Abstract
Azole resistance has been insufficiently investigated in the yeast Candida tropicalis. Here we determined the molecular mechanisms responsible for azole resistance in a clinical isolate of this pathogenic yeast. Antifungal susceptibility testing performed by a disk diffusion method showed resistance or markedly decreased susceptibility to azoles, which was confirmed by determination of MICs. Considering the relationship between azole susceptibility and the respiration reported for other yeast species, the respiratory activity of this isolate was investigated. Flow cytometry using rhodamine 123 and oxygraphy demonstrated an increased respiratory activity, which was not linked to an overexpression or increased number of copies of the mitochondrial genome. Among previously described resistance mechanisms, an increased activity of efflux pumps was investigated by flow cytometry using rhodamine 6G. However, the efflux of rhodamine 6G was lower in the resistant isolate than in susceptible ones. Likewise, real-time reverse transcription-PCR quantification of the expression of C. tropicalis MDR1 (CtMDR1), which encodes an efflux protein belonging to the major facilitator superfamily, did not show overexpression of this gene. In contrast, the resistant isolate overexpressed the CtERG11 gene coding for lanosterol 14α-demethylase. This was in agreement with the larger amount of ergosterol found in this isolate. Moreover, sequencing of CtERG11 showed a point mutation leading to a tyrosine substitution in the protein sequence, which might lead to decreased binding affinity for azoles. In conclusion, overexpression of CtERG11 associated with a missense mutation in this gene seemed to be responsible for the acquired azole resistance of this clinical isolate.
The incidence of life-threatening fungal infections due to Candida species has increased markedly over the past 2 decades. These opportunistic infections are particularly frequent in severely immunocompromised patients, such as organ or bone marrow transplant recipients, those receiving cytotoxic drugs for treatment of an underlying cancer, and patients with AIDS (19, 32). Currently, yeasts belonging to the Candida genus rank fourth or fifth among the causative agents of septicemia (5). To fight these infections, four main classes of antifungals are now available (22, 26): polyenes (mainly represented by amphotericin B) and azoles, which act on membrane ergosterol and its biosynthetic pathway, respectively; pyrimidine analogs represented by flucytosine (5FC), which, through its intracellular conversion to 5-fluorouracil, inhibits protein synthesis and DNA replication; and the recently commercialized caspofungin, which inhibits the synthesis of the cell wall β-glucan polysaccharides (8).
However, as a result of the extensive use of antifungals in prophylaxis or therapy of mycoses, the recovery of resistant yeast isolates has been increasingly reported (7, 12, 21). Additionally, a marked shift in the distribution of yeast species was observed (37). Although Candida albicans remains by far the most common species encountered, more than 30% of all yeast infections are due to non-C. albicans species, such as Candida glabrata, Candida tropicalis, Candida krusei, and Candida parapsilosis, which presently comprise, along with C. albicans, nearly 95% of clinical yeast isolates. For example, in the United States, the prevalence of C. albicans in septicemia has declined from 87% of the total yeast isolates in 1987 to a low of 31% in 1992, with concomitant increases of C. glabrata (from 2% to 26%), C. tropicalis (2% to 24%), and C. parapsilosis (9% to 20%) (23). Precise identification of these non-C. albicans species is required, considering the therapeutic hazard of some of these species, which can be naturally resistant or poorly susceptible to current antifungals. For example, C. krusei is naturally resistant to fluconazole and C. glabrata is susceptible only to high doses of this drug.
Among non-C. albicans species, C. tropicalis represents the third most common species isolated but the second in respiratory specimens. This yeast usually remains susceptible to polyenes and azole antifungals but is frequently resistant to 5FC (10). However, clinical isolates resistant to azoles, particularly to fluconazole, are increasingly reported (18, 33, 38).
