Abstract
The rate of occurrence of the extended-spectrum beta-lactamase (ESBL)-producing phenotype among Escherichia coli isolates in Tel Aviv is 12% (22). The aim of this study was to understand the molecular epidemiology of E. coli ESBL producers and to identify the ESBL genes carried by them. We studied 20 single-patient ESBL-producing E. coli clinical isolates. They comprised 11 distinct nonrelated pulsed-field gel electrophoresis (PFGE) genotypes: six isolates belonged to the same PFGE clone, four other clones included two isolates each, and six unrelated clones included only one isolate. All isolates produced various beta-lactamases with pIs ranging from 5.2 to 8.2, varying within similar PFGE clones. The most prevalent ESBL gene was blaCTX-M; 16 isolates carried blaCTX-M-2 and three carried a new ESBL gene designated blaCTX-M-39. Three strains carried blaSHV (two blaSHV-12 and one blaSHV-5), and two strains carried inhibitor-resistant ESBL genes, blaTEM-33 and blaTEM-30; 18 strains carried blaTEM-1 and eight strains carried blaOXA-2. Plasmid mapping and Southern blot analysis with a CTX-M-2 probe demonstrated that blaCTX-M-2 is plasmid borne. The wide dissemination of ESBLs among E. coli isolates in our institution is partly related to clonal spread, but more notably to various plasmid-associated ESBL genes, occurring in multiple clones, wherein the CTX-M gene family appears almost uniformly. We report here a new CTX-M gene, designated blaCTX-M-39, which revealed 99% homology with blaCTX-M-26, with a substitution of arginine for glutamine at position 225.
Extended-spectrum beta-lactamases (ESBLs) are common among Enterobacteriaceae in Israel (22). Twelve percent of Escherichia coli isolates in our institution have an ESBL-producing phenotype (22). Resistance to β-lactam agents in E. coli has been reported as early as 1941 (1), and is mostly mediated by plasmid-encoded β-lactamases, either broad-spectrum enzymes, such as TEM-1 or SHV-1 (2), or ESBLs (6). The prevalence and type of ESBL genes may vary between geographical areas (36). The most common ESBLs reported in E. coli isolates belong to the TEM and SHV groups (33, 36). In China, TEM-type enzymes were the main type of ESBLs found among E. coli EBSL-producing strains, followed by SHV- and CTX-M-type enzymes (37). In a recent report from Canada, the main group of ESBLs in E. coli was SHV, whereas only 6% of the ESBL producers carried blaTEM and blaCTX-M (20).
An increasing proportion of blaCTX-M genes in E. coli are being recognized in many countries (5, 11, 14, 37), with the OXA group much less frequently reported (26). Other, more rarely occurring ESBL families in E. coli include TLA-1, reported from Mexico City (29), and IBC, reported from Greece (34).
In order to understand the high prevalence of the ESBL-producing phenotype among E. coli isolates in our institution, we studied the genetic relatedness among a group of them, determined the ESBL enzymes produced by these strains, and analyzed several plasmids carrying these genes.
MATERIALS AND METHODS
Bacterial strains.
Twenty unique patient E. coli isolates possessing an ESBL-producing phenotype were analyzed. The ESBL phenotype was determined by the confirmatory disk diffusion assay using the clavulanic acid combination disk method (Oxoid, Hampshire, England) with both cefotaxime and ceftazidime (9). All strains were isolated at the Tel Aviv Sourasky Medical Center, a 1,200-bed tertiary-care, university-affiliated hospital, and were collected from May to December 2000.
Bacterial identification and MICs of the extended-spectrum cephalosporins cefotaxime, ceftazidime, cefepime, the monobactam antibiotic aztreonam, and the ureidopenicillin piperacillin were performed by means of an automated identification and microdilution system using an overnight panel (Microscan, Dade International, Inc., West Sacramento, CA), and the results were recorded and interpreted according to published guidelines. Escherichia coli 4107 (TEM-26, pI 5.6), Klebsiella oxytoca 4076 (K1, pI 6.5), E. coli 4075 (TEM-1, pI 5.4), Enterobacter cloacae 4080 (P99, pI 7.8), E. coli 4133 (SHV-1, pI 7.6), and E. coli DH5α/pCLL3414 (pI 9.0) were used as isoelectric focusing (IEF) standards. These strains, as well as E. coli J53 pMG267 (blaCTX-M-14), E. coli J53 R55 (blaOXA-3), E. coli J53 pMG203 (blaOXA-7), and E. coli carrying blaPER1 were used as positive controls in the PCR assays, and E. coli ATCC 25922 was used as a negative control. Since the clinical strain E. coli 1292 characterized in this study was determined to carry the ESBL gene blaCTX-M-2, it was used as a positive control for PCRs.
Pulsed-field gel electrophoresis analysis.
