Abstract
G protein-coupled receptor CCR5 is the main coreceptor for macrophage-tropic human immunodeficiency virus type 1 (HIV-1), and various small-molecule CCR5 antagonists are being developed to treat HIV-1 infection. It has been reported that such CCR5 antagonists, including TAK-779, bind to a putative binding pocket formed by transmembrane domains (TMs) 1, 2, 3 and 7 of CCR5, indicating the importance of the conformational changes of the TMs during virus entry. In this report, using a single-round infection assay with human CCR5 and its substitution mutants, we demonstrated that a new CCR5 antagonist, TAK-220, shares the putative interacting amino acid residues Asn252 and Leu255 in TM6 with TAK-779 but also requires the distinct residues Gly163 and Ile198 in TMs 4 and 5, respectively, for its inhibitory effect. We suggested that, together with molecular models of the interactions between the drugs and CCR5, the inhibitory activity of TAK-220 could involve direct interactions with amino acid residues in TMs 4, 5, and 6 in addition to those in the previously postulated binding pocket. The possible interaction of drugs with additional regions of the CCR5 molecule would help to develop a new small-molecule CCR5 antagonist.
Shortly after the identification of the human CD4 glycoprotein as the primary receptor for human immunodeficiency virus type 1 (HIV-1) (17, 29, 32, 36), it became apparent that CD4 was not sufficient to mediate infection (1, 16, 18-21, 27-29, 34). HIV-1 attachment is mediated by the interaction of the viral envelope glycoprotein gp120 and its receptor CD4 on target cells. This binding induces a conformational change in gp120 that exposes a binding site for a coreceptor, usually of either the chemokine receptor CCR5 or CXCR4 (5, 51). Despite the significant success achieved with antiretroviral combination therapies, the emergence of resistant viruses and the lack of patient compliance stemming from adverse side effects and complex regimens have resulted in many therapeutic failures (50). Therefore, the development of better-tolerated antiretroviral agents that function through novel mechanisms and lack cross-resistance to the existing drugs is essential for the future management of HIV infections.
The interaction of CCR5 with viral gp120 is critical for membrane fusion and virus entry because a blockade of such binding can inhibit HIV-1 infection efficiently (42). Viral glycoprotein and its receptor proteins are exposed at the viral and cell surfaces, respectively, and thus compounds that inhibit viral entry do not require high membrane permeability. In addition, coreceptors are encoded by cellular genes and so are not susceptible to mutations that would cause resistance to antiviral drugs. Therefore, virus entry is a promising target for the development of novel therapeutics (24), and a new generation of inhibitors of HIV-1 replication, based on the blockade of virus entry, is now in clinical trials and in various stages of development (10, 23, 30, 37, 40, 41, 45). These include (i) small-molecule CCR5 antagonists (TAK-779 [3], UK-427857 [9], SCH-D [4], and AK602 [35]) or a CXCR4 antagonist (AMD070 [44]), (ii) chemokine analogues with modified amino-termini (11), (iii) the receptor mimic, soluble CD4 (2), (iv) peptide fusion inhibitors T20 (31) and T1249 (14), and (v) neutralizing antibodies that bind gp120 or gp41 (13). Small-molecule CCR5 antagonists such as SCH-C (8, 25), UK-427857 (43), SCH-D (43), and AK602 (38) have been shown to cause viral load reductions after administration to HIV-1-infected individuals in phase IIa clinical trials (26).
Studies with SCH-C, the chemically related AD101, and the chemically unrelated TAK-779 have shown that all these CCR5 antagonists block the binding of gp120 to CCR5 (22, 49). The binding sites for SCH-C, AD101, and TAK-779 have been mapped to a pocket formed between transmembrane domains (TMs) 1, 2, 3, and 7 of CCR5 (22, 49), and the importance of putative structural changes in these TMs induced by the binding of drugs was proposed (22, 49). However, additional analysis of CCR5 structure supported the involvement of TM5 residue in the conformational change of the CCR5 receptor upon the binding of CCR5 antagonist SCH-C (8). The mechanisms of this and other changes in CCR5 structure induced by small-molecule CCR5 antagonists and their effects on virus entry remain to be elucidated.
In this study, we analyzed the TMs of CCR5 in terms of the inhibitory activity on virus entry by the new small-molecule CCR5 antagonist TAK-220 (26) and the structurally unrelated TAK-779. Mutational analysis revealed that amino acid residues Asn252 and Leu255 in TM6 were involved in the inhibitory activity of both TAK-220 and TAK-779. On the other hand, substitution mutations at Gly163 and Ile198 in TM4 and TM5, respectively, affected the activity of TAK-220 but not of TAK-779. From these data, we postulated a molecular model in which TAK-220 interacts with amino acid residues in TMs 4, 5, and 6 in addition to those in the previously defined binding pocket formed by TMs 1, 2, 3, and 7. These findings explain the high inhibitory activity of TAK-220 on HIV-1 infection and will provide valuable information for the development of a small-molecule CCR5 antagonist.
MATERIALS AND METHODS
Cells and culture conditions.
293T cells (293T/17; ATCC CRL 11268) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich Corp., St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (ICN Biomedicals, Inc., Aurola, OH), 100 U/ml penicillin, and 100 μg/ml streptomycin (D10). U87-CD4 cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health) were maintained in D10 supplemented with 300 μg/ml G418.
