Abstract
GW678248, a novel nonnucleoside reverse transcriptase inhibitor, has been evaluated for anti-human immunodeficiency virus activity in a variety of in vitro assays against laboratory strains and clinical isolates. When GW678248 was tested in combination with approved drugs in the nucleoside and nucleotide reverse transcriptase inhibitor classes or the protease inhibitor class, the antiviral activities were either synergistic or additive. When GW678248 was tested in combination with approved drugs in the nonnucleoside reverse transcriptase inhibitor class, the antiviral activities were either additive or slightly antagonistic. Clinical isolates from antiretroviral drug-experienced patients were selected for evaluation of sensitivity to GW678248 in a recombinant virus assay. Efavirenz (EFV) and nevirapine (NVP) had ≥10-fold increases in their 50% inhibitory concentrations (IC50s) for 85% and 98% of the 55 selected isolates, respectively, whereas GW678248 had a ≥10-fold increase in the IC50 for only 17% of these isolates. Thus, 81 to 83% of the EFV- and/or NVP-resistant viruses from this data set were susceptible to GW678248. Virus populations resistant to GW678248 were selected by in vitro dose-escalating serial passage. Resistant progeny viruses recovered after eight passages had amino acid substitutions V106I, E138K, and P236L in the reverse transcriptase-coding region in one passage series and amino acid substitutions K102E, V106A, and P236L in a second passage series.
Highly active antiretroviral therapy (HAART) combination therapeutic regimens have dramatically decreased the morbidity and increased the life expectancy of patients infected with human immunodeficiency virus (HIV). Nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) have become important components of combination therapies and have been shown to be effective at decreasing plasma viremia, are well tolerated, and may reduce the pill burden compared with that of other regimens (21). Unlike the nucleoside reverse transcriptase inhibitors (NRTIs), the NNRTIs do not require anabolism to the active triphosphate for activity and bind in a region of the HIV RT that is away from the catalytic site (8). Although very effective, the current NNRTIs have been described as having a “low genetic barrier” to resistance; i.e., the presence of one or two key mutations produces resistance, and the rate of cross-resistance to other NNRTIs is high (7, 10). Despite the high potential for resistance, NNRTIs are used extensively in first-line combination therapies. HAART regimens that combine the NNRTI efavirenz (EFV) with two NRTIs have been shown to be more effective than similar regimens that combine the protease inhibitor indinavir (IDV) with the same two NRTIs (26).
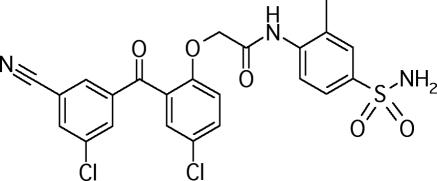
Recently, we have described the activities of a new class of NNRTIs, the benzophenones (4). Members of the benzophenone series showed low-nanomolar potencies against wild-type (WT) HIV type 1 (HIV-1) and a wide spectrum of NNRTI-resistant HIV strains, including strains containing Y181C and K103N mutations, which are often found in patients failing NNRTI-based antiviral therapy. From an extensive study of the benzophenone structure-activity relationship, we selected GW678248 (Fig. 1) for further development (K. R. Romines, G. A. Freeman, L. T. Schaller, J. R. Cowan, S. S. Gonzales, J. H. Tidwell, C. W. Andrews III, R. D. K. Stammers, R. J. Hazen, R. G. Ferris, S. A. Short, J. H. Chan, and L. R. Boone, submitted for publication). The initial biochemical and antiviral characterization of GW678248 indicates that this compound is a potent inhibitor of HIV RT, is active against a wide variety of NNRTI-resistant mutant strains, is only modestly affected by human serum proteins, and has a high selectivity index (8a).
FIG. 1.
Structure of GW678248.
This report describes the additional anti-HIV activities of GW678248, a novel benzophenone NNRTI. In this study, the activities of GW678248 in combination with approved anti-HIV agents, the sensitivities of 55 clinical isolates from NNRTI-experienced patients, and the genotypic and phenotypic patterns of resistant viruses selected during serial passage in the presence of GW678248 are reported.
(The results of this study were presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, 14 to 17 September 2003, Chicago, Ill.)
MATERIALS AND METHODS
Compounds.
GW678248, zidovudine (AZT), abacavir (ABC), stavudine (d4T), dideoxycytidine (ddC), didanosine (ddI), nevirapine (NVP), delavirdine (DLV), lamivudine (3TC), and amprenavir (APV) were synthesized by GlaxoSmithKline, Research Triangle Park, NC. The nucleotide prodrug tenofovir disoproxil fumarate (TDF) was purchased from the pharmacy; and the active drug substance, (R)-9-(2-phosphonylmethoxypropyl)-adenine (tenofovir [TFV]), was isolated in the Medicinal Chemistry Department, GlaxoSmithKline. The marketed protease inhibitors IDV, lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV) were obtained from the Medicinal Chemistry Department at GlaxoSmithKline.
Cell lines and primary cell cultures.
MT-4 cells, a human T-cell leukemia virus type 1 (HTLV-1)-transformed human T-cell line (19), were maintained as described previously (5). HeLa-CD4-LTR β-gal cells (15) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS) and under the selective pressure of Geneticin and hygromycin B. Normal donor peripheral blood mononuclear cells (PBMCs) were isolated from random buffy coats (35 to 40 ml of elutriated whole blood in anticoagulant from HIV-negative donors) received from the American Red Cross Carolina Division. PBMCs were isolated by density gradient centrifugation over Lymphocyte Separation Medium (catalog no. 50494/36427; Organon Teknika, Durham, NC) and stimulated by the addition of 1.25 mg phytohemagglutinin (PHA; catalog no. 34490; Amersham Biosciences, Piscataway, NJ) for 24 to 48 h (6).
Virus strains.
HIV-1 strain IIIB was derived from cell-free supernatants of cultures of the chronically infected cell line H93B (H9/HTLV-IIIB). HIV-1 strain HXB2 was derived from the molecular clone pHXB2-D (9). HIV-2 strain NIH-ZY-M, WT virus, was obtained from D. Zagury.
Construction of isogenic HIV-1 mutant virus for passage and virus sensitivity testing.