Bacterial DNA was prepared and cleaved with 20 U SpeI endonuclease (New England Biolabs, Boston, MA) as previously described (23). Agarose plugs were loaded onto a 1% agarose gel (BMA Products, Rockland, ME), prepared and run in 0.5× Tris-borate-EDTA buffer on a CHEF-DR III apparatus (Bio-Rad Laboratories, Inc., Hercules, CA). Electrophoresis was performed at 6 V/cm and 14°C. The running time was 23 h with pulse times ranging from 3 to 20 s. Gels were stained with ethidium bromide, destained in distilled water, and photographed in UV light using a Bio-Rad GelDoc camera (Bio-Rad). Pulsed-field gel electrophoresis (PFGE) DNA macrorestriction patterns were visually compared and interpreted according to the criteria established by Tenover et al. (32).
Isoelectric focusing and detection of beta-lactamases.
IEF was performed with crude lysates of cultures of E. coli clinical isolates, grown on tryptic soy broth (TSB; Biolife Italiana, Milan, Italy) and TSB supplemented with 10 μg/ml ceftriaxone (Sigma). Cultures were harvested, and cell extracts were prepared by sonication. Protein content in the cell extracts was determined with Bradford reagent (Bio-Rad). Detection of beta-lactamase activity and determination of pI were performed by IEF electrophoresis according to the method of Matthew (17), using an LKB Multiphor II Electrophoresis System apparatus on prepared PAGplates (pH 3.5 to 9.5; Amersham Biosciences, Buckinghamshire, United Kingdom). Beta-lactamases with known pIs (5.4, 5.6, 6.5, 7.6, 7.8, and 9) were electrophoresed in parallel as controls. Beta-lactamase activity was revealed with nitrocefin (0.5 mg/ml, Calbiochem-Novabiochem Corp., San Diego, CA).
Detection of ESBL genes by PCR.
The presence of blaCTX-M, blaTEM, blaSHV, and blaOXA genes in the E. coli isolates was determined by PCR. Bacterial cell lysates were used as DNA templates. Primers used for the PCR assays are listed in Table 1. Screening for the presence of blaCTX-M-2 genes was performed using CTX-M-2-specific primers. CTX-M degenerate primers were used to detect blaCTX-M-9 and related genes. For detection of genes belonging to the blaCTX-M-10 group, CTX-M-10-specific primers were used. Three sets of primers were used to detect the presence of OXA-type genes in individual reactions (Table 1). The PCR conditions were as follows: 15 min at 95°C, 40 cycles of 1 min at 94°C, 2 min at 55°C, and 3 min at 72°C, and 10 min at 72°C. PCRs were performed with Hot-StarTaq DNA polymerase (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The resulting PCR products were analyzed in a 1% agarose gel.
TABLE 1.
Primers used for PCR amplification for the detection of bla genes
| bla genea | Sequenceb | Product length (bp) | Reference |
|---|---|---|---|
| TEM Full | F: KACAATAACCCTGRTAAATGC | 936 | 28 |
| R: AGTATATATGAGTAAACTTGG | |||
| SHV Full | F: TTTATCGGCCYTCACTCAAGG | 930 | 28 |
| R: GCTGCGGGCCGGATAACG | |||
| OXA-2 Full | F: ATGGCAATCCGAATCTTCG | 828 | This study |
| R: TTATCGCGCAGCGTCCGAG | |||
| CTX-M-2 | F: ATGATGACTCAGAGCATTCG | 884 | 3, 31 |
| R: TTATTGCATCAGAAACCGTG | |||
| CTX-M-deg | F: CGYTTTSCIATGTGCAG | 550 | 28 |
| R: ACCGCRATATCRTTGGT | |||
| CTX-M-10 | F: GCAGCACCAGTAAAGTGATGG | 524 | 25, 31 |
| R: GCGATATCGTTGGTGGTACC | |||
| FEC-1 | F: GCGATAACGTGGCGATGAATAAGC | 407 | This study |
| R: GTTGAGGCTGGGTGAAGTAAGTGA | |||
| CTX-M-25 | F: CACACGAATTGAATGTTCAG | 924 | This study |
| R: TCACTCCACATGGTGAGT | |||
| CTX-M-8 | F: ATGATGAGACATCGCGTTAAG | 864 | This study |
| R: CGGTGACGATTTTCGCGGCAG | |||
| OXA-1 | F: ACACAATACATATCAACTTCGC | 813 | 27, 31 |
| R: AGTGTGTTTAGAATGGTGATC | |||
| OXA-10 | F: CGTGCTTTGTAAAAGTAGCAG | 651 | 31 |
| R: CATGATTTTGGTGGGAATGG | |||
| PER-1 | F: ATGAATGTCATTATAAAAGC | 925 | 24 |
| R: AATTTGGGCTTAGGGCAGAA |
CTX-M-deg, degenerate primers.
K is G or T; R is A or G; Y is C or T; S is G or C; I is inosine.