Plasmids CCR5 and mutagenesis of the CCR5 coding sequence.
The human CCR5 (huCCR5) cDNA was amplified by PCR from a human spleen cDNA library (Toyobo, Osaka) and subcloned into the pcDNA3.1 expression vector (Stratagene, La Jolla, CA) between the BamHI and XbaI sites. The nucleotide sequence of the coding region of amplified huCCR5 was completely matched to that of the already published huCCR5 (GenBank accession number, XM_030397). Point mutants of huCCR5 in pcDNA3.1 were constructed by PCR-site-directed mutagenesis using specific primers. For the murine-type mutations in the second extracellular loop (ECL2), the following primer pairs were used: 5′-TCAATTCTGGAAGAGTTTCCAGACATTAA-3′ and 5′-TTAATGTCTGGAAACTCTTCCAGAATTGA-3′ for N192S, 5′-CCAGACATTAAAGATGGTCATCTTGGGGC-3′ and 5′-GCCCCAAGATGACCATCTTTAATGTCTGG-3′ for I198M, and 5′-ACACACTCAGTATTATTTCTGGAAGAATT-3′ and 5′-AATTCTTCCAGAAATAATACTGAGTGTGT-3′ for Y184H, S185T, and Q188Y. Underlines indicate the codons for mutated amino acids. The constructs were sequenced to verify the presence of the correct mutations.
Generation of the recombinant viruses.
pNL4-3-Luc+-Env−Rev− is similar to the HIV-1 luciferase reporter vector described by Chen et al. (15). To construct the vector, a stop codon was introduced near the 5′ end of env using an oligonucleotide in the KpnI site (nucleotide [nt] 6343) of proviral clone pNL4-3, and the gag-pol coding region was deleted by removing nucleotides from the SpeI site (nt 1507) through the EcoRI site (nt 5743). The firefly luciferase gene was then inserted into the nef gene by removing the BamHI (nt 8021) to XhoI (nt 8443) fragment. Envelope (Env)-defective recombinant HIV-1 pseudotype particles (HIV-1-luciferase reporter virus) capable of single-round infection and bearing the firefly luciferase gene were generated by cotransfecting 0.2 μg pNL4-3- Luc+-Env−Rev−, 0.4 μg Env expression vector for the HIV-1 JR-FL Env protein, 0.6 μg Gag/Pol expression vector, and 0.8 μg Rev expression vector into 293T cells using six-well plates, Lipofectamine, and Lipofectamine PLUS reagent (Invitrogen Corp., Carlsbad, CA). Cells were harvested 48 h posttransfection. Viral supernatants were collected by centrifugation at 3,500 rpm with a TOMY TS-38LB rotor (TOMY, Japan) at 4°C for 10 min and frozen at −80°C until used. The amount of virus present in the supernatants was quantified by measuring p24gag using a commercial enzyme-linked immunosorbent assay (HIV-1 p24 antigen ELISA kit, RETRO-TEK, ZeptoMetrix Corp., Buffalo, NY).
Env pseudotype assay of HIV-1 infection.
The infection assay with HIV-1-luciferase reporter virus was performed as follows. U87-CD4 cells were seeded in 24-well plates (0.5 × 105 cells/well to 0.6 × 105 cells/well) in D10 and transfected with plasmids to express wild-type or mutant CCR5 and luciferase derived from Renilla reniformis using Lipofectamine 2000 reagent (Invitrogen Corp., Carlsbad, CA). The next day, transfected cells were infected with HIV-1-luciferase reporter virus (10 to 20 ng of p24Gag) in a total volume of 250 μl with or without CCR5 antagonists. After 2 h, cells were washed twice with phosphate-buffered saline (PBS) and then 1 ml D10 was added to the wells. After 2 d of additional culture, cells were washed with PBS and resuspended with 200 μl of the lysis buffer packaged in the commercially available Dual-Luciferase assay kit (Promega, Madison, WI) as described below to isolate total cytoplasmic cellular proteins. The cell suspension was shaken gently at room temperature for 5 min. The supernatant fraction was recovered by centrifugation at 14,000 rpm in a KUBOTA RA-50J rotor (KUBOTA, Japan) at 4°C for 5 min. Luciferase activity in 10 μl lysates was assayed using the Dual-Luciferase assay system and a luminometer (Lumat LB6501; Berthold, Wildbad, Germany). The transfection efficiency of CCR5s was normalized against cotransfected luciferase derived from Renilla reniformis. The entry of Env-pseudotyped HIV-1 reporter viruses into the transfected U87-CD4 cells in the presence and absence of CCR5 antagonists was determined by quantifying firefly luciferase expression. Luciferase activity was directly proportional to viral entry, as confirmed by serial dilution of the input virus in the absence of antagonists (data not shown). Total protein concentrations were determined with the Coomassie Plus protein assay reagent kit (Pierce Biotechnology, Rockford, IL).
Modeling of huCCR5-ligand complexes.
