isolated, and the presence and integrity of the plasmid were confirmed by vector- and insert-specific PCR amplification and restriction enzyme analysis (data not shown). Expression of ScpB by pBEC101 on individual transformants was confirmed by extracting cell surface proteins with mutanolysin and subsequent Western blot analysis with anti-C5a peptidase monoclonal antibody F1. All analyzed transformants containing pBEC101 expressed surface C5a peptidase. Figure 5 shows a representative Fn binding assay comparing one of the complemented clones, BEC971, to wild-type COH1 and the mutant TOH97. As expected, TOH97 demonstrated 50% less binding to Fn than COH1, and BEC971 bound to Fn at wild-type levels. Binding of all clones containing pBEC101 was comparable to or better than that of the wild-type strain.
FIG. 5.
Fn binding of radiolabeled COH1 (▪), TOH97 (⧫), and mutant BEC971 containing the complementation plasmid pBEC101 (▴). Microtiter plates were coated with four different concentrations of Fn, and adherence of radiolabeled bacteria was performed as described in Materials and Methods. Points represent means of triplicates, and standard deviations are indicated. pBEC101 expressed ScpB in the complementing strain BEC971 and restored the binding to immobilized Fn to at least wild-type levels.
These data demonstrate in trans complementation by the recombinant scpB gene, restoring Fn binding, and provide further evidence that ScpB mediates GBS adherence to Fn.
DISCUSSION
Attachment of GBS to host cell receptors is hypothesized to be an important initial step in colonization and subsequent infection. However, the adhesins contributing to the binding of epithelial cell and/or host ECM molecules are poorly understood. To date the only characterized adhesin of S. agalactiae is the laminin binding protein (Lmb) (36), although binding to laminin is not observed with all strains (43).
Some of the host molecules that serve as substrates for bacterial binding have recently been identified. They include cytokeratin 8 (44), laminin (36), and solid-phase (but not soluble) Fn (43).
In this study, we have used genomic peptide libraries displayed on the surface of bacteriophage to isolate Fn binding proteins of GBS. Phage display has been successfully used to isolate and characterize surface proteins of other gram-positive organisms, such as S. aureus (17, 51), S. dysgalactiae (45), S. epidermidis (27), and S. equi (22). One advantage of phage display compared to other types of expression libraries is that the screening process is replaced by panning, which allows fast identification of peptides from a complex library that demonstrate binding to a desired molecule.
We constructed a phage display library of a type Ia strain of GBS and isolated those phages which contained GBS peptides that mediate binding to immobilized Fn (Fig. 1). Some of the isolated peptides shared homology to other gram-positive proteins which have been previously implicated in adherence and virulence (Table 2). However, this approach also picks unrelated molecules in addition to potentially relevant ones. One particular clone, though, showed high homology to the C5a peptidase of S. agalactiae. The scpB gene is 97% identical between GAS and GBS (7), and furthermore, a homolog has been identified in GGS (8). C5a peptidase is a serine protease which cleaves the complement component C5a between His-67 and Lys-68 in the polymorphonuclear leukocyte binding site, destroying its chemoattractant function. Many GBS express the surface-associated serine protease, although it is not functional in all strains (4). Due to its ubiquitous expression, Bohnsack et al. (4) suggested that C5a peptidase might have a second, unknown function. Stafslien and Cleary (37) hypothesized a potential role for C5a peptidase from GAS (ScpA) as an adhesin or invasin, based on its relatively large size and very limited substrate specificity. For example, the Hap serine protease in Haemophilus influenzae has been shown to mediate bacterial adherence to epithelial cells (14, 38), providing a precedent for a serine protease to exhibit more than one function.
Our in vitro binding studies with the recombinant C5a peptidase protein and the peptide fragment expressed as recombinant GST fusion proteins indicated that ScpB does bind to immobilized Fn. However, rScpB-PDF (peptide fragment) did not display the same high binding affinity for Fn as the whole rScpB (Fig. 3). This could be explained by a different protein conformation of the fragment when fused with GST compared to the whole protein. Alternatively, it is also possible that a second binding domain exists, which is absent in the fragment but confers higher affinity on the entire protein.
Using the C5a peptidase mutant TOH97, we confirmed the role of this protease in binding Fn. This nonpolar scpB mutation was shown previously to destroy peptidase activity against C5a (J. F. Bohnsack, personal communication). The location of the mutation within scpB specifically interrupted the predicted Fn binding domain. Here we show that this mutant binds Fn 50% less than the wild-type strain. However, binding in the presence of recombinant ScpB had no effect on Fn binding of the wild-type or mutant strain (data not shown). The mutation had no effect on other phenotypic characteristics known for GBS, including capsular polysaccharide. Complementation of the mutation in trans with the wild-type gene expressed on a plasmid restored Fn binding. Since the scpB mutation did not completely inhibit adherence to Fn, our results also suggest that other bacterial factors may be involved in Fn binding.