While resistance mechanisms to 5FC have been extensively investigated in C. tropicalis, little information is available about the mechanisms responsible for its azole resistance. Resistance mechanisms have been studied principally in C. albicans (4) and C. glabrata (3, 30, 36). Three main mechanisms are usually described (29): (i) increased efflux of the azole drug, due to the overexpression of efflux pumps such as the ATP-binding cassette (ABC) transporters, which use ATP as the source of energy, and the major facilitator superfamily (MFS) membrane transporters, which use a proton gradient; (ii) overexpression of the ERG11 gene, coding for the azole target lanosterol 14α-demethylase; and (iii) a point mutation in the ERG11 sequence which leads to a decreased affinity of azoles for their target and, therefore, to an azole resistance. These three mechanisms can be found separately, but they can also be combined, contributing to a step-by-step acquisition of azole resistance (17).
Concerning C. tropicalis, only one strain resistant to azole antifungals has been studied (1). For this strain, azole resistance was due to an increased efflux of azole drugs linked to an overexpression of the C. tropicalis MDR1 gene (CtMDR1), which encodes an MFS protein, whereas a gene homologue of C. albicans CDR1, encoding an ABC protein, was not overexpressed. However, azole resistance of this strain was induced in vitro by successive passages on fluconazole-containing agar plates. In the present study, we determined the molecular mechanisms responsible for azole resistance in a clinical isolate of C. tropicalis obtained from a urine sample of a patient previously treated with miconazole for inguinal candidiasis.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Four isolates of C. tropicalis were used throughout this study. The clinical isolate designated 40301893 was isolated in our hospital laboratory in 2003 from a 77-year-old woman receiving antibiotics for septic shock after heart surgery. Two weeks later, the patient presented an inguinal mycosis, which was treated with local applications of miconazole. However, due to a persistent fever, antifungal treatment was changed 2 weeks later to fluconazole (100 mg/day, intravenously), and a urine sample was collected, which permitted the isolation of C. tropicalis. Identification of the yeast isolate was performed using the ID32C test strip (bioMérieux, Marcy l'Etoile, France), and antifungal susceptibility testing, performed using the Fungitest strip (Bio-Rad, Marnes-la-Vallée, France), revealed a resistance to azoles. Treatment was therefore changed, 1 week later, to amphotericin B intravenously, and a new urine sample collected after 48 h of treatment was found to be sterile. As no matched susceptible isolate was available, two other clinical isolates, designated 91.4959 and 91.1328, isolated from urine samples from two distinct patients in 1991, as well as a reference strain, IHEM 10264, from the Institute of Hygiene and Epidemiology-Mycology section (Brussels, Belgium), were used as controls.
All isolates were maintained by biweekly passages on yeast extract-peptone-glucose (YEPD) agar plates containing yeast extract (5 g/liter), peptone (10 g/liter), glucose (20 g/liter), chloramphenicol (0.5 g/liter), and agar (20 g/liter). They were preserved by lyophilization and by freezing at −80°C in 20% (wt/vol) glycerol. In addition, the three clinical isolates were deposited at the Institute of Hygiene and Epidemiology-Mycology Culture Collection and are publicly available under the accession numbers 21232, 21233, and 21234 (for isolates 91.1328, 91.4959, and 40301893, respectively).
Antifungal susceptibility testing.
Susceptibility to polyene and azole drugs was determined by a disk diffusion method on Casitone agar (Bacto-Casitone, 9 g/liter; glucose, 20 g/liter; yeast extract, 5 g/liter; chloramphenicol, 0.5 g/liter; agar, 18 g/liter; pH 7.2) using Neosensitabs tablets from Rosco Diagnostic (Taastrup, Denmark) as previously described (2, 3, 6). Results were confirmed on RPMI 1640-glucose agar which contained RPMI 1640 (Sigma Aldrich Ltd., St. Quentin Fallaviers, France) at 10.4 g/liter, 3-[N-morpholino]-propanesulfonic acid hemisodium salt at 165 mM, l-glutamine at 2 mM, chloramphenicol at 1 g/liter, glucose at 20 g/liter, and agar at 15 g/liter. The diameters of the inhibition zones were measured after incubation for 48 h at 37°C. According to the manufacturer's recommendations, isolates were considered to be resistant when the diameter of the inhibition zone was less than 11 mm for azoles or 9 mm for polyenes and considered to be susceptible when the diameter was greater than 20 mm for azoles or 15 mm for polyenes. All experiments were performed in triplicate, and results, which are expressed as mean diameters, were compared using the Wilcoxon-Mann-Whitney test.