A panel of isogenic viruses that possessed mutations of interest in the RT-coding region was prepared from HIV-1 strain HXB2. Specific amino acid substitutions in the HXB2 RT were generated by site-directed mutagenesis of the RT DNA carried by plasmid pRT2. The codon changes, which yielded K103N, Y181C, V106A, V106I, P236L, and V106I-P236L mutations, were verified by nucleoside sequence determination of the entire RT-coding region on both DNA strands. Isogenic recombinant viruses were recovered following cotransfection of MT-4 cells with linearized, mutant RT plasmids and molecular clone HXB2ΔRTBstII from which RT was deleted (14). The recombinant progeny virus was expanded in MT-4 cells and harvested, the titer was determined, and the sequence was verified.
MT-4 cell assay.
Anti-HIV activity and compound-induced cytotoxicity were measured in parallel by means of a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS)-based procedure in MT-4 cells. Aliquots of the test compounds were serially diluted in medium (RPMI 1640, 10% [vol/vol] FBS, and 10 μg/ml gentamicin) in 96-well plates. Exponentially growing MT-4 cells were harvested and centrifuged at 192 × g for 10 min in a Jouan centrifuge. Cell pellets were resuspended in fresh medium (RPMI 1640, 20% [vol/vol] FBS, 20% [vol/vol] interleukin-2 [IL-2], and 10 μg/ml gentamicin) to a density of 5 × 105 cells/ml. Cell aliquots were infected by the addition of HIV-1IIIB diluted to give a viral inoculum of 100× 50% tissue culture infective doses (TCID50s) per well. A similar cell aliquot was diluted with medium to provide a mock-infected control. Cell infection was allowed to proceed for 1 h at 37°C in a tissue culture incubator with a humidified 5% CO2 atmosphere. After incubation, the virus-treated cell suspensions were diluted sixfold with fresh medium, and 125 μl of the cell suspension was added to each well of the plate containing prediluted compound. Compound GW678248 was tested over final concentration ranges of 0.5 to 500 nM. The plates were then placed in a tissue culture incubator at 37°C with humidified 5% CO2 for 5 days. HIV-induced cytopathic effects were assessed by a CellTiter 96 MTS staining method (catalog no. G3581; Promega, Madison, WI). The optical density at 492 nm was measured by using a microplate absorbance reader (catalog no. 20-300; Tecan, Research Triangle Park, NC).
Combination antiviral activity assay in MT-4 cells.
Anti-HIV activity and compound-induced cytotoxicity were measured in parallel in an MT-4 cell assay. Aliquots of GW678248 were serially diluted vertically in a 96-well master assay plate, in medium (RPMI 1640, 10% [vol/vol] FBS, and 10 μg/ml gentamicin), at concentrations that were 40-fold higher than the final assay concentration. Approved HIV inhibitors were diluted horizontally across master assay plates, also at concentrations that were 40-fold higher than the final assay concentration. Checkerboard-style dilutions were arranged so that every concentration of GW678248 was tested in the presence and the absence of every concentration of the approved HIV inhibitor. Anti-HIV activity tests were performed in triplicate assays with each combination, and direct cytotoxicity tests were a single readout for each combination.
PBMC assay.
PHA- and IL-2-stimulated PBMCs were pelleted at 192 × g for 10 min in a Jouan centrifuge and resuspended to 4 × 106 cells/ml in RPMI 1640, 20% (vol/vol) FBS, 10% (vol/vol) IL-2, and 10 μg/ml gentamicin; and 100 μl was distributed to 96-well tissue culture plates. Test compounds were titrated fourfold into RPMI 1640 20% (vol/vol) FBS, 10% (vol/vol) IL-2, and 10 μg/ml gentamicin at four times the final assay concentration. Fifty microliters of titrated inhibitor was dispensed onto the 100 μl of PBMCs and incubated at 37°C in 5% CO2 for 1 h. Fifty microliters of diluted HIV-1IIIB was then added to each well, and the contents of the plates were thoroughly mixed. GW678248 was tested over a final concentration range of 0.003 to 5,000 nM. The plates were placed in a humidified incubator at 37°C with 5% CO2 for 7 days. On day 7 of the assay, 50 μl of the culture supernatants was transferred to a new 96-well plate. The RT levels in the supernatants were measured by the method of Schwartz et al. (24).
Drug combinations (deviations from dosewise additivity).
Fifty percent inhibitory concentrations (IC50s) were calculated by curve fitting of the data to the Hill equation by using a nonlinear least-squares curve-fitting program based on the Marquardt-Levenberg algorithm (18). The interaction of each pair of compound combinations was analyzed by the methods described by Selleseth et al. (25), which provide an estimate of the strength of any interaction and of its statistical significance. Synergy and antagonism are defined as deviations from dosewise additivity, which results when two drugs interact as if they were the same drug. Values for the average deviation from additivity in the range of −0.1 to −0.2 indicate a weak synergy of the interaction, and values that approach −0.5 indicate a strong synergy of the interaction. Conversely, values of +0.1 to +0.2 indicate that a weak antagonism exists between the treatments.
Series one passage with WT HIV-1HXB2.
Selection for resistant variants was performed by sequential passage of HIV-1 strain HXB2 in escalating concentrations of GW678248. For the initial passage, GW678248 was present at 1 nM, approximately the IC50 in MT-4 cells. A total of 4 × 107 MT-4 cells was resuspended in 500 μl of cell culture medium containing HIV-1HXB2 (100 TCID50s per culture). Following 1 h of virus adsorption, the virus-cell suspension was brought to a final volume of 24 ml by the addition of RPMI 1640 medium containing 10% (vol/vol) FBS (1.6 × 106 MT-4 cells/ml). Five hundred microliters of the infected cell suspension was added per well in a 48-well tissue culture plate containing 500 μl of diluted test compound. Six parallel lineages were prepared for each test compound. Cultures were incubated at 37°C in a 5% CO2 humid atmosphere. Samples of the cell supernatant were collected from each culture at 2- to 4-day intervals and monitored for the levels of RT. Cultures containing approximately 125,000 RT cpm/30 μl or more were harvested. After centrifugation at 192 × g for 10 min in a Jouan centrifuge, the supernatants were collected for further passage, titer determination, nucleotide sequencing, and sensitivity testing. For the subsequent passages, fresh MT-4 cells (4 × 105) were infected with 300 μl of culture medium containing virus from the previous passage (regardless of the virus titer) and cultured in the presence of the compound at concentrations that were increased twofold compared with that in the previous passage. This procedure was repeated with increasing concentrations of GW678248 for eight passages.