Cloning and sequencing of bla genes.
blaCTX-M-2, blaCTX-M-39 and blaOXA-2 complete gene PCR products were ligated into pGEM-T easy PCR cloning vector and transformed into a competent cell of E. coli JM109 according to the manufacturer's instructions (Promega, WI). Transformed colonies were selected on LB agar plates supplemented with ampicillin (100 μg/ml). Plasmids were isolated using the Rapid Plasmid miniprep system (Marligen Biosciences) and digested with EcoRI restriction enzyme (New England Biolabs) to confirm the presence of the insert. Sequencing of cloned genes was performed using SP6 and T7 promoter primers. Sequencing of the other PCR products was performed using the same primers used for PCR amplifications (listed in Table 1).
Sequences were analyzed with an ABI PRISM 3100 genetic analyzer (PE Biosystems), using the DNA sequencing analysis software and 3100 data collection software version 1.1. The nucleotide and the deduced protein sequences were analyzed and compared using the software available via the Internet at the NCBI web site (http://www.ncbi.nlm.nih.gov/).
Plasmid isolation and Southern blot analysis.
Plasmid DNA was isolated from 20 E. coli strains using the QIAGEN plasmid DNA midi kit (QIAGEN, Hilden, Germany). For Southern blot analysis, plasmids were digested with ApaI restriction endonuclease (New England Biolabs), electrophoresed, and transferred to a Hybond N+ membrane (Amersham Biosciences, Buckinghamshire, United Kingdom). The blaCTX-M-2 gene, labeled with digoxigenin, using the digoxigenin high prime DNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany), was used as a probe. Hybridization and detection were performed according to the manufacturer's instructions.
Nucleotide sequence accession number.
The blaCTX-M-39 gene nucleotide sequence appears in the GenBank nucleotide sequence database under accession no. AY954516.
RESULTS
Antimicrobial susceptibilities and the ESBL-producing phenotype.
The confirmatory ESBL-producing phenotype assay identified all 20 strains as ESBL producers when cefotaxime with and without clavulanic acid were used as substrates, but only 1 of 20 when ceftazidime with and without clavulanic acid were used. An antibiotic susceptibility pattern (Table 2) showed that three isolates (15%) had an MIC above the breakpoints for all cephalosporins (≥64 μg/ml for cefotaxime and ≥32 μg/ml for ceftazidime and cefepime), and that 17 strains were susceptible to at least one of these agents; two isolates (10%) were susceptible to cefotaxime, four (20%) to aztreonam, six (30%) to cefepime, and nine (45%) to ceftazidime.
TABLE 2.
MICs of E. coli isolates
| Strain | MICa (μg/ml)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | PIP | TZP | A/S | AZT | CTX | CAZ | CEP | IMP | |
| 1020 | >16 | >64 | <16 | <8/4 | >16 | >32 | 4 | >16 | <4 |
| 1438 | >16 | >64 | <16 | >16/8 | >16 | >32 | 4 | >16 | <4 |
| 1241 | >16 | >64 | <16 | >16/8 | >16 | >32 | >16 | >16 | <4 |
| 1254 | >16 | >64 | <16 | >16/8 | >16 | >32 | 16 | >16 | <4 |
| 1131 | >16 | >64 | <16 | <8/4 | >16 | >32 | 8 | >16 | <4 |
| 1482 | >16 | >64 | <16 | <8/4 | >16 | >16 | 16 | >16 | <4 |
| 1027 | >16 | >64 | <16 | 16/8 | 8 | >32 | 4 | 16 | <4 |
| 1031 | >16 | >64 | 64 | >16/8 | >16 | >32 | 4 | >16 | <4 |
| 1227 | >16 | >64 | <16 | >16/8 | >16 | >32 | 4 | >16 | <4 |
| 1266 | >16 | >64 | >64 | >16/8 | >16 | >32 | >16 | >16 | <4 |
| 1292 | >16 | >64 | >64 | 16/8 | >16 | >32 | >16 | >16 | <4 |
| 1204 | >16 | >64 | <16 | 16/8 | >16 | >32 | 16 | >16 | <4 |
| 1393 | >16 | >64 | <16 | <8/4 | <8 | 32 | >16 | >16 | <4 |
| 1430 | >16 | >64 | <16 | <8/4 | <2 | <4 | <2 | <8 | <4 |
| 1455 | >16 | >64 | 64 | <8/4 | >16 | 16 | >16 | <8 | <4 |
| 1299 | >16 | >64 | >64 | 16/8 | >16 | >32 | 16 | <8 | <4 |
| 1313 | >16 | >64 | <16 | >16/8 | <2 | <4 | <2 | >16 | <4 |
| 1466 | >16 | >64 | >64 | 16/8 | <2 | 32 | <2 | <8 | <4 |
| 1326 | >16 | >64 | 64 | <8/4 | >16 | >32 | 16 | <8 | <4 |
| 1356 | >16 | >64 | 64 | 16/8 | >16 | >32 | 16 | >16 | <4 |
AMP, ampicillin; A/S, ampicillin-sulbactam; AZT, aztreonam; CTX, cefotaxime; CAZ, ceftazidime; CEP, cefepime; IMP, imipenem; PIP, piperacillin; TZP, piperacillin-tazobactam.