Amino acid sequences of huCCR5 and bovine rhodopsin were aligned by Clustal W (48) with some manual adjustments. According to the alignment, a homology model of huCCR5 was constructed based on the bovine rhodopsin crystal structure (Protein Data Bank code, 1F88 [6]) by using the SCWRL program (12). Only the TMs were modeled, and the conformations of the conserved residues were fixed to the crystal structure. After some manual adjustments to remove large steric hindrances, the whole structure was subjected to energy minimization over 500 steps with the steepest descent minimizer and then 3,000 steps with the conjugate gradient minimizer to a maximum gradient of 0.1 kcal/mol-1Å-1, using the Discover-CVFF force field (version 2.98, Accelrys Inc., San Diego, CA). During the minimization procedure, the following conditions were adopted. The dielectric constant was set to 4r, where r is the distance between two interacting atoms. The backbone atoms were tethered with a force constant of 300 kcal/Å2 to prevent a large movement from the initial positions.
The initial structures of TAK-220 and TAK-779 were constructed with Insight II (version 2000.1, Accelrys Inc., San Diego, CA) and subjected to energy minimization with Discover. Each of the structures was manually docked into the huCCR5 model considering the interactions between the amine moiety of the ligand and Glu283 as well as the interactions between the aromatic moiety of the ligand and the hydrophobic pocket of the receptor that consists of residues of TM3, TM5, and TM6, including Ile198. The complex structures were energy minimized, as mentioned above, to obtain the final models.
Detection of the surface receptor expression by FACS analysis.
293T cells (2.5 × 105) were transfected using Lipofectamine and PLUS reagents with 1 μg of expression plasmid for wild-type CCR5 or mutant constructs and allowed to express for 36 h. Prior to antibody staining, transfected cells were lifted with DMEM and washed twice with fluorescence-activated cell sorter (FACS) staining buffer (PBS containing 5% bovine serum albumin). Cells were resuspended in 100 μl of the FACS staining buffer containing 5 μl of phycoerythrin-conjugated monoclonal anti-human CCR5 (3A9; PharMingen, San Diego, CA) and allowed to incubate on ice for 30 min. After washing once with 1 ml of the FACS staining buffer, FACS analysis was performed with a Becton Dickinson FACScan flow cytometer using CellQuest software (Becton Dickinson, San Jose, CA). Live cells stained with phycoerythrin-conjugated anti-mouse immunoglobulin G (PharMingen, San Diego, CA) were gated by light scatter, and 50,000 cells were acquired for each receptor.
Statistical analysis.
Data are means and standard errors of the mean (SEM) and were used to perform statistical analyses by the Student’s t test for impaired observations. A P value of <0.01 or <0.05 was considered significant. Fifty percent inhibitory concentrations were calculated using the SAS system procedure NLIN, which produces least squares estimates of the parameters of a nonlinear model (logistic model).
Reagents.
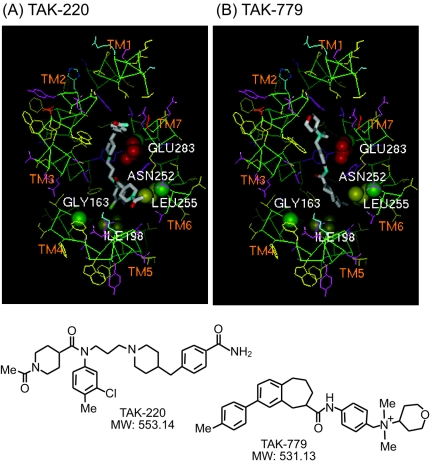
The newly designed nonpeptidic molecule TAK-220, 1-acetyl-N-[3-[4-(4-carbamoylbenzyl)-1-piperidyl]propyl]-N-(3-chloro-4-methylphenyl)piperidine-4-carboxamide (Mr = 553.14), and TAK-779, (N,N-dimethyl-N-(4-[[[2-(4-methylphenyl)-6,7-dihydro-5H-benzocycloheptan-8-yl]carbonyl]amino]benzyl)tetrahydro-2H-pyran-4-aminium chloride (Mr = 531.13) (Fig. 7), were synthesized by Takeda Pharmaceutical Company Ltd.
FIG. 7.
Molecular models of TAK-220-huCCR5 and TAK-779-huCCR5 complexes and chemical structures of the antagonists. The binding pocket viewed from the extracellular side of the receptor is shown for each complex. The key binding-site residues are represented by Corey-Pauling-Koltun (CPK) models with residue names. Main chain atoms are colored green, and acidic, basic, hydrophilic, and hydrophobic side chains are colored red, cyan, magenta, and yellow, respectively. TAK-220 and TAK-779 are color-coded for each atom (white for carbon, red for oxygen, and cyan for nitrogen).
RESULTS
Inhibitory effects of TAK-220 on huCCR5 mutated to muCCR5 sequence.