Series two passage with HXB2 containing WT or site-directed RT mutations conferring NNRTI resistance.
Selection for drug-resistant HIV variants was performed by sequential passage of HIV-1HXB2 strains in escalating concentrations of GW678248. The WT and strains containing site-directed RT mutations associated with NNRTI resistance (12) were used as the starting virus. The selection of mutations in the starting virus was based on key mutations that confer resistance to current NNRTIs (e.g., K103N, Y181C, and V106A) or, where identified, as mutations that were selected by passage in the presence of GW678248 in the initial passage series (e.g., V106I, P236L, and V106I-P236L). For the first passage, either 0.5 or 1 nM compound was chosen as the starting concentration, based on the sensitivity of the nonpassaged mutant viruses to GW678248 in a HeLa MAGI (multinuclear activation of galactosidase indicator) assay system. In HeLa assays, the mean IC50 values for GW678248 ranged from 0.7 to 1.1 nM against K103N, Y181C, V106I, or P236L virus. For viruses with the V106A or V106I-P236L mutation, the IC50 values for GW678248 were 3.4 and 7 nM, respectively. The IC50s were not determined for these strains individually in the MT-4 system prior to the start of passage. A total of 2 × 106 MT-4 cells were resuspended in 100 μl of culture medium containing HIV-1 (titrated to result in the addition of 100 TCID50s per culture). The virus suspension was brought to a final volume of 10 ml by the addition of RPMI 1640 medium containing 10% (vol/vol) FBS with or without compound. The cultures were incubated in 25-cm2 tissue culture flasks at 37°C in a 5% CO2 humid atmosphere. Samples of the cell supernatant were collected from each culture at 2- to 4-day intervals and monitored for the levels of RT. Cultures containing approximately 125,000 RT cpm/30 μl or more were harvested. After centrifugation at 192 × g for 10 min in a Jouan centrifuge, the supernatants were collected for further passage, titer determination, nucleotide sequencing, and sensitivity testing. If the cultures did not contain RT levels high enough for harvest, fresh medium and compound were added and incubation was continued. For the subsequent passages, fresh MT-4 cells were infected with 100 μl of culture medium containing virus (regardless of the virus titer) and cultured in the presence of GW678248, typically at twofold increased concentrations. Exceptions were made when virus breakthrough failure led to the restart of a passage at the same or a lower concentration than the previous passage.
Virus infectivity titration in MT-4 cells.
The infectivity titers of serially passaged virus were determined in MT-4 cells. Seven serial fourfold dilutions of passage supernatants containing virus, ranging from 1:16 through 1:65,536, in a volume of 125 μl were titrated in triplicate in 96-well culture plates. Uninfected MT-4 cells were pelleted at 192 × g for 10 min in a Jouan centrifuge and resuspended in RPMI 1640 medium with 15% (vol/vol) FBS and 10% (vol/vol) IL-2 at a concentration of 8.3 × 106 cells/ml. A 125-μl aliquot of cell suspension was added to the wells, which contained diluted virus. The plates were incubated for 5 days at 37°C in a humid 5% CO2 atmosphere. HIV-induced cytopathic effects were assessed by an MTS staining method. The numbers of TCID50 per ml for each virus were determined by the Spearman-Karber method, as described previously (1).
Drug sensitivity determination.
The drug sensitivities of HXB2 and virus passaged in the presence of GW678248 were determined in a modified MT-4 cell assay. MT-4 cells (1 × 106 per incubation) were incubated with 300 TCID50s of test HIV for 1 h at 37°C. Following incubation, the virus-cell suspension was diluted 10-fold with RPMI 1640 medium with 15% (vol/vol) FBS and 10% (vol/vol) IL-2. Subsequently, 125 μl of the virus-cell suspension was plated per well of a 96-well microtiter plate containing 125 μl of serially diluted test compound. The compounds were serially diluted 1:3.16 in the series two experiments. The plates were incubated for 5 days at 37°C. HIV-induced cytopathic effects were assessed by an MTS staining method.
Genotype analysis of passaged virus.
The DNA sequences of the entire protease (PR)-coding region (codons 1 to 99) and the first 235 codons of the RT-coding region were determined from passaged virus supernatants. Viral RNA was extracted from cell-free supernatants by lysis in guanidinium isothiocyanate, followed by alcohol precipitation (17). Target cDNA was generated by RT-PCR with the ViroSeq HIV-1 genotyping system (Celera Diagnostics, Alameda, CA). Sequencing primers used for the RT-coding region were 5′-AATTTTCCCATTAGTCCTATTGAAACTGTACCAG and 5′-CCCCACTAACTTCTGTATGTCATTGAC; the primers used for the PR coding region were 5′-GCCGATAGACAAGGAACTGTATCC and 5′-TGAAAAATATGCATCACC. Amplicon sequencing was performed at the GlaxoSmithKline Core Sequencing Facility by using PRISM FS dye terminator chemistry and an ABI 377 automated sequencer, with analysis performed by use of the Sequencher 3.0 DNA analysis program. When mixed viral populations were present, an isolate with a ratio of the mutant electropherogram peak size to the WT electropherogram peak size greater than 70% was designated a mutant.
Phenotypic susceptibilities of 55 viruses obtained from NNRTI-experienced patients.
Fifty-five viruses obtained from NNRTI-experienced patients were selected to be assayed for their susceptibilities to GW678248, EFV, or NVP. Data regarding resistance mutations in the PR- and RT-coding regions were available for each virus. Viruses were selected for study based on the presence of mutations at codons 103, 106, 181, and/or 188 in the HIV RT. Phenotypic assays were performed by scientists at ViroLogic (South San Francisco, CA) by previously published methods (20). Briefly, a replication defective retroviral vector containing a luciferase expression cassette inserted within a deleted region of the envelope (env) gene was constructed by using an infectious molecular clone of HIV-1. Resistant test vectors were constructed by incorporating amplified PR and/or RT regions from 55 patient viruses into the retroviral vector by using ApaI and PinAI restriction sites and conventional cloning methods. The virus stocks used for drug susceptibility testing were produced by cotransfecting human embryonic kidney 293 cell cultures with resistant test vector plasmid DNA and an expression vector encoding the Env proteins of amphotropic murine leukemia virus 4070A. To measure susceptibilities to the RT inhibitors, viral stocks generated in the absence of drug were harvested and used to infect fresh 293 cell cultures in plates containing serial dilutions of the test NNRTI. Replication was monitored by measuring luciferase expression in infected target cells approximately 48 h after infection.