Our experiments with truncated C5a peptidase suggest that a peptide domain (aa 116 to 227) within ScpB mediated binding to Fn. This sequence is part of the catalytic domain of scpB, including two of the active residues, Asp-130 and His-193 (Fig. 2). Ser-512 was not found to be part of the Fn binding domain. Sequence analysis of the C5a peptidase revealed that the protease contains two amino acid triplet sequences Arg-Gly-Asp (RGD), one in the N-terminal half and one in the C-terminal half of the protein. This tripeptide has been shown to mediate cell adhesion to many proteins and was originally identified in Fn (32). However, our data show that neither of the RGD motifs is part of the binding peptide identified by phage display. This result is in agreement with a previous report by Tamura and Rubens (43) showing that peptides containing the sequence RGD do not inhibit the binding of GBS to Fn. Therefore, adherence of GBS C5a peptidase to Fn must be mediated by another mechanism which is independent of the RGD motif.
Fn is a large glycoprotein of eukaryotic extracellular matrices and plasma and has been shown to be the binding substrate for a variety of pathogenic bacteria, such as S. pyogenes (9) and S. aureus (20). Respiratory epithelial cells, such as A549, produce Fn on their surface (data not shown), and the lung is one of the main sites of GBS colonization. If Fn is important for the binding of GBS to epithelial cells, the ScpB− mutant TOH97 should display a reduced level of adherence to this cell line. In preliminary experiments TOH97 demonstrated reduced adherence to A549 cells compared to the wild type (data not shown).
The results of a study by Cheng et al. (6) show that recombinant ScpB binds Fn and may mediate invasion of A549 cells in part, confirming our results. Taken together, these observations indicate that GBS adherence to epithelial cells is mediated in part by the interaction of C5a peptidase and Fn.
Based on our observations, we speculate that ScpB may facilitate S. agalactiae persistence and colonization of epithelial surfaces, which is a prerequisite for subsequent disease. Hendrixson and St. Geme (14) have demonstrated that the Hap serine protease in H. influenzae promotes bacterial interaction with epithelial cells and possibly degrades host proteins. Our report and that of Cheng et al. (6) demonstrate a second example of a serine protease showing two functions: in this case, adherence to a host molecule and evasion of the host immune system. We are continuing to characterize the Fn binding domain of ScpB and pursuing the role of this protease in the pathogenesis of GBS infections.
Acknowledgments
This work was supported by a Feodor-Lynen Fellowship from the Alexander von Humboldt Foundation (Germany) to C.B. and by the National Institutes of Health Streptococcal Initiative, grants N01-AI-75326 and AI-30068 to C.E.R.
We thank Patrick Cleary and Qi Cheng for helpful discussions of the results and Anne Clancy for critical review of the manuscript.
Editor: E. I. Tuomanen
REFERENCES
- 1.Altschul, S. F., T. L. Maden, A. A. Schaffer, J. Zhang, Z. Zhang, W. W. Miller, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. J., and M. S. Edwards. 1991. Group B streptococcal infections, p. 820-881. In J. S. Remington and J. O. Klein (ed.), Infectious diseases of the fetus and newborn infant. W. B. Saunders Co., Philadelphia, Pa.
- 3.Bohnsack, J. F., K. W. Mollison, A. M. Buko, J. W. Ashworth, and H. R. Hill. 1991. Group B streptococci inactivate complement component C5a by enzymatic cleavage at the C-terminus. Biochem. J. 273:635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohnsack, J. F., S. Takahashi, L. Hammitt, D. V. Miller, A. A. Aly, and, E. E. Adderson. 2000. Genetic polymorphism of group B streptococcus scpB alters functional activity of a cell-associated peptidase that inactivates C5a. Infect. Immun. 68:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 6.Chaffin, D. O., S. B. Beres, H. H. Yim, and, C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Cheng, Q., D. Stafslien, S. S. Puroshothaman, and P. Cleary. 2002. The group B streptococcal C5a protease is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed]
- 7.Chmouryguina, I., A. Suvorov, P. Ferrieri, and P. P. Cleary. 1996. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 64:2387-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleary, P. P., J. Peterson, C. Chen, and C. Nelson. 1991. Virulent human strains of group G streptococci express a C5a peptidase enzyme similar to that produced by group A streptococci. Infect. Immun. 59:2305-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtney, H. S., I. Ofek, W. A. Simpson, D. L. Hasty, and E. H. Beachey. 1986. Binding of Streptococcus pyogenes to soluble and insoluble fibronectin. Infect. Immun. 53:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtney, H. S., Y. Li, J. B. Dale, and D. L. Hasty. 1994. Cloning, sequencing and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn 917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, C. A., S. R. Talay, G. Molinari, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 13.Hanski, E., and M. G. Caparon. 1992. Introduction of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, into heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrixson, D. R., and J. W. St. Geme III. 1998. The Haemophilus influenza Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 15.Hill, H. R., J. F. Bohnsack, E. Z. Morris, N. H. Augustine, C. J. Parker, P. P. Cleary, and J. T. Wu. 1988. Group B streptococci inhibit the chemotactic activity of the fifth component of complement. J. Immunol. 141:3551-3556. [PubMed] [Google Scholar]