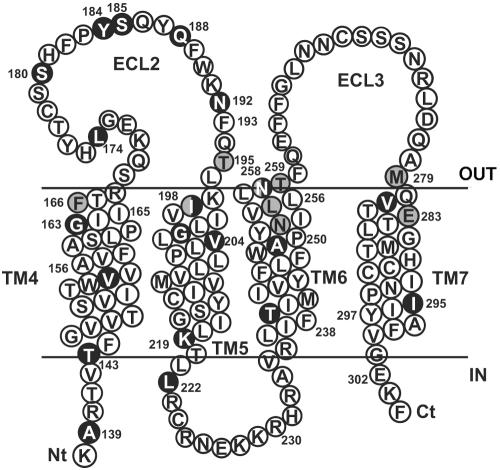
A new CCR5 antagonist, TAK-220, strongly inhibits the replication of CCR5-using (R5) HIV-1 clinical isolates with 50% inhibitory concentrations (IC50s) ranging from 0.55 to 1.7 nM in peripheral blood mononuclear cells from healthy donors (47). TAK-220 also inhibits the regulated on activation, normal T-cell expressed and secreted (RANTES) binding to huCCR5 with an IC50 of 3.5 nM (47). However, the drug was eventually inactive on the inhibition of RANTES binding to the mouse CCR5 (muCCR5) molecule (IC50 > 10,000 nM; data not shown). These results suggested that species-specific sensitivity of TAK-220 to CCR5 resulted from the difference in amino acid sequences between huCCR5 and muCCR5. Therefore, to identify the target sites of TAK-220, we introduced substitution mutations in huCCR5 so that amino acid residues specific for huCCR5 were converted to those of muCCR5 (Fig. 1).
FIG. 1.
Schematic representation of the huCCR5 and mutated amino acid residues. The extracellular face is above the membrane, which is indicated by solid lines, whereas the intracellular face is below the membrane. The model is based on the Billick et al. and Paterlini studies (8, 39). Gray circles denote residues that are substituted by Ala. Black circles denote the different amino acid residues between huCCR5 and muCCR5 sequences.
At first, we focused on the amino acid sequence in ECL2 and its vicinity, since it was demonstrated that TAK-220 strongly inhibited the binding of a monoclonal antibody, 45531.111, which recognizes the carboxyl-terminal half of ECL2, including Tyr184 to Phe189 (33, 47). Furthermore, the ECL2s of human and mouse CCR5 differ by only six amino acids, five of which are located in this region. Accordingly, a stretch of carboxyl-terminal amino acids in ECL2 of huCCR5 was replaced by that of muCCR5, resulting in mutants ECL2m1 and ECL2m2. The former mutant included five substitutions (Y184H, S185T, Q188Y, N192S, and I198M), and the latter mutant retained the first four substitutions except Ile198, which was located in the extracellular boundary of TM5.
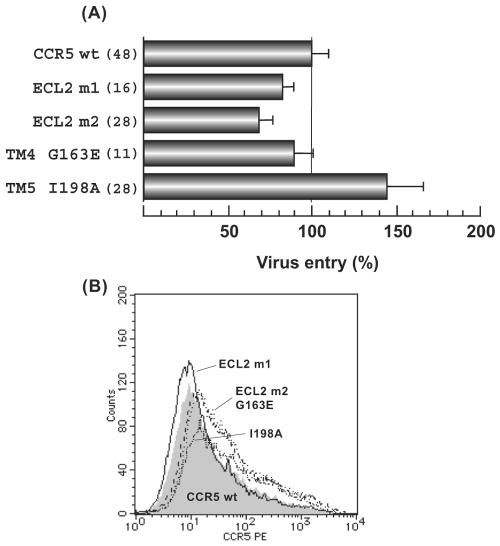
Because muCCR5 was reported to be unable to support the infection of macrophage-tropic HIV-1 (7), we first examined the infectivity of recombinant HIV-1-luciferase reporter virus with the expression of mutated huCCR5. As shown in Fig. 2, cell surface expressions of both ECL2 m1 and ECL2 m2 were comparable to that of wild-type huCCR5 in 293T cells (Fig. 2B). For the FACS analysis, wild-type and mutant huCCR5s were expressed in 293T cells instead of the U87-CD4 cells that were used for infection assay because U87-CD4 cells were too adhesive to be removed from culture dishes without trypsin treatment. These two mutants could support recombinant virus infection (virus infectivity was higher than 70% of that of wild-type huCCR5), indicating that insensitivity of muCCR5 to macrophage-tropic HIV-1 was not due to the differences in amino acid sequences between huCCR5 and muCCR5 in ECL2 and its vicinity (Fig. 2A).
FIG. 2.
Entry of pseudotyped reporter viruses into U87-CD4 expressing mouse-type mutated huCCR5 or wild-type (wt) huCCR5. (A) U87-CD4 cells transiently expressing wild-type huCCR5 or huCCR5 mutants were infected with HIV-1JR-FL Env-pseudotyped reporter viruses. Luciferase activity (in relative light units [RLU]) was measured 48 h postinfection using a dual luciferase assay kit and standardized against wild-type huCCR5 and huCCR5 mutant expression levels with Renilla reniformis luciferase activity. The coreceptor activity of each mutant, expressed as a percentage of wild-type coreceptor activity, is calculated by using the following formula: (mutant luciferase RLU/wt luciferase RLU) × 100%. Data are shown as average ± SEM from multiple experiments, and the number of experiments is indicated in parentheses. (B) Cell surface expressions of wild-type huCCR5 and huCCR5 mutants on 293T cells are represented. FACS analysis was performed as described in Materials and Methods. A shaded area indicates the expression of wild-type huCCR5 and lines indicate huCCR5 mutants. The amino acid substitutions for ECL2m1 were Y184H, S185T, Q188Y, N192S, and I198M, and the substitutions for ECL2m2 were Y184H, S185T, Q188Y, and N192S.