RESULTS
Antiviral activity of GW678248 in MT-4 cells and human PBMCs.
A comparison of the activities of the NNRTI compound GW678248 against WT HIV-1 or HIV-2 in MT-4 cell and PBMC assays is shown in Table 1. These data indicate that GW678248 inhibits WT HIV-1 replication in MT-4 cells and PBMCs with IC50 values of 1.0 and 0.4 nM, respectively. The anti-HIV-1 activity of GW678248 is essentially equivalent to the activity of EFV, 18-fold greater than that of DLV, and more than 90-fold greater than that of NVP against WT virus. Neither GW678248 nor the other NNRTIs tested were active against HIV-2.
TABLE 1.
Antiviral activities of GW678248, DLV, NVP, or EFV in MT-4 cell or PBMC assays in vitro
| Treatment | Cell line | Anti-HIV IC50 (nM)a
|
|
|---|---|---|---|
| HIV-1IIIBb | HIV-2c | ||
| GW678248 | MT-4 cells | 1.0 ± 0.6 | >50 |
| PBMCs | 0.4 ± 0.35 | NDd | |
| DLV | MT-4 cells | 18 ± 6 | ND |
| PBMCs | ND | ND | |
| NVP | MT-4 cells | 99 ± 32 | >10,000 |
| PBMCs | ND | ND | |
| EFV | MT-4 cells | 1.6 ± 0.8 | >50 |
| PBMCs | 2.1 ± 0.5 | ND | |
Values are means ± standard deviations.
The number of independent determinations in MT-4 cells was 4 for GW678248 and DLV, 43 for NVP, and 12 for EFV. In PBMCs the number of independent determinations was three for GW678248 and EFV.
The number of independent determinations was four for GW678248, NVP, and EFV.
ND, not determined.
Antiviral activities of GW678248 in combination with other marketed anti-HIV agents.
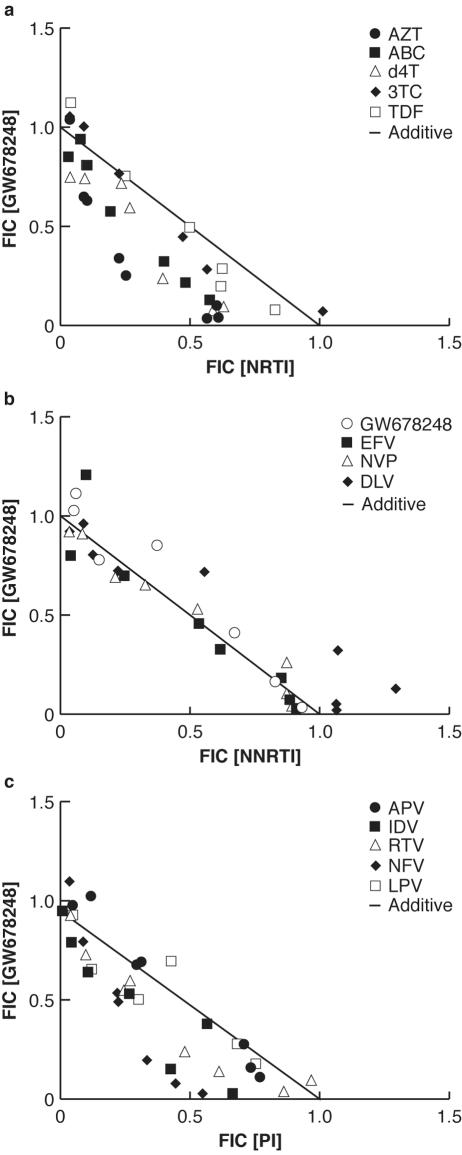
Table 2 presents the values for the deviation from additivity for combinations of GW678248 and currently approved anti-HIV-1 agents. These effects are further detailed in the graphical presentation of the isobolograms for the interaction of GW678248 with the other agents in Fig. 2.
TABLE 2.
Inhibition of HIV-1 strain IIIB by GW678248 in combination with other marketed anti-HIV agents in MT-4 cells
| Compound | Deviation from additivitya
|
Interaction with GW678248 | ||
|---|---|---|---|---|
| Mean | SD | P(t) | ||
| GW678248 | −0.023 | 0.106 | 0.387 | Additive |
| AZT | −0.464 | 0.077 | 3E-5 | Synergistic |
| d4T | −0.251 | 0.050 | 0.0004 | Synergistic |
| TFV | −0.190 | 0.068 | 0.004 | Synergistic |
| ddC | 0.042 | 0.050 | 0.152 | Additive |
| ddI | −0.050 | 0.057 | 0.133 | Additive |
| ABC | −0.203 | 0.074 | 0.006 | Synergistic |
| 3TC | −0.159 | 0.110 | 0.045 | Synergistic |
| EFV | 0.026 | 0.074 | 0.318 | Additive |
| NVP | −0.091 | 0.068 | 0.055 | Additive |
| DLV | 0.130 | 0.067 | 0.015 | Slightly antagonistic |
| IDV | −0.276 | 0.056 | 5E-4 | Synergistic |
| LPV | −0.215 | 0.078 | 0.006 | Synergistic |
| NFV | −0.206 | 0.089 | 0.009 | Synergistic |
| RTV | −0.292 | 0.033 | 9E-6 | Synergistic |
| APV | −0.035 | 0.107 | 0.336 | Additive |
| SQV | −0.047 | 0.098 | 0.264 | Additive |
Values are mean ± standard deviations from three determinations. P(t), probability, as determined by the t test, that the deviation from additivity is actually equal to zero.
FIG. 2.
Isobolograms of the inhibition of HIV-1 by GW678248 in combination with several marketed anti-HIV agents in MT-4 cells. (a) In combination with NRTIs; (b) in combination with NNRTIs; (c) in combination with protease inhibitors (PIs). FIC, fractional inhibitory concentration.
In the current series of experiments, tests in which GW678248 (Table 2) was used as both the horizontally and the vertically diluted agent show that no artifactual synergistic or antagonistic effects were seen.