MICs of amphotericin B, ketoconazole, fluconazole, and voriconazole were determined by the Etest procedure by following the manufacturer's recommendations (AB Biodisk, Solna, Sweden) on Casitone agar and RPMI 1640-glucose agar plates. MICs were read after 48 h at 37°C as the drug concentration at which the inhibition ellipse intercepted the scale on the antifungal strip. For clotrimazole, which was not available as an Etest strip, MICs were determined by microdilution assay by following the NCCLS M27A protocol in Casitone and RPMI 1640-glucose broths inoculated with 103 blastoconidia/ml (200 μl per well of the microtiter plates). Clotrimazole was dissolved in dimethyl sulfoxide to reach final concentrations ranging from 0.1 to 250 μg/ml. After incubation for 48 h at 37°C, the absorbance at 595 nm was read. The MIC90 was defined as the lowest concentration of the drug that inhibited growth by at least 90% compared with the growth of the drug-free control. All of these experiments were performed in triplicate. Results, which are expressed as mean values, were analyzed using the Wilcoxon-Mann-Whitney test.
Characterization of the respiratory activity.
The respiratory status of the cells was determined by flow cytometry after incubation with rhodamine 123, a fluorescent dye that incorporates into the mitochondrion in a transmembrane potential-dependent manner (25). Blastoconidia from 48-h-old cultures in YEPD broth were harvested by centrifugation and washed in distilled water. Cells were resuspended in 0.15 M phosphate-buffered saline (PBS) at pH 7.2. Cells (2 × 106/ml) were incubated for 30 min at 37°C in the presence of 10 μg/ml rhodamine 123. To inhibit the electron flow of the respiratory chain, cells were first incubated with 1 mM sodium azide for 2 h at 37°C before the addition of rhodamine 123. After cells were washed in cold PBS, the fluorescence of 10,000 cells was quantified with a FACScan flow cytometer (BDIS Europe, Erembodegem, Belgium) and the data were analyzed with the Cellquest software from BDIS.
The respiratory activity of the four isolates was also determined by oxygraphy using stationary growth-phase blastoconidia (48 h at 37°C) from YEPD agar plates. After being washed in sterile distilled water and enumerated by hemocytometer counting, cells (109/ml) were suspended in 10 mM potassium phosphate buffer, pH 7.4, containing 20 mM glucose. Oxygen consumption was measured at 37°C with an Oxytherm oxygraph (Hansatech Instruments Ltd., Norfolk, England) on 108 cells. Respiration activities are expressed as nanomoles of oxygen consumed per milliliter per minute.
Analysis of the mitochondrial genome was performed by quantification of the number of copies and of the expression of the mitochondrial CtCYTb gene, which encodes cytochrome b, by real-time PCR and real-time reverse transcription-PCR as described below.
Sterol analysis.
Sterols were extracted from 50 mg lyophilized cells grown to stationary phase in YEPD broth. Extraction was performed by saponification at 90°C for 1 h in methanolic 40% (wt/vol) KOH in which pyrogallol was added. After cooling and addition of water, the unsaponified fraction was removed by the addition of 3 volumes of heptane. The UV spectrum was determined after desiccation of the heptanic fraction, and the amount of ergosterol was evaluated by determining the maximum absorbance at 281.5 nm (31). The sterol species of the heptanic fraction were then separated by gas chromatography with an AT-1 capillary column (25 m by 0.32 mm; Alltech Canada Biotechnology Centre Inc., Guelph, Canada) and finally identified by their relative retention times.
Flow cytometric analysis of the efflux of rhodamine 6G.