Genetic relatedness.
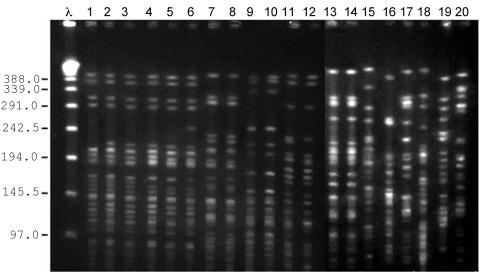
Genetic relatedness analysis showed that the 20 E. coli isolates belonged to 11 unrelated PFGE clones (Fig. 1). One of these clones (type A) comprised six genetically related strains: three strains were isolated from patients hospitalized in the same ward during a 3-month period and their period of hospitalization overlapped, suggesting an epidemiological cluster. Two other strains were isolated from patients from different wards and at different times, and had no apparent epidemiological relation to each other or to the other cases. The sixth strain was isolated from a patient who had been seen in the emergency ward, and had never been hospitalized before, suggesting that the isolate was acquired in the community. PFGE analysis also showed four other clusters of two genetically related isolates each, as well as six isolates that possessed a unique PFGE pattern (Fig. 1 and Table 3).
FIG. 1.
Pulsed-field gel electrophoresis of the 20 E. coli strains after SpeI digestion. Eleven distinct clones were identified. λ lane, molecular size markers of lambda ladder. Lanes 1 to 6, clone A. Lanes 7 to 8, clone C. Lanes 9 and 10, clone I. Lanes 11 and 12, clone D. Lanes 13 and 14, clone K. Lanes 15 to 20, clones F, G, H, J, B, and E, respectively.
TABLE 3.
Characterization of β-lactamase genes in E. coli clinical isolatesa
| E. coli isolate | PFGE type | pI | Detection of bla genes by PCR
|
|
|---|---|---|---|---|
| blaCTX-M | Additional broad and extended bla genes | |||
| 1020 | A | 5.4; 7.9 | CTX-M-2 | TEM-1; OXA-2 |
| 1438 | A | 5.4; 7.9 | CTX-M-2 | TEM-1 |
| 1241 | A | 5.4; 6.5; 7.6; 7.9 | CTX-M-2 | TEM-1 |
| 1254 | A | 5.4; 7.9 | CTX-M-2 | TEM-1; OXA-2 |
| 1131 | A | 5.4; 7.6; 7.8; 7.9 | CTX-M-2 | TEM-1 |
| 1482 | A | 5.4; 7.6; 7.9 | CTX-M-2 | TEM-1 |
| 1027 | B | 5.4; 7.7; 7.9 | CTX-M-2 | TEM-1; OXA-2 |
| 1031 | C | 5.4; 6.5; 7 7.5; 7.9 | CTX-M-2 | TEM-33 |
| 1227 | C | 5.4; 7.9 | CTX-M-2 | TEM-1; OXA-2 |
| 1266 | D | 5.4; 7.8; 8.2 | NI | SHV-12 |
| 1292 | D | 5.4; 5.6; 7.7; 7.9; 8.2 | CTX-M-2 | TEM-1; SHV-12; OXA-2 |
| 1204 | E | 5.2; 7.7; 7.9 | CTX-M-2 | TEM-30, OXA-2 |
| 1393 | F | 5.4; 6.8; 7.0 | CTX-M-39 | TEM-1 |
| 1430 | G | 5.4; 6.0; 6.8; 7.0 | CTX-M-39 | TEM-1 |
| 1455 | H | 5.4; 8.2 | NI | TEM-1, SHV-5 |
| 1299 | I | 5.4; 7.7; 7.9 | CTX-M-2 | TEM-1, OXA-2 |
| 1313 | I | 5.4; 6.5; 7.9 | CTX-M-2 | TEM-1, OXA-2 |
| 1466 | J | 5.4; 6.0; 6.8; 7.0 | CTX-M-2b; CTX-M-39 | TEM-1 |
| 1326 | K | 5.4; 7.8; 7.9 | CTX-M-2 | TEM-1 |
| 1356 | K | 5.4; 7.9 | CTX-M-2 | TEM-1 |
ESBL genes and the pI of CTX-M-2, 7.9, are marked in bold letters; CTX-M-39 and the proposed pI 6.8 to 7.0 are underlined. NI, not identified by PCR, but hybridized with CTX-M-2 probe in the Southern analysis.
blaCTX-M-2 gene detected in this strain by PCR, but a beta-lactamase with pI of 7.9 was not observed.