In combination testing with NRTIs (Table 2 and Fig. 2a), the activity of GW678248 was found to be strongly synergistic with the effects of AZT; synergistic with the effects of ABC, 3TC, TFV, or d4T; and additive with the effects of ddC or ddI. With NNRTIs (Fig. 2b), the activity of GW678248 was additive with the effects of EFV or NVP and slightly antagonistic to the effects of DLV (P < 0.05). Combination testing of GW678248 with the protease inhibitors (Table 2) APV and SQV resulted in additive anti-HIV effects, while testing with IDV, RTV, LPV, or NFV resulted in synergistic effects.
Phenotypic susceptibilities of 55 viruses obtained from NNRTI-experienced patients.
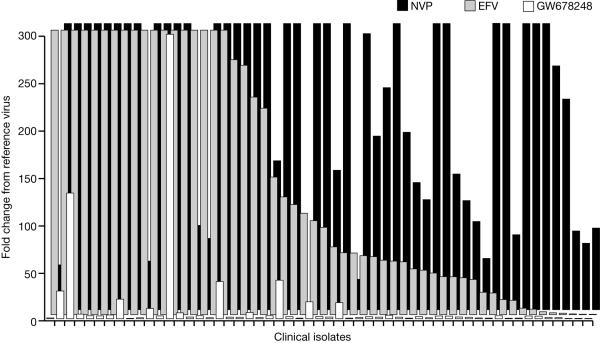
The ability of GW678248 to inhibit HIV-1 strains derived from patients who have experienced HIV-1 RNA rebound while taking combination regimens that included an NNRTI was assessed. The patient viruses used in this study had a mean of 13 protease mutations (range, 9 to 23 protease mutations) and 21 RT mutations (range, 5 to 33 RT mutations). The viral genotypes for selected loci in the RT of the 55 clinical isolate viruses are listed in Table 3, and the phenotypes of the viruses are presented in Fig. 3. Eighty-five percent (47 of 55) of the viruses had a mean change in the EFV IC50 >10-fold, and 98% of the viruses (54 of 55) had a mean change in the NVP IC50 of >10-fold. Eighty-three percent of the viruses (46 of 55) were susceptible to GW678248, with a change in the IC50 ≤10-fold. One of the 55 patient viruses had an IC50 >0.5 μM for GW678248 (range for the WT reference virus, 0.00035 to 0.0022 μM); the remaining 54 viruses had a mean IC50 of 0.0062 μM for GW678248 (range, 0.0006 to 0.109 μM). Fifty-six percent of the viruses (31 of 55) had <2.5-fold changes in IC50 values to GW678248. The six viruses with K103N as the only NNRTI resistance mutation had a mean fold change in EFV IC50 of 43, yet they were sensitive to GW678248, with a mean fold change in the GW678248 IC50 of 1.9. Similarly, the three viruses with Y181C as the only NNRTI resistance mutation had a mean fold change in the NVP IC50 of 328 and a mean fold change in the GW678248 IC50 of 1.89. Of the 47 patient viruses with >10-fold changes in the EFV IC50, 38 (81%) had ≤10-fold changes in the GW678248 IC50. Of the 54 patient viruses with >10-fold changes in the NVP IC50, 45 viruses (83%) had ≤10-fold changes in the GW678248 IC50. Seven of the eight patient viruses most resistant to GW678248 had mutations at codon 106 or 188. The four viruses with the Y188L mutation had a mean fold change in the GW678248 IC50 of 27, the two viruses with the V106A mutation had a mean fold change in the GW678248 IC50 of 353, and the single virus with the V106I mutation had a 20.7-fold change in the GW678248 IC50. Thus, 81 to 83% of the EFV- and/or NVP-resistant viruses from this data set were susceptible to GW678248.
TABLE 3.
Genotypic amino acid changes at several loci of interest in the reverse transcriptase-coding region of 55 clinical isolate viruses obtained from NNRTI-experienced patients
| NRTI-associated mutations
|
NNRTI-associated mutations
|
||
|---|---|---|---|
| Amino acid locus | No. (%) of isolates with mutation | Amino acid locus | No. (%) of isolates with mutation (%) |
| M41 | 35 (63) | A98 | 12 (22) |
| D67 | 30 (54) | L100 | 6 (11) |
| K70 | 12 (22) | K103 | 29 (53) |
| L210 | 24 (44) | V106 | 3 (5) |
| T215 | 42 (76) | V108 | 11 (20) |
| K219 | 14 (25) | V179 | 15 (27) |
| M184 | 34 (62) | Y181 | 24 (44) |
| Y188 | 4 (7) | ||
| G190 | 17 (31) | ||
| P225 | 1 (2) | ||
FIG. 3.
Phenotypic susceptibilities to efavirenz, nevirapine, and GW678248 of 55 clinical isolate viruses obtained from NNRTI-experienced patients.
In vitro serial passage of HIV-1 in the presence of dose-escalating concentrations of GW678248.
In the first series of dose-escalating serial passage experiments, six parallel lineages of HIV-1 strain HXB2 were carried for eight passages to select for resistance to GW678248. The results of genotypic analysis of the six viral lineages at passage 8 (64 nM GW678248) indicate that all lineages selected for V106I, E138K, and P236L. One of six lineages yielded a 50-50 mixture at residue 41 of Met and Leu. The significance of detection of an M-L mixture at residue 41 in one sample is unknown, and reproducibility will be monitored in future studies. This mutation was not seen in the second series of passage experiments (see below). Isolates from passages earlier than passage 8 were unavailable for genotype analysis, and therefore, temporal sequence data could not be obtained.
In a second series of HIV-1 serial passage experiments, either the WT or strains of HIV-1 containing NNRTI-resistance related mutations were used to initiate the passages (Table 4). The selection of the starting genotypes was based on key mutations that confer resistance to current NNRTIs (e.g., K103N, Y181C, and V106A) or specific mutations identified as being among those selected in the presence of GW678248 in the series one passage (e.g., V106I, P236L, and V106I-P236L).
TABLE 4.
