Abstract
β-Lactamase TLA-2 is encoded by a 47-kb plasmid isolated from an unidentified bacterial strain from a wastewater treatment plant. TLA-2 is an Ambler class A β-lactamase that shares 52% amino acid identity with CGA-1 from Chryseobacterium gleum and 51% with TLA-1 from Escherichia coli. The enzyme hydrolyzes mostly cephalosporins.
Most of the clavulanic acid-inhibited expanded-spectrum Ambler class A β-lactamases (ESBL) (1) are either derivatives of narrow-spectrum TEM- and SHV-type β-lactamases or CTX-M-type β-lactamases (3, 11). Plasmid-carried ESBL genes are found in Enterobacteriaceae and more rarely in Pseudomonas aeruginosa (13). In addition to the so-called classical ESBL, a series of other Ambler class A (1) ESBL are known as BES, GES, PER, SFO, TLA-1, and VEB, with a quite large distribution of PER- and VEB-type enzymes in P.aeruginosa (6, 14, 15, 19, 20, 23).
A survey was previously performed by Szczepanowski et al. (24) to analyze bacterial populations in the activated-sludge compartment of a wastewater treatment plant for the presence of plasmids conferring erythromycin resistance to the host bacterium. Plasmids were extracted and introduced into Escherichia coli strain DH5α by transformation. A 47-kb plasmid (pRSB101) was sequenced, and it was shown to carry a class A β-lactamase gene, named blaTLA-2 (24).
The purpose of the present study was to analyze the properties of the novel β-lactamase TLA-2.
The entire blaTLA-2 gene was amplified by PCR with primers TLA-2A (5′-TCCCTGGAGCACTTATGAAT-3′) and TLA-2B (5′-ATTAAGGATAAACTCATCCGC-3′), which were designed from the nucleotide sequence under GenBank/EMBL accession no. AJ698325. The gene was cloned into the pPCRBluntII-TOPO plasmid and expressed in E. coli DH10B (Invitrogen, Life Technologies, Cergy-Pontoise, France), giving rise to the E.coli DH10B(pTLA-2) recombinant strain. Antibiotic susceptibility testing by disk diffusion (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) showed that E. coli DH10B(pTLA-2) displayed a typical clavulanic acid-inhibited ESBL phenotype (data not shown).
MICs of β-lactams were determined by using the broth microdilution technique according to the Clinical and Laboratory Standards Institute recommendations (9). MICs of β-lactams showed that this β-lactamase gene, once expressed from a multicopy plasmid, conferred resistance or reduced susceptibility to cephalosporins, with a major effect on ceftazidime and aztreonam (Table 1). Surprisingly, MICs of amino-, carboxy-, and ureidopenicillins were only slightly modified by the expression of the β-lactamase TLA-2 (Table 1).
TABLE 1.
MICs of β-lactams for E. coli DH10B(pTLA-2) and E. coli DH10B
| β-Lactam(s) | MIC (μg/ml) for E. coli:
|
|
|---|---|---|
| DH10B(pTLA-2) | DH10B | |
| Amoxicillin | 8 | 2 |
| Amoxicillin + CLAa | 2 | 2 |
| Ticarcillin | 8 | 2 |
| Ticarcillin + CLA | 2 | 2 |
| Piperacillin | 2 | 1 |
| Piperacillin + TZB | 1 | 1 |
| Cephalothin | 64 | 2 |
| Cefuroxime | 16 | 0.5 |
| Cefoxitin | 16 | 1 |
| Ceftazidime | 64 | 0.06 |
| Ceftazidime + CLA | 0.5 | 0.06 |
| Ceftazidime + TZB | 0.5 | 0.06 |
| Cefotaxime | 1 | 0.06 |
| Cefotaxime + CLA | 0.06 | 0.06 |
| Cefotaxime + TZB | 0.06 | 0.06 |
| Cefepime | 0.25 | 0.06 |
| Cefepime + CLA | 0.06 | 0.06 |
| Cefepime + TZB | 0.06 | 0.06 |
| Moxalactam | 1 | 0.12 |
| Aztreonam | 4 | 0.12 |
| Aztreonam + CLA | 0.12 | 0.12 |
| Aztreonam + TZB | 0.12 | 0.12 |
| Imipenem | 0.12 | 0.12 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
As previously described, the blaTLA-2 gene was not embedded in a class 1 integron but it was bracketed by 145-bp direct repeats of unknown function (24). The blaVEB-1 gene has been identified in P. aeruginosa 10.2, also bracketed by direct repeats, but it is not structurally related to blaTLA-2 (2).