Identification of beta-lactamase activity by IEF.
All 20 E. coli strains showed at least two distinct IEF bands indicating the presence of beta-lactamases. The approximate pI values of the beta-lactamases that were revealed with IEF and inhibited by clavulanic acid ranged from 5.4 to 8.2 (Table 3). Nineteen of 20 isolates (95%) had a clavulanic acid-inhibited beta lactamase with a pI of 5.4 (consistent with TEM-1) and one isolate (1204) had a distinct band with a pI of 5.2. Fifteen isolates (75%) showed a clavulanic acid-inhibited beta-lactamase with a pI of 7.9 consistent with CTX-M-2, and all three isolates with the new enzyme, CTX-M-39, possessed nitrocefin-positive bands with a pI value of 6.8 to 7.0, suggesting this as the putative pI value. Additional beta-lactamases identified are detailed in Table 3.
PCR and sequence analysis for detection of ESBL genes.
At least one ESBL gene was found in each of the 20 E. coli isolates with the ESBL-producing phenotype. Sixteen isolates harbored one family type of ESBL gene and four isolates harbored two types (Table 3). The major ESBL group found was CTX-M; 16 isolates possessed CTX-M-2 (80%) and three isolates possessed a new CTX-M enzyme, designated CTX-M-39. Two additional ESBL groups that were found were TEM and SHV. Due to the abundance of CTX-M-type enzymes in the E. coli clones studied, a more detailed genetic analysis was performed on this group of genes.
CTX-M genes.
Sequencing of the cloned entire open reading frames (ORFs) of the blaCTX-M-positive isolates identified them as blaCTX-M-2, correlating with a pI of 7.9 obtained by IEF. PCR using primers specific for blaFEC-1, of the CTX-M family, amplified additional blaCTX-M products from three strains. Sequencing of these genes revealed that they were identical to each other and similar to blaCTX-M-25 from E. coli (GenBank accession number AF518567) (21) and blaCTX-M-26 from Klebsiella pneumoniae (accession number AY157676) (7). PCR with primers specific for the entire ORFs of blaCTX-M-25 and blaCTX-M-26, amplified products in all three strains. PCR product from strain 1466 was cloned into the pGEM-T easy vector and sequenced. Sequencing revealed 99% homology with blaCTX-M-26, with a substitution of argininge for glutamine at position 225. This new enzyme was designated CTX-M-39 by the Lahey Clinic site (http://www.lahey.org).
PCR screening for blaCTX-M-9, blaCTX-M-10, blaCTX-M-8 and related genes was negative.
Plasmid DNA.
The majority of the E. coli clones examined carried numerous plasmids. Hybridization with a blaCTX-M-2 probe revealed the presence of the corresponding gene in plasmid DNA originating from 18 of 20 clones. Clones 1393 and 1430, possessing blaCTX-M-39, did not hybridize with this probe. Plasmid DNA from clones 1266 and 1455, which yielded negative PCR results with four pairs of primers specific for the CTX-M gene family, hybridized positively with the CTX-M-2 probe.
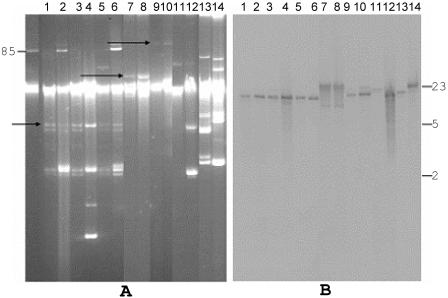
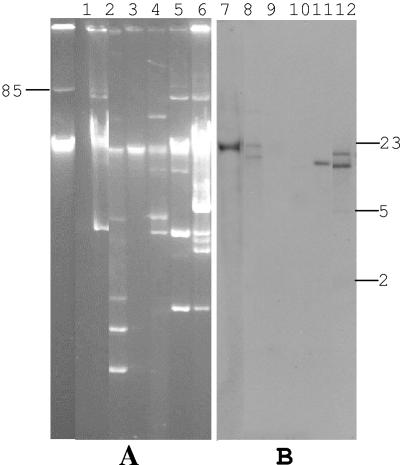
In order to examine the location of blaCTX-M, Southern analysis on plasmid DNA was performed. A uniform-sized plasmid was revealed from the six isolates belonging to clone A (Fig. 2 A, lanes 1 to 6). In these isolates, similar sized DNA fragments were hybridized with blaCTX-M-2 (Fig. 2B, lanes 1 to 6), suggesting the presence of blaCTX-M-2 on the same plasmid. Plasmid profiles were similar within the same genetic clone (clone C and clone I), but differed between clones (Fig. 2A). Hybridization with a blaCTX-M-2 probe within each clone showed the same pattern (Fig. 2A, lanes 7 to 8 and 9 to 10, and B, lanes 7 to 8 and 11 to 12), although the two strains belonging to clone C possessed different beta-lactamase genes (Table 3). Clones D and K showed different hybridization patterns with blaCTX-M-2 probe. Plasmid profiles of the six unique E. coli strains varied (Fig. 3A), and we did not find similarities in their hybridization patterns (Fig. 3B).