Virus sensitivity testing
| Initiating virus | Maximum GW678248 concn (nM) | Passage no. | Genotype | Drug sensitivity (IC50 [nM])a
|
||||
|---|---|---|---|---|---|---|---|---|
| GW678248b | NVPb | EFVc | AZTc | APVc | ||||
| HXB2 | 0 | Not passaged | WT | 0.89 | 194 | 0.2 | 131 | 78 |
| WT | 128 | 10 | K102E | >500 | 3,900 | 0.1 | 151 | 70 |
| V106A | ||||||||
| P236L | ||||||||
| K103N | 64 | 8 | K103N | >500 | >250,000 | >50 | 134 | 94 |
| V106A | ||||||||
| Y181C | 128 | 9 | V90I | >500 | >250,000 | >50 | 14 | 89 |
| K101E | ||||||||
| Y181C | ||||||||
| F227C | ||||||||
| M230L | ||||||||
| V106A | 64 | 9 | V106A | NDd | ND | ND | ND | ND |
| E138K | ||||||||
| P225H | ||||||||
| D237N | ||||||||
| V106I | 128 | 10 | V106I | 400 | >250,000 | 2.8 | 121 | 58 |
| I132L | ||||||||
| E138K | ||||||||
| Y181C | ||||||||
| P236L | 128 | 8 | V106A | >500 | 5640 | 0.2 | 181 | 321 |
| E138K | ||||||||
| I142T | ||||||||
| G196R | ||||||||
| P236L | ||||||||
| V106I-P236L | 512 | 10 | V106I | ND | ND | ND | ND | ND |
| Y188L | ||||||||
| P236L | ||||||||
The “greater than”symbol indicates that there was less than 50% inhibition at the highest concentration tested.
Values are the result of two determinations.
Values are the result of one determination.
ND, not determined, as virus was present at an insufficient titer to perform the virus sensitivity assays.
WT virus passaged in the presence of up to 128 nM GW678248 developed three mutations, K102E, V106A, and P236L, after 10 passages (Table 4). V106A emerged rapidly at passage 2 (2 nM) and was retained throughout all subsequent passages. P236L emerged at passage 5 (16 nM), and K102E appeared at passage 8 (32 nM) as a K-E mixture at passages 8 and 9, followed by only as an E at passage 10. Notably, in this passage series, V106A emerged rather than V106I, and there was no selection for a mutation at locus 138 compared with the selection for a mutation at locus 138 in the virus generated in the first serial passage series.
In the series that was initiated with virus containing K103N, the only additional mutation selected was V106A, which emerged at passage 5 (8 nM). K103N was retained for the duration of the passage series.
In the series that was initiated with virus containing Y181C, the mutations V90I and K101E emerged in passage 4 (4 nM), and M230L emerged in passage 5 (8 nM) as a M-L mixture but only as an L from passage 6 onwards. Finally, F227C accumulated at passage 8 (64 nM). Y181C was retained for the duration of the passage series.
In the passage series that was initiated with virus containing V106A, the drug pressure resulted in the emergence of D237N at passage 2 (2 nM) as a mixture of D-N but only as an N from passage 3 onwards. The appearance of P225H was detected at passage 4 (8 nM) and was maintained for the duration of the series. By passage 7 (16 nM; note that 32 nM was present at passage 6) the E138K mutation emerged. The initial V106A mutation was maintained for the duration of the passage series.
In the passage series that was initiated with virus containing V106I, which was chosen because of selection in the series one passage study, the first additional mutation to be selected was E138K at passage 3 (4 nM). This mutation was also selected in the series one passage study. Interestingly, in the presence of these mutations, continued passage led to the selection of Y181C by passage 6 (32 nM), as a mixture of Y-C in passages 6 and 7, but only as a C by passage 9. Continued passage led to the emergence of I132L at passage 8 (64 nM). V106I remained throughout the passage series.
In the series that was initiated with the virus containing P236L, a G196R mutation was selected at passage 3 (4 nM) and E138K plus I142T was selected at passage 5 (16 nM). With the exception of I142T not being detected in the passage 7 sample, these mutations were maintained for the duration of the passage series. V106A was selected in passage 6 (32 nM) as a V-A mixture but as an A from passage 7 onwards. The K173Q mutation was observed only in passage 7 (64 nM). The significance of the transient appearance of K173Q is unknown, and reproducibility will be monitored in future studies. The P236L mutation was maintained throughout this passage series.
In the series that was initiated with the mutant with the V106I and P236L double mutation, only one additional mutation (Y188L) was selected at passage 7 (64 nM).
Drug sensitivity assays of viruses selected in the series two passages were performed on the last passage in each series (Table 3). These viruses were passaged once in the absence of compound before assay. The virus selected from the passage initiated with WT HIV was resistant to GW678248 and cross-resistant to NVP but not to EFV. The selected virus was sensitive to AZT and APV, as expected.
The viruses recovered from passages initiated with the various NNRTI-resistant mutants were resistant to GW678248, cross-resistant to NVP, and variably cross-resistant to EFV, depending on the specific mutations selected. The viruses selected from the passages initiated with the NNRTI-resistant mutants were sensitive to the nucleoside inhibitor AZT or the protease inhibitor APV, with the one exception of a fourfold elevated APV IC50 against the virus selected from the P236L series. The significance of this finding is uncertain and will be further explored and monitored for reproducibility in future studies.
Passage of HIV-1 strains that contain RT mutations conferring resistance to NVP or EFV at the initiation did not result in viruses that had regained sensitivity as a consequence of accumulating resistance to GW678248.
The sensitivity of isogenic, site-directed mutant viruses in HeLa MAGI assay with many of these selected mutations, alone and in combination, have been examined and are reported separately (8a). The viruses containing the triple mutation V106I, E138K, and P236L showed a greatly diminished sensitivity to GW678248 (165-fold less sensitive), while they were sensitive to NVP and less than 3-fold more resistant to EFV.
DISCUSSION
NNRTI-based HAART, especially regimens containing EFV, are becoming the standard of treatment. Several studies have demonstrated the equivalence or superiority and low toxicity of NNRTI-based HAART compared with those of protease inhibitor-based HAART (26; R. Levy, D. Labriola, and N. Ruiz, Eighth Conference on Retroviruses and Opportunistic Infections, abstr. 325, 2001; K. Tashima, S. Staszewski, M. Nelson, A. Rachlis, D. Skiest, R. Stryker, L. Bessen, V. Wirtz, S. Overfield, and D. Sahner, Fifteenth International AIDS Conference, abstr. TuPeB4547, 2004), as well as a prolonged time to virologic failure (26). The emergence of resistance to anti-HIV drugs, even when they are given as HAART, has necessitated the development of new, highly potent antiretroviral agents active against drug-resistant viral strains. In a separate study we have shown that GW678248 is a potent inhibitor of both WT and NNRTI-resistant HIV-1 in HeLa-CD4 MAGI cells and merits further development (8a). These results were also presented, in part, at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy (R. J. Hazen, R. Harvey, R. Ferris, K. Creech, M. St. Clair, K. Romines, G. Freeman, L. Schaller, J. Chan, R. Dornsife, and L. Boone, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. H-445, 2003). We have shown in the present study that GW678248 has anti-HIV-1 potency similar to that of EFV and superior to that of NVP in both PBMC and MT-4 cell assays. Like the other NNRTIs tested, GW678248 was not active against HIV-2. This absence of activity against HIV-2 is a common characteristic of the NNRTI class, although there are reports that NNRTI has activity against certain strains of HIV-2 (22, 27).