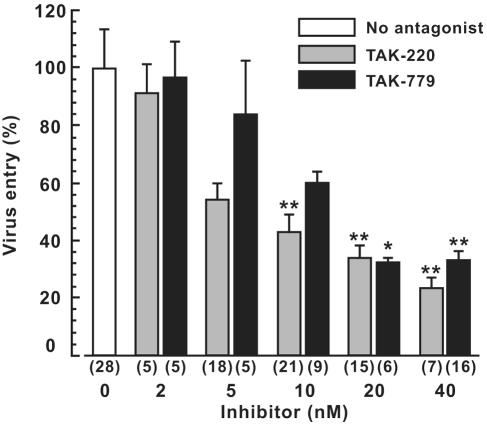
We next examined the infectivity of recombinant HIV-1-luciferase reporter virus on target cells expressing these mutants in the presence of CCR5 antagonists. As shown in Fig. 3, both TAK-220 and TAK-779 inhibited recombinant virus infection to target cells expressing wild-type huCCR5 in a dose-dependent manner. IC50s estimated from the inhibition curve were 7.7 nM for TAK-220 and 17 nM for TAK-779. When the ECL2m1 mutant was expressed, TAK-220 was almost completely unable to inhibit virus entry, whereas it inhibited infection to cells with the ECL2m2 mutant. Thus, this suggested that Ile198 has an important role for the inhibitory activity of TAK-220. Consistent with this result, the alanine substitution at Ile198 (I198A) abolished the inhibitory effect of TAK-220 (Fig. 4).
FIG. 3.
Dose-dependent entry inhibition by TAK-220 and TAK-779. A pseudotyped reporter virus infection assay was performed as described in the legend of Fig. 2A except for the addition of CCR5 antagonists (TAK-220 or TAK-779) at indicated concentrations. Data are shown as average ± SEM from multiple experiments, and the number of experiments is indicated in parentheses. Asterisks indicate a statistically significant decrease relative to virus entry without CCR5 antagonists (*, P <0.05; **, P <0.01; Student's t test).
FIG. 4.
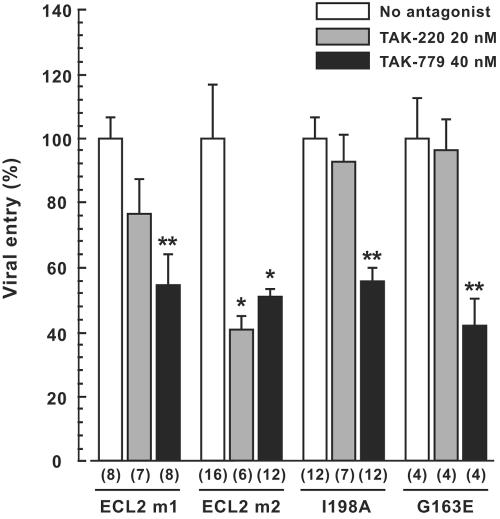
Effects of mouse-type mutations in huCCR5 on entry inhibition of pseudotyped reporter virus by TAK-220 and TAK-779. U87-CD4 cells transiently expressing wild-type huCCR5 or huCCR5 mutants were infected as described in Fig. 2A except for the presence or absence of CCR5 antagonists (TAK-220 or TAK-779) at the concentrations indicated. Results are expressed as in Fig. 3.
In contrast, both ECL2m1 and ECL2m2 mutants were sensitive to the inhibitory effect of TAK-779, and the infectivity of cells expressing the I198A mutant, whose expression on the cell surface was similar to that of the wild type (Fig. 2B), was also abrogated by TAK-779.
In addition to the difference at Ile198, muCCR5 contained Glu163 instead of Gly163 in TM4 of huCCR5. This was interesting to note because the insensitivity of African green monkey CCR5 to HIV-1 infection was attributed to an amino acid difference at this position (46). Therefore, mouse-type substitution at Gly163 (G163E) was introduced in huCCR5 and the effect of CCR5 antagonists was examined. As shown in Fig. 2, G163E mutation did not result in the loss of either infectivity (Fig. 2A) or cell surface expression (Fig. 2B). However, the mutation abolished the inhibitory effect of TAK-220 but did not affect that of TAK-779 (Fig. 4). Taken together, the data obtained by mouse-type substitution mutation indicated that TAK-779 and TAK-220 may interact with huCCR5 molecules through distinct amino acid residues and that those in TM4 and TM5 may be involved in the inhibitory activity of TAK-220.
Inhibitory effects of TAK-220 and TAK-779 on huCCR5 with alanine (Ala) substitutions in TMs.
Systematic Ala-scanning mutagenesis in TMs of huCCR5 revealed that small-molecule CCR5 antagonists could access a putative binding pocket formed by TMs 1, 2, 3, and 7 (22). However, the data presented above strongly suggested the involvement of other TMs, such as TMs 4 and 5, in the binding of TAK-220. Therefore, Ala substitutions were introduced into the same amino acid residues as previously analyzed (22) in TMs 4, 5, 6, and 7 of the huCCR5 construct (Fig. 1).