FIG. 2.
Electrophoresis of plasmid DNA of strains belonging to clones A, C, I, D and K (panel A), and Southern blot hybridization of the plasmid DNAs using CTX-M-2 probe (panel B). Panel A, plasmid pAFF2 (85 to 90 kb), used as a molecular size marker. Lanes 1 to 6, plasmid DNA of six strains belonging to PFGE type-A (strains 1020, 1438, 1241, 1254, 1131, and 1482, respectively). Lanes 7 and 8, 9 and 10, 11 and 12, and 13 and 14, plasmid DNA from pairs of strains belonging to the same PFGE type, strains 1031 and 1227 (clone C); strains 1299 and 1313 (clone I); strains 1266 and 1292 (clone D); and strains 1356 and 1326 (clone K), respectively. The arrows indicate plasmids that are common within the same clone. Panel B, Southern blot analysis of the plasmid DNAs digested with ApaI restriction enzyme, using blaCTX-M-2 as a probe. DNA size standards (kb) are shown on the right.
FIG. 3.
Electrophoresis of plasmid DNA of six unique PFGE types (panel A), and Southern blot analysis of the plasmid DNAs digested with ApaI, using blaCTX-M-2 as a probe (panel B). Plasmid pAFF2 (85 to 90 kb) was used as a molecular size marker. Panel A, lanes 1 to 6, uncut plasmid DNA isolated from strains 1027 (clone B), 1204 (clone E), 1393 (clone F), 1430 (clone G), 1455 (clone H), and 1466 (clone J), respectively. Panel B, Southern blot analysis of the plasmid DNAs digested with ApaI enzyme, using blaCTX-M-2 as a probe. DNA size standards (kb) are shown on the right.
Other bla genes.
Nineteen isolates (95%) possessed a beta-lactamase with a pI of 5.4. Sequencing of the entire ORFs of blaTEM PCR products from 17 strains revealed blaTEM-1. Additional blaTEM genes found were inhibitor-resistant genes; E. coli 1031 possessed blaTEM-33 (16), and E. coli 1204 possessed blaTEM-30 (35), corresponding to the IEF band with a pI of 5.2.
Three of the 20 isolates possessed blaSHV (Table 3). These strains showed a positive IEF band with a pI of 8.2, corresponding to blaSHV-12 or blaSHV-5 (6). Sequencing of the entire ORF of these genes identified two as blaSHV-12 and one as blaSHV-5.
PCR with primers for the entire ORF of the class D oxacillinase blaOXA-2 yielded 828-bp products in eight strains. Cloning and sequencing in both directions identified them as blaOXA-2. Four of these eight isolates possessed a beta-lactamase with a pI of 7.7, corresponding to this gene (12). This band was missing in the other four blaOXA-2-carrying clones, possibly due to a lack or a low level of expression of OXA-2 by these strains. PCRs with primers specific for blaOXA-4 and blaOXA-10 were negative. PCR performed with primers specific for blaPER-1 was negative.
Discussion
The increasing incidence of ESBLs among Enterobacteriaceae is a growing problem. In this study we attempted to understand the molecular epidemiology of ESBL-producing E. coli isolates in an urban tertiary-care teaching hospital in Israel.
Twenty single-patient isolates were examined based on the ESBL producer phenotype, and all were found to have at least one ESBL gene. Three different ESBL genes were found (blaCTX-M, blaSHV, and blaTEM) and the most prevalent ESBL type was CTX-M (80%). We report here the presence of a new CTX-M enzyme, designated CTX-M-39, closely related to CTX-M-25 and CTX-M-26 (21). The antibiotic susceptibility profiles of the E. coli isolates showed a higher MIC of cefotaxime than of the other broad-spectrum cephalosporins, correlating with previous reports on cefotaximases (3). CTX-M-type ESBLs are geographically widespread and their increasing rate of occurrence has been reported in other countries as well (4).
The molecular epidemiology of these CTX-M enzymes among E. coli strains in our institution is complex. We found clonal diversity but also PFGE clusters; the dominant strain accounted for 30% of the cases, however only half of these appeared to be epidemiologically linked, suggesting that in our hospital setting, patient-to-patient transmission may be responsible for only the minority of the cases of E. coli, and is not the main mechanism of dissemination of ESBL producers.