In combination with the marketed anti-HIV-1 agents, the activity of GW678248 was found to be additive or synergistic with the activities of the other agents, with the exception of slight antagonism with DLV. The slight antagonism seen with DLV is not considered detrimental to the development of GW678248, as the treatment guidelines do not recommend the use of two NNRTIs in combination. It is not clear why we have observed both additive and synergistic activities with GW678248 in combination with different members within a single drug class. The effects of another NNRTI in development, TMC125, has been found to be additive with the effects of APV, IDV, LPV, NFV, RTV, or SQV in an MT-4 cell assay (2). The effect of EFV has been found to be additive to synergistic with the effect of NFV in a yield reduction assay with HIV-1 RF and MT-2 cells (16). In this test, the degree of synergy between EFV and NFV increased when the combination index was determined at either the IC75 or the IC90 compared with additive effects when the combination index was determined at the IC50. Thus, in one assay system, it was possible to demonstrate various degrees of interaction between EFV and NFV that ranged from additivity to synergy at 50, 75, and 95% inhibition levels. While it may be tempting to speculate that in vitro synergy would predict that a certain combination might be more beneficial than one which is only additive, there are other more important factors which determine the suitability of using antiretroviral drugs in combination, including pharmacokinetics, interactions with drug-metabolizing enzymes, and toxicities. Many of these factors can be evaluated only in animal or human studies, and the efficacy of a combination regimen can be evaluated only in clinical trials. The greatest value of the in vitro combination studies is to identify the antagonism of the antiviral activity that would indicate that certain drugs should not be used clinically in combination. In this regard, the in vitro combination antiviral data support clinical testing of GW678248 with other approved antiretroviral drugs otherwise suitable for use in combination with NNRTIs.
Tests with 55 viruses obtained from NNRTI treatment-experienced patients showed a high degree of in vitro resistance to either EFV or NVP. These same viruses were found to be 81 to 83% susceptible to in vitro inhibition by GW678248. While most patients receiving NNRTI-containing HAART regimens have sustained antiviral responses, rebounds in plasma viral loads have been observed in some patients concomitant with the emergence of resistant strains of HIV-1. Reduced susceptibility to NVP is often associated with Y181C and Y188L substitutions (23), while reduced susceptibility to EFV is often associated with K103N, G190S, and Y188L substitutions. Viruses with K103N or Y188L substitutions exhibit reduced susceptibilities to all of the presently available NNRTIs (3). The retention of the in vitro activity of GW678248 at low nanomolar concentrations against a panel of viruses exhibiting high levels of resistance from NNRTI-experienced patients suggests a clinical utility for GW678248 as a follow-on therapy.
The results from in vitro serial passage experiments with either WT virus or virus with the K103N mutation as the starting virus indicate that GW678248 selects for V106A, among other mutations. In a companion paper (8a), using a panel of viruses with site-directed mutations identified in the series one serial passage experiment, we were able to demonstrate the contribution of each mutation to the overall resistance pattern of the virus. The data demonstrate that single mutations at V106I, E138K, or P236L caused very little loss of sensitivity to GW678248. The double mutation V106I and P236L or the triple mutation V106I, E138K, and P236L was required to cause high levels of resistance to GW678248. The presence of the K103N mutation at the initiation of passage does not seem to alter the pattern of GW678248-induced mutation and may prevent the selection of the P236L mutation seen with WT virus passage. Evidence in the literature supports the fact that the P236L mutation may be of minor concern in the clinic. The P236L mutation is rapidly selected in in vitro serial passage in the presence of DLV, yet it is infrequently observed in the viruses from patients receiving DLV therapy (11), where the K103N mutation is commonly observed. It is thought that the replicative fitness of viruses containing the K103N mutation is higher than that of viruses containing the P236L mutation, accounting for the dominance of the K103N mutation in the patient under therapy. The start of the passage with Y181C did not lead to a substitution at V106 in the resistant virus but, rather, led to other mutations of V90I, K101E, F227C, and M230L, in addition to the maintenance of Y181C. This virus sample demonstrated cross-resistance to all three NNRTIs tested. The contribution of each mutation in this virus to resistance has not been determined. The M230L mutation is generally associated with virologic failure of treatment with all NNRTIs (13), but it is seen in only approximately 1% of the NNRTI-treated patient population. In our panel of 55 viruses from NNRTI-experienced patients, no instance of the M230L mutation was seen. Passages starting with WT virus or virus with the V106I or P236L mutation resulted in resistance to GW678248 and cross-resistance with NVP but not EFV. The activity of EFV on viruses with mutations at the locus at position 106 or 236 suggests that EFV may be a suitable follow-on agent following GW678248 therapy where viruses with these mutations have been selected. None of the 55 treatment-experienced patient viruses in our study had the P236L mutation.
Currently, a prodrug of GW678248, designated GW695634, is in phase II clinical trials for the evaluation of its safety and efficacy and is intended for use in combination therapeutic regimens with other antiretroviral drugs.
Acknowledgments
All authors were employees of GlaxoSmithKline at the time that this research was conducted. The research presented in this paper was funded by GlaxoSmithKline.
We thank D. Paulsen for her expertise in extraction of viral RNAs and sequencing procedures.
REFERENCES
- 1.The AIDS Clinical Trials Group Virology Technical Advisory Committee and the Division of AIDS, National Institutes of Allergy and Infectious Diseases. 1994. HIV drug susceptibility assay. ACTG virology manual for HIV laboratories. RES 4-5. Division of AIDS, National Institutes of Allergy and Infectious Diseases, Bethesda, Md.