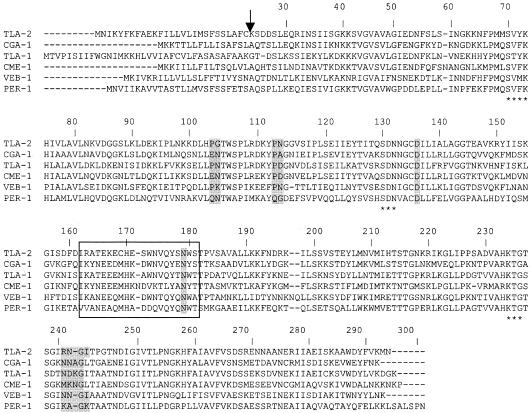
The TLA-2 β-lactamase has features of Ambler class A β-lactamases (1, 24) (Fig. 1). TLA-2 is distantly related to other β-lactamases and shares the highest amino acid sequence identity with Ambler class A β-lactamases of gram negatives such as CGA-1 (52%) from Chryseobacterium gleum (4) and with TLA-1 (51%), encoded by plasmid RZA92 from E. coli R170 (23). TLA-2 is weakly related to the other ESBL (Fig. 1). Detailed analysis of the protein sequence of TLA-2 indicated that it might belong to a subgroup of ESBL that, in addition to CGA-1 and TLA-1, also includes CEP-A (21), CFX-A (16), CME-1 (22), PER-1 (15), TLA-1 (23), and VEB-1 (20). TLA-2 shares 39% amino acid identity with PER-1, 33% with CEP-A, and 45% with CME-1 (data not shown).
FIG. 1.
Amino acid sequence comparison of representative class A β-lactamases. The residues highlighted in gray are specific to the β-lactamases of the PER-1 subgroup as suggested by Tranier et al. (25), whereas those of the Ω-loop sequence are boxed. Asterisks indicate the conserved motifs SXXK, SDN, and KTG of Ambler class A β-lactamases (1). The arrow indicates the cleavage site for the leader peptide of TLA-2.
In order to perform a biochemical analysis of TLA-2, enzyme purification was carried out by ion-exchange chromatography as previously described (17). Briefly, 8 liters of an E. coli DH10B(pTLA-2) culture in Trypticase soy broth was pelleted, resuspended, and disrupted by sonication in 60 ml 100 mM sodium phosphate buffer (pH 7). The protein extracts obtained were loaded onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech) with a 20 mM Tris-HCl buffer (pH 8.8). The β-lactamase recovered in the flowthrough was subsequently dialyzed against 50 mM sodium phosphate buffer (pH7.2), loaded onto an S-Sepharose column preequilibrated with the same buffer, and eluted with a linear NaCl gradient (0to 500 mM). The fractions containing the highest β-lactamase activity, as determined by nitrocefin test (Oxoid, Dardilly, France), were pooled and dialyzed overnight against 50 mM sodium phosphate buffer (pH 7).
The protein purification rate and the relative molecular mass of the purified β-lactamase TLA-2 were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis as previously described (17). Purification of the enzyme was difficult due to low-level biosynthesis. A specific activity of 75 μmol min−1 mg of protein−1 was determined with 100 μM cephalothin as a substrate, at 30°C in 100 mM sodium phosphate buffer, for the purified β-lactamase TLA-2. The purification coefficient was calculated to be 35-fold, and its purity was estimated to be >90%. The mature protein had a relative molecular mass of ca. 30 kDa. Isoelectric focusing analysis (5) identified a β-lactamase with a pI value of 8.8. N-terminal Edman sequencing was performed on an Applied Biosystems Procise 494HT, as previously described (12). N-terminal amino acid sequencing of the mature protein revealed a cleavage site of the deduced blaTLA-2 gene product between residues 22 and 23 (C-K) (Fig. 1).
The purified β-lactamase TLA-2 was used for kinetic measurements performed at 30°C in 100 mM phosphate buffer as previously described (10, 18). The kcat and Km values were determined by analyzing β-lactam hydrolysis under initial-rate conditions with a UV spectrophotometer by using the Eadie-Hoffstee linearization of the Michaelis-Menten equation, as previously described (10, 18). The kinetic parameters of the TLA-2 β-lactamase revealed its activity against restricted and expanded-spectrum cephalosporins (Table 2). The enzyme showed the highest level of activity against cephalothin (kcat value of 90 s−1). However, the low affinity (high Km value) reduced the catalytic efficiency (kcat/Km) of TLA-2 against cephalothin. TLA-2 showed an uncommonly high affinity for expanded-spectrum cephalosporins, with Km values greatly lower than those of most ESBL (4-fold lower than that of VEB-1 for cefotaxime and 10-fold lower than that of VEB-1 for ceftazidime) (14, 15, 19, 20, 22, 23). Overall, TLA-2 had a similar hydrolysis profile compared to that of TLA-1, except for aztreonam for which no detectable hydrolysis was observed with TLA-2. TLA-2 had no detectable activity also against amoxicillin, ticarcillin, cefoxitin, and imipenem. For these substrates, determinations of 50% inhibitory concentrations (IC50) or Ki were performed with cephalothin (100 μM) as substrate. TLA-2 was strongly inhibited by ampicillin (Ki value of 1.8 μM), ticarcillin (Ki value of 25 nM), or aztreonam (Ki value of 0.1 μM). Imipenem (IC50 value of 3.5 μM) and cefoxitin (Ki value of 1.7 μM) were also good inhibitors of TLA-2 activity, as observed for GES-1 (19) and PER-1 (15). These high affinities may explain the increased MICs of penicillins and aztreonam observed for E. coli DH10B(pTLA-2) despite lack of significant hydrolysis. Surprisingly, TLA-2 was not inhibited or was weakly inhibited in vitro by β-lactam inhibitors (Ki value of 300 μM for clavulanic acid and IC50 of >100 μM for tazobactam and 40 μM for sulbactam) although MICs of cephalosporins were significantly lowered by the addition of β-lactam inhibitors. This characteristic could not be explained by any known mutation present in inhibitor-resistant TEM (IRT) β-lactamases (8). Residues 69 (Met) and 276 (Asn) of TEM-1 β-lactamase are conserved in TLA-2, whereas they are not in IRT enzymes, while other residues conferring resistance in IRT enzymes (8), found in TLA-2 β-lactamase, were also found in other ESBL of the PER-1 subgroup, which remain susceptible to β-lactam inhibitors. The apparent discrepancy between in vitro and in vivo susceptibilities remains to be explained.