Among the various clones we studied, multiple plasmids were found to carry blaCTX-M-2. Although we found blaCTX-M-2-carrying plasmids common to individual clones, a given clone could produce various combinations of beta-lactamases. For example, all six isolates belonging to clone A produced CTX-M-2, while OXA-2 was produced by only two of six. In the case of clone I, by contrast, both isolates possessed the same blaCTX-M-2-carrying plasmid and produced the same enzymes. These data suggest that ESBL spread in our institution is due to dissemination of transferable genetic elements between strains on the one hand, and to spread of ESBL-carrying clones on the other.
OXA-2, belonging to Ambler class D beta-lactamases (12), was found in 40% of the strains studied. The presence of OXA-type enzymes in Enterobacteriaceae has been described infrequently. OXA-4 and OXA-7 were reported in E. coli strains from Sao Paulo, Brazil (18), as was OXA-30, an OXA-1 derivative originally isolated in Hong Kong (13, 30). Forty percent of our E. coli strains were found to carry blaOXA-2, reported previously in Pseudomonas aeruginosa in France (accession number AJ295229) (13). A similar enzyme was also isolated from Salmonella spp. in France (accession number AJ311891) (10), and from an Enterobacter aerogenes strain in Venezuela (accession number U13380) (8). Ours is the first report on the natural presence and wide dissemination of OXA-2 in E. coli.
Beta-lactamases belonging to the TEM group were identified in two isolates, both inhibitor-resistant enzyme derivatives, TEM-33 and TEM-30, which have been previously reported in E. coli isolates from different countries in Europe (15, 19). The existence of CTX-M-2, inhibited by clavulanic acid, in the strains carrying these enzymes, enabled the phenotypic identification of these strains as ESBL producers.
The high proportion (12%) of ESBL producers among the E. coli isolates in our institution and the complex molecular epidemiology of various clones with diverse types of ESBL genes are alarming. Our findings suggest the existence of horizontal transfer of ESBL genes, leading to an epidemiological pattern that poses a challenge to the infection control team.
Acknowledgments
We thank Karen Bush and Anne Marie Queenan, R. W. Johnson Pharmaceutical Research Institution, Raritan, NJ, George Jacoby, Lahey Clinic, Burlington, MA, and Haluk Vahaboglu, Kocaeli University, Istanbul, Turkey, for providing us with the ESBL-producing bacterial strains used in the IEF and PCR experiments as positive controls.
This work was supported by a grant from the Israel-United States Binational Science Foundation.
REFERENCES
- 1.Abraham, E. P., and E. Chain. 1940. An enzyme from bacteria able to destroy penicillin. Nature 146:837. [PubMed] [Google Scholar]
- 2.Ambler, R. P. 1980. The structure of β-lactamases. Phil. Trans. R. Soc. Lond. B 289:321-331. [DOI] [PubMed] [Google Scholar]
- 3.Bauerfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bou, G. M., M. Cartelle, D. M. Tomas, D. Canle, F. Molina, R. Moure, J. M. Eiros, and A. Guerrero. 2002. Identification and broad dissemination of the CTX-M-14 beta-lactamase in different Escherichia coli strains in the northwest area of Spain. J. Clin. Microbiol. 40:4030-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenwald, N. P., G. Jevons, J. M. Andrews, J. H. Xiong, P. M. Hawkey, and R. Wise. 2003. An outbreak of a CTX-M-type beta-lactamase-producing Klebsiella pneumoniae: the importance of using cefpodoxime to detect extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 51:195-196. [DOI] [PubMed] [Google Scholar]
- 8.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter, M. W., K. J. Oakton, M. Warner, and D. M. Livermore. 2000. Detection of extended-spectrum beta-lactamases in klebsiellae with the Oxoid combination disk method. J. Clin. Microbiol. 38:4228-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casin, I., J. Breuil, A. Brisabois, et al. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded β-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 11.Chanawong, A., F. Hannachi M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale, J. W., D. Godwin, D. Mossakowska, P. Stephenson, and S. Wall. 1985. Sequence of the OXA-2 β-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 191:39-44. [DOI] [PubMed] [Google Scholar]
- 13.Dubois, V., C. Arpin, C. Quentin, J. Texier-Maugein, L. Poirel, and P. Nordmann. 2003. Decreased susceptibility to cefepime in a clinical strain of Escherichia coli related to plasmid- and integron-encoded OXA-30 β-Lactamase. Antimicrob. Agents Chemother. 47:2380-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henquell, C., D. Sirot, C. Chanal, C. De Champs, P. Chatron, B. Lafeuille, P. Texier, J. Sirot, and R. Cluzel. 1994. Frequency of inhibitor-resistant TEM β-lactamases in Escherichia coli isolates from urinary tract infections in France. J. Antimicrob. Chemother. 34:707-714. [DOI] [PubMed] [Google Scholar]