- 2.Andries, K., H. Azijn, T. Thielemans, D. Ludovici, M. Kukla, J. Heeres, P. Janssen, B. De Corte, J. Vingerhoets, R. Pauwels, and M. de Bethune. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Burnville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutation selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, J. H., G. A. Freeman, J. H. Tidwell, K. R. Romines, L. T. Schaller, J. R. Cowan, S. S. Gonzales, G. S. Lowell, C. W. Andrews, D. J. Reynolds, M. St. Clair, R. J. Hazen, R. G. Ferris, K. L. Creech, G. B. Roberts, S. A. Short, K. Weaver, G. W. Koszalka, and L. R. Boone. 2004. Novel benzophenones as non-nucleoside reverse transcriptase inhibitors of HIV-1. J. Med. Chem. 47:1175-1182. [DOI] [PubMed] [Google Scholar]
- 5.Daluge, S. M., D. J. M. Purifoy, P. M. Savina, M. H. St. Clair, N. R. Parry, I. K. Dev, P. Novak, K. M. Ayers, J. E. Reardon, G. B. Roberts, J. A. Fyfe, M. R. Blum, D. R. Averett, R. E. Dornsife, B. A. Domin, R. Ferone, D. A. Lewis, and T. A. Krenitsky. 1994. 5-Chloro-2′,3′-dideoxy-3′-fluorouridine (935U83), a selective anti-human immunodeficiency virus agent with an improved metabolic and toxicological profile. Antimicrob. Agents Chemother. 38:1590-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daluge, S. M., S. S. Good, M. B. Faletto, W. H. Miller, M. H. St. Clair, L. R. Boone, M. Tisdale, N. R. Parry, J. E. Reardon, R. E. Dornsife, D. R. Averett, and T. A. Krenitsky. 1997. 1592U89, a novel carbocyclic nucleoside analog with potent, selective anti-human immunodeficiency virus activity. Antimicrob. Agents Chemother. 41:1082-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2003. Drug resistance mutations in HIV-1. Top. HIV Med. 11:92-96. [PubMed] [Google Scholar]
- 8.Esnouf, R., J. Ren, C. Ross, Y. Jones, D. Stammers, and D. Stuart. 1995. Mechanism of inhibition of reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 2:303-308. [DOI] [PubMed] [Google Scholar]
- 8a.Ferris, R. G., R. J. Hazen, G. B. Roberts, M. H. St. Clair, J. H. Chan, K. R. Romines, G. A. Freeman, J. H. Tidwell, L. T. Schaller, J. R. Cowan, S. A. Short, K. L. Weaver, D. W. Selleseth, K. R. Moniri, and L. R. Boone. 2005. Antiviral activity of GW678248, a novel benzophenone nonnucleoside reverse transcriptase inhibitor. Antimicrob. Agents Chemother. 49:4046-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, A. G., E. Collalti, L. Ratner, R. C. Gallo, and F. Wong-Stall. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262-265. [DOI] [PubMed] [Google Scholar]
- 10.Fumero, E., and D. Podzamczer. 2003. New patterns of HIV-1 resistance during HAART. Clin. Microbiol. Infect. 9:1077-1084. [DOI] [PubMed] [Google Scholar]
- 11.Gerondelis, P., R. H. Archer, C. Palaniappan, R. C. Reichman, P. J. Fay, R. A. Bambara, and L. M. Demeter. 1999. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end and DNA 3′-end-directed RNase H activities. J. Virol. 73:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V.A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society—USA panel. Clin. Infect. Dis. 37:1722-1723. [DOI] [PubMed] [Google Scholar]
- 13.Huang, W., N. T. Parkin, Y. S. Lie, T. Wrin, R. Haubrich, S. Deeks, N. Hellman, C. J. Petropoulos, and J. M. Whitcomb. 2000. A novel HIV-1 RT mutation (M230L) confers NNRTI resistance and dose-dependent stimulation of replication. Antivir. Ther. 5(Suppl. 3):24 (abstr. 30). [Google Scholar]
- 14.Kellam, P., and B. A. Larder. 1994. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 38:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, R. W., R. M. Labe, C. D. Reid, and S. K. Erickson-Viitanen. 2002. Potency of nonnucleoside reverse transcriptase inhibitors (NNRTIs) used in combination with other human immunodeficiency virus NNRTIs, NRTIs, or protease inhibitors. Antimicrob. Agents Chemother. 46:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 18.Mager, M. E. 1972. Data analysis in biochemistry and biophysics. Academic Press, Inc., New York, N.Y.
- 19.Miyoshi, I., I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata, and Y. Hinuma. 1981. Type C virus particles in a cord T-cell line derived by co-cultivating normal cord leukocytes and human leukaemic T cells. Nature 294:770-771. [DOI] [PubMed] [Google Scholar]
- 20.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000.A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Portsmouth, S., N. Imami, A. Pires, J. Stebbing, J. Hand, M. Nelson, F. Gotch, and B. G. Gazzard. 2004. Treatment of primary HIV-1 infection with nonnucleoside reverse transcriptase inhibitor-based therapy is effective and well tolerated. HIV Med. 5:26-29. [DOI] [PubMed] [Google Scholar]
- 22.Ren, J., L. E. Bird, P. P. Chamberlain, G. B. Stewart-Jones, D. I. Stuart, and D. K. Stammers. 2002. Structure of HIV-2 reverse transcriptase at 2.35-Å resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 99:14410-14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schinazi, R., B. Larder, and J. Mellors. 2000. Mutations in retroviral genes associated with drug resistance. 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 24.Schwartz, O., Y. Henin, V. Marechal, and L. Montagnier. 1988. A rapid and simple colorimetric test for the study of anti-HIV agents. AIDS Res. Hum. Retrovir. 4:441-448. [DOI] [PubMed] [Google Scholar]
- 25.Selleseth, D. W., C. L. Talarico, T. Miller, M. W. Lutz, K. K. Biron, and R. J. Harvey. 2003. Interactions of 1263W94 with other antiviral agents in inhibition of human cytomegalovirus replication. Antimicrob. Agents Chemother. 47:1468-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, and N. M. Ruiz. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N. Engl. J. Med. 341:1865-1873. [DOI] [PubMed] [Google Scholar]
- 27.Witvrouw, M., C. Pannecouque, K. Van Laethem, J. Desmyter, E. De Clercq, and A.-M. Vandamme. 1999. Activity of non-nucleoside reverse transcriptase inhibitors against HIV-2 and SIV. AIDS 13:1477-1483. [DOI] [PubMed] [Google Scholar]