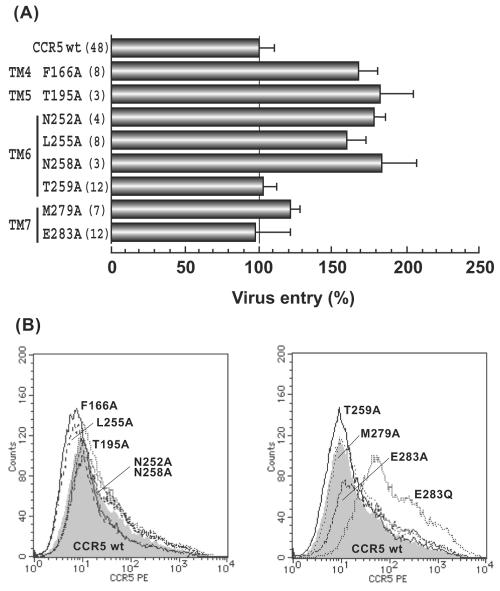
Mutant huCCR5 molecules with Ala substitution, F166A (TM4), T195A (TM5), N252A (TM6), L255A (TM6), N258A (TM6), T259A (TM6), M279A (TM7), and E283A (TM7), were tested. All of these mutants were expressed at the same level as that of wild-type huCCR5 (Fig. 5B) and were confirmed to have little or no effect on recombinant HIV-1-luciferase reporter virus infection (Fig. 5A) and then were analyzed for the inhibitory activity of TAK-220 and TAK-779.
FIG. 5.
Entry of reporter viruses into U87-CD4 cells expressing Ala substituted huCCR5. (A) Infection of pseudotyped reporter viruses to U87-CD4 cells transiently expressing wild-type huCCR5 or huCCR5 mutants and measurement of the luciferase activity were the same as described in Fig. 2A. Results are expressed as in Fig. 3. (B) Cell surface expression of wild-type huCCR5 and Ala substituted huCCR5s on 293T cells is shown. FACS analysis was performed as described in Materials and Methods, and other details are the same as for Fig. 2B. PE, phycoerythrin.
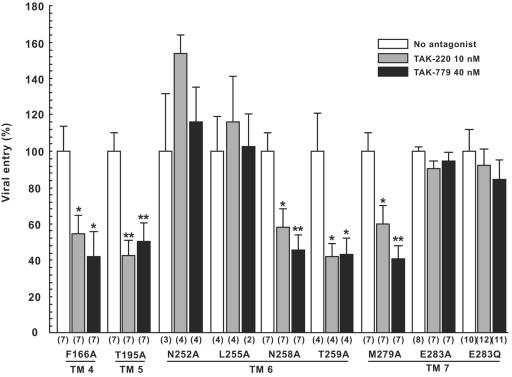
The F166A mutation in TM4, T195A in TM5, N258A and T259A in TM6, and M279A in TM7 had little effect on antiviral activity of both TAK-220 and TAK-779, and both drugs inhibited virus infection at a degree comparable to that of wild-type huCCR5 (Fig. 6). On the other hand, N252A and L255A mutations in TM6 and E283A in TM7 completely impaired the inhibitory activity of both inhibitors (Fig. 6). These data strongly supported the hypothesis that both TAK-220 and TAK-779 interacted with the same amino acid residues to inhibit entry of R5 HIV-1 to target cells. Especially, E283 has been postulated as a key anchor amino acid residue whose acidic moiety is involved in electrostatic interaction with the ammonium group of various small-molecule CCR5 antagonists (22, 49). In fact, when the acidic moiety of E283 was converted to glutamine (E283Q), inhibitory activity of both TAK-220 and TAK-779 was abolished (Fig. 6).
FIG. 6.
Effects of Ala substitutions in huCCR5 on entry inhibition of pseudotyped reporter virus by TAK-220 and TAK-779. U87-CD4 cells transiently expressing wild-type huCCR5 or huCCR5 mutants were infected as described in Fig. 2A except for the presence or absence of CCR5 antagonists (TAK-220 or TAK-779) at the concentrations indicated. Luciferase activity was measured as described in Fig. 2A. Results are expressed as in Fig. 3.
Distinct interaction of TAK-220 and TAK-779 with huCCR5 in molecular modeling.
From the results outlined above, distinct features of TAK-220 in the context of binding to CCR5 were revealed. Mutations in TM4 (G163E) and TM5 (I198A) demonstrated the involvement of different amino acid residues in the binding of TAK-220 and TAK-779, whereas Ala substitutions in TM6 (N252A and L255A) and TM7 (E283A) indicated overlapping amino acid residues between the binding sites for TAK-220 and TAK-779. To confirm this notion, molecular models of TAK-220-huCCR5 and TAK-779-huCCR5 complexes were constructed as described in Materials and Methods. As shown in Fig. 7, TAK-220 and TAK-779 occupy the same binding pocket surrounded by the TM with E283 in TM7 as an anchor residue and were located near the key residues (N252 and L255) in TM6 suggested by mutagenesis studies.
In contrast, the model predicted that TAK-220 extends its chain to a cavity composed of TM4 and TM5 to get closer to Gly163 and Ile198 than TAK-779 does. The interaction between TAK-220 and Gly163 in TM4 might not be direct, but the distance should be close enough to affect binding when the glycine residue was converted to glutamic acid.
DISCUSSION
In this study, we showed that TAK-220, a new small-molecule CCR5 antagonist, interacted with overlapping sites but also distinct sites from those of already reported inhibitors such as TAK-779 and AD101 (22, 49).