- 16.Henquell, C., C. Chanal, D. Sirot, R. Labia, and J. Sirot. 1995. Molecular characterization of nine different types of mutants among 107 inhibitor-resistant TEM β-lactamases from clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 39:427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthew, M., M. Harris, M. J. Marshall, and G. W. Rose. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 18.Medeiros, A. A., M. Cohenford, and G. A. Jacoby. 1985. Five novel plasmid-determined β-lactamases. Antimicrob. Agents Chemother. 27:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miro, E., F. Navarro, B. Mirelis, M. Sabate, A. Rivera, P. Coll, and G. Prats. 2002. Prevalence of clinical isolates of Escherichia coli producing inhibitor-resistant beta-lactamases at a university hospital in Barcelona, Spain, over a 3-year period. Antimicrob. Agents Chemother. 46:3991-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, S. Paton, and the Canadian Hospital Epidemiology Committee of the Canadian Nosocomial Infection Surveillance Program, Health Canada. 2004. Ambler Class-A extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munday, C. J., D. A. Boyd, N. Brenwald, M. Miller, J. M. Andrews, R. Wise, M. R. Mulvey, and P. M. Hawkey. 2004. Molecular and kinetic comparison of the novel extended-spectrum beta-lactamases CTX-M-25 and CTX-M-26. Antimicrob. Agents Chemother. 48:4829-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navon-Venezia, S., O. Hammer-Munz, D. Schwartz, D. Turner, B. Kuzmenko, and Y. Carmeli. 2003. Occurrence and phenotypic characteristics of extended-spectrum beta-lactamases among members of the family Enterobacteriaceae at the Tel Aviv Medical Center (Israel) and evaluation of diagnostic tests. J. Clin. Microbiol. 41:155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noller, A. C., M. C. McEllistrem, O. Colin Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordmann, P., and T. Naas. 1994. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver, A., J. C. Perez-Diaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., P. Gerome, C. De Champs, J. Stephanazzi, T. Naas, and P. Nordmann. 2002. Integron-located oxa-32 gene cassette encoding an extended-spectrum variant of OXA-2 beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quellette, M., L. Bissonnette, and P. H. Roy. 1997. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1β-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlesinger, Y., S. Navon-Venezia, I. Chmelnitsky, O. Hammer-Münz, A. Leavitt, H. S. Gold, M. J. Schwaber, and Y. Carmeli. 2005. Characterization of extended-spectrum beta-lactamases (ESBLs) among Enterobacter isolates in Tel Aviv. Antimicrob. Agents Chemother. 49:1150-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva, J., C. Aguilard, G. Ayala, M. A. Estrada, U. Garza-Ramos, R. Lara-Lemus, and L. Ledezma. 2000. TLA-1: a new plasmid-mediated extended-spectrum beta-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siu, L. K., J. Y. Lo, K. Y. Yuen, P. Y. Chau, M. H. Ng, and P. L. Ho. 2000. β-Lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like β-lactamase, OXA-30. Antimicrob. Agents Chemother. 44:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steward, C. D., J. K. Rasheed, S. K. Hubert, J. W. Biddle, P. M. Raney, G. J. Anderson, P. P. Williams, K. L. Brittain, A. Oliver, J. E. McGowan, Jr., and F. C. Tenover. 2001. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards extended-spectrum beta-lactamase detection methods. J. Clin. Microbiol. 39:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenover, F. C., P. M. Raney, P. P. Williams, J. K. Rasheed, J. W. Biddle, A. Oliver, S. K. Fridkin, L. Jevitt, and J. E. McGowan, Jr. 2003. Evaluation of the NCCLS extended-spectrum beta-lactamase confirmation methods for Escherichia coli with isolates collected during Project ICARE. J. Clin. Microbiol. 41:3142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzelepi, E., C. Magana, E. Platsouka, D. Sofianou, O. Paniara, N. J. Legakis, A. C. Vatopoulos, and L. S. Tzouvelekis. 2003. Extended-spectrum beta-lactamase types in Klebsiella pneumoniae and Escherichia coli in two Greek hospitals. Int. J. Antimicrob. Agents 21:285-288. [DOI] [PubMed] [Google Scholar]
- 35.Vedel, G., A. Belaaouaj, L. Gilly,. R. Labia, A. Philippon, P. Névot, and G. Paul. 1992. Clinical isolates of Escherichia coli producing TRI β-lactamases: novel TEM-enzymes conferring resistance to β-lactamase inhibitors. J. Antimicrob. Chemother. 30:449-462. [DOI] [PubMed] [Google Scholar]
- 36.Winokur, P. L., R. Canton, J. M. Casellas, and N. Legakis. 2001. Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin. Infect. Dis. 32:94-103. [DOI] [PubMed] [Google Scholar]
- 37.Xiong, Z., D. Zhu, F. Wang, Y. Zhang, R. Okamoto, and M. Inoue. 2002. Investigation of extended-spectrum beta-lactamase in Klebsiellae pneumoniae and Escherichia coli from China. Diagn. Microbiol. Infect. Dis. 44:195-200. [DOI] [PubMed] [Google Scholar]