TABLE 2.
Kinetic parameters of the purified β-lactamase TLA-2a
| β-Lactam | kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| Benzylpenicillin | 5 | 15 | 330 |
| Cephalothin | 90 | 260 | 350 |
| Cefuroxime | 0.5 | 10 | 50 |
| Cefotaxime | 0.5 | 10 | 50 |
| Ceftazidime | 5 | 50 | 100 |
| Cefpirome | 0.5 | 30 | 15 |
| Cefepime | 0.1 | 10 | 10 |
| Cefsulodin | 0.1 | 10 | 10 |
Data are means from three independent experiments. Standard deviations were within 15%.
According to its functional properties, TLA-2 could be included in group 2e of the Bush-Jacoby-Medeiros classification scheme (7), since it exhibits good catalytic efficiencies toward most cephalosporins and not toward penicillins, but it lacks sensitivity to β-lactam inhibitors, which does not fit the criteria of this class of β-lactamases. This report extends the variety of Ambler class A ESBL (1) that may be identified in a waterborne environment. Although there is not any evidence yet for diffusion of TLA-2 in the clinical setting, the fact that this enzyme can degrade expanded-spectrum cephalosporins and is encoded by a mobilizable plasmid might anticipate future clinical relevance.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA 3539), Université Paris XI, Paris, France, and the European Community (6th PCRD, LSHM-CT-2003-503-335). L.P. is a researcher from the INSERM (Paris, France).
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J.-M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Lévesque, G. Tiraby, and S. G. Wakey. 1991. A standard numbering scheme for class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, D., D. Girlich, T. Naas, S. Nagarajan, and P. Nordmann. 2004. Functional and structural characterization of the genetic environment of an extended-spectrum β-lactamase blaVEB gene from a Pseudomonas aeruginosa isolate obtained in India. Antimicrob. Agents Chemother. 48:3284-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellais, S., T. Naas, and P. Nordmann. 2002. Molecular and biochemical characterization of Ambler class A extended-spectrum beta-lactamase CGA-1 from Chryseobacterium gleum. Antimicrob. Agents Chemother. 46:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellais, S., L. Poirel, T. Naas, D. Girlich, and P. Nordmann. 2000. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 44:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. Sampaio, C. Chanal, D. Sirot, C. De Champs, J. L. Viallard, R. Labia, and J. Sirot. 2000. A novel class A extended-spectrum β-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob. Agents Chemother. 44:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaibi, E. B., D. Sirot, G. Paul, and R. Labia. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing, 15th informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 10.Cornish-Bowden, A. (ed.). 1995. Graphs of the Michaelis-Menten equation, p. 30-37. In Fundamentals of enzyme kinetics. Portland Press, Inc., Seattle, Wash.
- 11.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto, Y., and M. Inoue. 1999. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordmann, P., E. Ronco, T. Naas, C. Duport, Y. Michel-Briand, and R. Labia. 1993. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippon, L. N., T. Naas, A.-T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2195-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers, M. B., A. C. Parker, and C. J. Smith. 1993. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob. Agents Chemother. 37:2391-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossolini, G. M., N. Franceschini, L. Lauretti, B. Caravelli, M. L. Riccio, M. Galleni, J.-M. Frère, and G. Amicosante. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaCME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob. Agents Chemother. 43:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva, J., C. Aguilar, G. Ayala, M. A. Estrada, U. Garza-Ramos, R. Lara-Lemus, and L. Ledezma. 2000. TLA-1: a new plasmid-mediated extended-spectrum β-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczepanowski, R., I. Krahn, B. Linke, A. Goesmann, A. Pühler, and A. Schlüter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 25.Tranier, S., A. T. Bouthors, L. Maveyraud, V. Guillet, W. Sougakoff, and J. P. Samama. 2000. The high resolution crystal structure for class A β-lactamase PER-1 reveals the bases for its increase in breadth of activity. J. Biol. Chem. 275:28075-28082. [DOI] [PubMed] [Google Scholar]