TAK-220 was initially characterized by the ability to inhibit bindings of ECL2 specific anti-CCR5 monoclonal antibody 45531.111 and RANTES to CCR5-expressing cells (47). We performed the present study to elucidate how TAK-220 interferes with the HIV-1 envelope glycoprotein gp120 binding to coreceptor CCR5 to inhibit the fusion/entry of HIV-1. For this purpose, we carried out the mapping studies using a panel of point mutants of huCCR5.
Previous studies on the interaction between huCCR5 and CCR5 antagonists implicated that some conformational changes in the TMs were required in the process of HIV-1 entry (8, 49). In addition, based on the result shown in Fig. 4, we focused on TMs 4, 5, and 6 and analyzed the effects of point mutations in these regions on the inhibitory activity of TAK-779 and TAK-220.
Mutational analysis in our study revealed that Ala substitutions at Asn252 and Leu255 in TM6 induced resistance to both TAK-220 and TAK-779. This result strongly suggested the interaction of these amino acid residues with CCR5 antagonists. However, a model previously proposed by Dragic et al. (22), in which TAK-779 binds to the cavity formed by TMs 1, 2, 3, and 7, excluded TM6 as well as TM4 and TM5 from the binding sites for TAK-779. This conclusion was partially based on the result that the inhibitory effect of TAK-779 on viral entry was not observed in cells expressing huCCR5 with mutations at Asn252 and Leu255 in TM6. The reason for this discrepancy is unclear, but it could be due to a difference in the concentration of drugs used in the assay. Because we used a lower concentration of drugs to avoid the binding saturation for both the wild type and the mutant (40 nM for TAK-779 and 10 to 20 nM for TAK-220; 200 nM for TAK-779 in reference 22), a decrease in the sensitivity to drugs could become more evident. In fact, when an excess amount of drugs (100 nM) was applied in our recombinant virus entry assay, both drugs exhibited the inhibitory effect on the virus infection to cells expressing these mutants (N252A and L255A) (data not shown). Furthermore, consistent with our result, the extensive computer analysis of the binding of TAK-779 to huCCR5 actually predicted the interaction of these two amino acid residues (Asn252 and Leu255) in TM6 with TAK-779 (39).
On the other hand, the substitutions at Gly163 and Ile198 in TM4 and TM5, respectively, abolished the sensitivity to TAK-220 but not to TAK-779. Such an extended specificity for inhibitory activity was also reported in the case of another drug, SCH-C, with the Ile198 mutations, although it was attributed to an indirect effect of drug binding (8). In our computer-assisted model, however, TAK-220 could be located in the vicinity of these two residues in TM4 and TM5. This interaction was also predicted in the molecular model by Paterlini (39). Therefore, it is likely that TAK-220 interacts with the region of amino acid residues, including Gly163 and Ile198, as shown in Fig. 7A, to prevent conformational changes necessary for viral fusion/entry, resulting in the antiviral effect. Further analysis with additional mutants, however, would be required to distinguish between conformational effects of mutants on the binding pocket versus direct effects on the binding site.
Nevertheless, it would be reasonable to speculate that the involvement of additional amino acid residues in the binding of TAK-220 to huCCR5 should enhance the affinity of the drug to the binding pocket of the coreceptor molecule. In fact, inhibitory activity of TAK-220 on huCCR5-mediated HIV-1 infection appears to be higher than that of TAK-779 (26).
Although there are available regimens consisting of antiretroviral combination therapies, the need for new classes of antiretroviral drugs has become apparent from the spreading of resistant viruses and the long-term toxicity (50). In this respect, since coreceptors for virus entry are encoded by cellular genes and are not susceptible to mutations that would cause resistance to antiviral drugs, CCR5 antagonists which target the different parts of the entry process should be quite useful for multiple-drug combination therapy with other antiretroviral drugs. The results that TAK-220 and a chemically unrelated TAK-779 recognize the overlapping but not identical residues in the binding pocket of huCCR5 predicts additional compounds with different binding specificities. Further analysis of the putative conformational changes in the TMs during the fusion/entry process will provide valuable information for the development of CCR5 antagonists with a distinct mode of binding.
Acknowledgments
We thank S. Imamura for his helpful discussion.