- 16.Hoogenboom, H. R., A. D. Griffith, K. S. Johnson, D. J. Chiswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobsson, K., and L. Frykberg. 1995. Cloning of ligand-binding domains of bacterial receptors by phage display. BioTechniques 18:878-885. [PubMed] [Google Scholar]
- 18.Kreikemeyer, B., S. R. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 19.Kuypers, J. M., L. M. Heggen, and C. E. Rubens. 1989. Molecular analysis of a region of the group B streptococcus chromosome involved in type III capsule expression. Infect. Immun. 57:3058-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuypers, J. M., and R. A. Proctor. 1989. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect. Immun. 57:2306-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lindmark, H., and B. Guss. 1999. SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect. Immun. 67:2383-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madoff, L. C., J. L. Michel, and D. L. Kasper. 1991. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect. Immun. 59:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguin, E., P. Duwaat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, T. R., C. E. Rubens, and C. B. Wilson. 1988. Lung antibacterial defense mechanisms in infant and adult rats: implications for the pathogenesis of group B streptococcal infections in neonatal lung. J. Infect. Dis. 157:91-100. [DOI] [PubMed] [Google Scholar]
- 26.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rhode, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson, M., L. Frykberg, J.-I. Flock, L. Pei, M. Lindberg, and B. Guss. 1998. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nizet, V., R. L. Gibson, E. Y. Chi, P. E. Framson, M. Hulse, and C. E. Rubens. 1996. Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect. Immun. 64:3818-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nizet, V., P. Ferrieri, and, C. E. Rubens. 2000. Molecular pathogenesis of group B streptococcal disease in newborns, p. 180-221. In D. L. Stevens and E. L. Kaplan (ed.). Streptococcal infections, clinical aspects, microbiology and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 30.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 31.Pei, Z., and M. J. Blaser. 1993. PEB1, the major cell binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J. Biol. Chem. 268:18717-18725. [PubMed] [Google Scholar]
- 32.Pierschbacher, M. D., and E. Ruoslahti. 1984. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309:30-33. [DOI] [PubMed] [Google Scholar]
- 33.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott, J., and G. Smith. 1990. Searching for peptide ligands with an epitope library. Science 249:386-390. [DOI] [PubMed] [Google Scholar]
- 35.Simpson, W. A., and E. H. Beachey. 1983. Adherence of group A streptococci to fibronectin on oral epithelial cells. Infect. Immun. 39:275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spellerberg, B., E. Rodzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stafslien, D. K., and P. P. Cleary. 2000. Characterization of the streptococcal C5a peptidase using a C5a-green fluorescent protein fusion protein substrate. J. Bacteriol. 182:3254-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenza IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 39.Sugano, N., H. Tanaka, K. Ito, and S. Murai. 1997. Arg-Gly-Asp (RGD) peptides inhibit Streptococcus mitis to adhere to fibronectin. J. Nihon Univ. Sch. Dent. 39:154-155. [DOI] [PubMed] [Google Scholar]
- 40.Talay, S. R., P. Valentin-Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura, G. S., J. M. Kuypers, S. Smith, H. Raff, and C. E. Rubens. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura, G. S., and C. E. Rubens. 1994. Pathogenesis of group B streptococcal infections. Curr. Opin. Infect. Dis. 7:317-322. [Google Scholar]
- 43.Tamura, G. S., and C. E. Rubens. 1995. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol. Microbiol. 15:581-589. [DOI] [PubMed] [Google Scholar]
- 44.Tamura, G. S., and A. Nittayajarn. 2000. Group B streptococci and other gram-positive cocci bind to cytokeratin 8. Infect. Immun. 68:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasi, J., L. Frykberg, L. E. Carlsson, M. Lindberg, and, B. Guss. 2000. M-like proteins of Streptococcus dysgalactiae. Infect. Immun. 68:294-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 47.Westerlund, B., and T. K. Korhonen. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687-694. [DOI] [PubMed] [Google Scholar]
- 48.Wexler, D. E., R. D. Nelson, and P. P. Cleary. 1983. Human neutrophil chemotactic response to group A streptococci: bacteria-mediated interference with complement-derived factors. Infect. Immun. 39:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winram, S. B., G. S. Tamura, and C. E. Rubens. 1998. In vitro systems for investigating group B streptococcal: host cell and extracellular matrix interaction. Methods Cell Sci. 20:191-201. [Google Scholar]
- 50.Yim, H. H., and C. E. Rubens. 1998. Site-specific homologous recombination mutagenesis in group B streptococci. Methods Cell Sci. 20:13-20. [Google Scholar]
- 51.Zhang, L., K. Jacobsson, J. Vasi, M. Lindberg, and L. Frykberg. 1998. A second IgG-binding protein in Staphylococcus aureus. Microbiology 144:985-991. [DOI] [PubMed] [Google Scholar]