This study was partly supported by Matching Fund Subsidy for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M. C. Gauduin, R. A. Koup, J. S. McDougal, et al. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res. Hum. Retroviruses 11:533-539. [DOI] [PubMed] [Google Scholar]
- 3.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayes, M., X. Rabasseda, and J. R. Prous. 2004. Gateways to clinical trials. Methods Find. Exp. Clin. Pharmacol. 26:295-318. [PubMed] [Google Scholar]
- 5.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 6.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billick, E., C. Seibert, P. Pugach, T. Ketas, A. Trkola, M. J. Endres, N. J. Murgolo, E. Coates, G. R. Reyes, B. M. Baroudy, T. P. Sakmar, J. P. Moore, and S. E. Kuhmann. 2004. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 78:4134-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair, W., and M. Perros. 2004. 5th Antiviral Drug Discovery and Development Summit. Expert Opin. Investig. Drugs 13:1065-1069. [DOI] [PubMed] [Google Scholar]
- 10.Blair, W. S., P. F. Lin, N. A. Meanwell, and O. B. Wallace. 2000. HIV-1 entry—an expanding portal for drug discovery. Drug Discov. Today 5:183-194. [DOI] [PubMed] [Google Scholar]
- 11.Blanpain, C., F. Libert, G. Vassart, and M. Parmentier. 2002. CCR5 and HIV infection. Recept. Channels 8:19-31. [PubMed] [Google Scholar]
- 12.Bower, M. J., F. E. Cohen, and R. L. Dunbrack, Jr. 1997. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J. Mol. Biol. 267:1268-1282. [DOI] [PubMed] [Google Scholar]
- 13.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 14.Castagna, A., P. Biswas, A. Beretta, and A. Lazzarin. 2005. The appealing story of HIV entry inhibitors: from discovery of biological mechanisms to drug development. Drugs 65:879-904. [DOI] [PubMed] [Google Scholar]
- 15.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 17.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 18.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 19.Deng, H. K., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 20.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 21.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 22.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. USA 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Souza, M. P., J. S. Cairns, and S. F. Plaeger. 2000. Current evidence and future directions for targeting HIV entry: therapeutic and prophylactic strategies. JAMA 284:215-222. [DOI] [PubMed] [Google Scholar]
- 24.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 25.Este, J. A. 2002. Sch-351125 and Sch-350634. Schering-Plough. Curr. Opin. Investig. Drugs 3:379-383. [PubMed] [Google Scholar]
- 26.Este, J. A. 2003. Virus entry as a target for anti-HIV intervention. Curr. Med. Chem. 10:1617-1632. [DOI] [PubMed] [Google Scholar]
- 27.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 29.James, W., R. A. Weiss, and J. H. Simon. 1996. The receptor for HIV: dissection of CD4 and studies on putative accessory factors. Curr. Top. Microbiol. Immunol. 205:137-158. [DOI] [PubMed] [Google Scholar]
- 30.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 31.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 32.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 33.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 34.Liao, F., G. Alkhatib, K. W. Peden, G. Sharma, E. A. Berger, and J. M. Farber. 1997. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 185:2015-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda, K., H. Nakata, Y. Koh, T. Miyakawa, H. Ogata, Y. Takaoka, S. Shibayama, K. Sagawa, D. Fukushima, J. Moravek, Y. Koyanagi, and H. Mitsuya. 2004. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J. Virol. 78:8654-8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDougal, J. S., M. S. Kennedy, J. M. Sligh, S. P. Cort, A. Mawle, and J. K. Nicholson. 1986. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 231:382-385. [DOI] [PubMed] [Google Scholar]
- 37.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 38.Nakata, H., K. Maeda, T. Miyakawa, S. Shibayama, M. Matsuo, Y. Takaoka, M. Ito, Y. Koyanagi, and H. Mitsuya. 2005. Potent anti-R5 human immunodeficiency virus type 1 effects of a CCR5 antagonist, AK602/ONO4128/GW873140, in a novel human peripheral blood mononuclear cell nonobese diabetic-SCID, interleukin-2 receptor γ-chain-knocked-out AIDS mouse model. J. Virol. 79:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterlini, M. G. 2002. Structure modeling of the chemokine receptor CCR5: implications for ligand binding and selectivity. Biophys. J. 83:3012-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pohlmann, S., and R. W. Doms. 2002. Evaluation of current approaches to inhibit HIV entry. Curr. Drug Targets Infect. Disord. 2:9-16. [DOI] [PubMed] [Google Scholar]
- 41.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 42.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 43.Rusconi, S., A. Scozzafava, A. Mastrolorenzo, and C. T. Supuran. 2004. New advances in HIV entry inhibitors development. Curr. Drug Targets Infect. Disord. 4:339-355. [DOI] [PubMed] [Google Scholar]
- 44.Schols, D. 2004. HIV co-receptors as targets for antiviral therapy. Curr. Top. Med. Chem. 4:883-893. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz, M. K., and T. N. Wells. 2002. New therapeutics that modulate chemokine networks. Nat. Rev. Drug Discov. 1:347-358. [DOI] [PubMed] [Google Scholar]
- 46.Siciliano, S. J., S. E. Kuhmann, Y. Weng, N. Madani, M. S. Springer, J. E. Lineberger, R. Danzeisen, M. D. Miller, M. P. Kavanaugh, J. A. DeMartino, and D. Kabat. 1999. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J. Biol. Chem. 274:1905-1913. [DOI] [PubMed] [Google Scholar]
- 47.Takashima, K., H. Miyake, N. Kanzaki, Y. Tagawa, X. Wang, Y. Sugihara, Y. Iizawa, and M. Baba. 2005. Highly potent inhibition of human immunodeficiency virus type 1 replication by TAK-220, an orally bioavailable small molecule CCR5 antagonist. Antimicrob. Agents Chemother. 49:3474-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. W. Clader, J. R. Tagat, S. McCombie, B. Baroudy, J. P. Moore, T. P. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volberding, P. A. 1999. Advances in the medical management of patients with HIV-1 infection: an overview. AIDS 13(Suppl. 1):S1-S9. [PubMed] [Google Scholar]
- 51.Weiss, R. A. 2002. HIV receptors and cellular tropism. IUBMB Life 53:201-205. [DOI] [PubMed] [Google Scholar]