The activity of efflux pumps was evaluated by flow cytometry as previously described (3) by measuring the efflux of rhodamine 6G, a fluorescent dye which uses the same membrane transporter as azoles in some yeast species (14). Briefly, yeast cells (107) of each isolate grown in YEPD were suspended in 1 ml of culture medium containing 100 μM rhodamine 6G (Sigma Aldrich Ltd.). After incubation for 30 min at 30°C with constant shaking, the uptake of rhodamine 6G was stopped by cooling the tubes on ice. The reaction mixtures were then 40-fold diluted in cold sterile PBS, and the fluorescence of the cells was immediately quantified at 535 nm with a FACScan flow cytometer (BDIS). The cells were then washed three times in cold YEPD medium to remove excess rhodamine 6G, and efflux was evaluated after an additional incubation of 15 min at 30°C in the same medium by measuring the fluorescence of the cells after a 40-fold dilution in PBS. The data presented correspond to fluorescence frequency distribution histograms (number of fungal cells versus relative fluorescence intensity expressed as arbitrary units on a logarithmic scale).
For some experiments, the intracellular accumulation of the fluorescent dye was also measured, as described above, using cells previously incubated for 2 h in the presence of 1 mM potassium cyanide.
CtERG11 gene sequencing.
Five pairs of oligonucleotide primers presented in Table 1 were designed with the WebPrimer program (http://seq.yeastgenome.org/cgi-bin/web-primer) from the C. tropicalis ATCC 750 CtERG11 gene sequence (GenBank accession number M23673) and synthesized by Eurogentec (Seraing, Belgium). Genomic DNA was extracted from each isolate using the DNeasy Plant minikit (QIAGEN Inc., Valencia, Calif.) and used as the template for PCR amplification (5 min of denaturation at 94°C, followed by 30 cycles of 30 s at 94°C for denaturation, 40 s at 50°C for annealing, and 50 s at 72°C for elongation, and by a final elongation step of 10 min at 72°C). PCR products were purified with the High Pure PCR products purification kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's recommendations. They were then semiquantified by agarose gel electrophoresis and used as the template for sequencing PCR, which was performed with the Dye Terminator cycle sequencing quick start kit (Beckman Coulter Inc., Fullerton, Calif.). The sequencing PCR products were then purified through G50 Sepharose columns (Amersham Biosciences) and, finally, sequenced on a CEQ8000 DNA analysis system (Beckman Coulter) with the aforementioned forward and reverse primers.
TABLE 1.
Oligonucleotides used for CtERG11 gene sequencing and for the evaluation of CtERG11, CtMDR1, and CYTb gene expression
| Gene name (gene product) | GenBank accession no. | Primer | Nucleotide sequence (5′ to 3′) | Nucleotide coordinatesa |
|---|---|---|---|---|
| CtERG11 (lanosterol | M23673 | ERG11-1F | TCTGACATGGTGTGTGTGTG | +1 to +20 |
| 14α-demethylase) | ERG11-1R | ATTGATGCCATCAATGGCAG | +659 to +678 | |
| ERG11-2F | ATCCCACAGGCTTATTTGAAA | +568 to +588 | ||
| ERG11-2R | GGTCTCTTTCCTTGGTTTTG | +1170 to +1189 | ||
| ERG11-3F | TGCTGAAGAAGCTTATACCC | +981 to +1000 | ||
| ERG11-3R | CAAGGAATCAATCAAATCTCTC | +1603 to +1622 | ||
| ERG11-4F | GGTGGTCAACATACTTCTGC | +1561 to +1580 | ||
| ERG11-4R | AGCAGGTTCTAATGGTAAGG | +2171 to +2190 | ||
| ERG11-5F | AAACGGTGATAAGGTTCCAG | +2127 to +2146 | ||
| ERG11-5R | TCCCAAGACATCAAACCCTG | +2733 to +2752 | ||
| CtACT (β-actin) | AJ389059 | ACT F | TTTACGCTGGTTTCTCCTTGCC | +399 to +420 |
| ACT R | GCAGCTTCCAAACCTAAATCGG | +699 to +720 | ||
| CtMDR1 (MFS multidrug transporter) | AF194419 | MDR1 F | TAAAGCAGGCTGGAGATGGA | +327 to +346 |
| MDR1 R | ACAACCTCCAACTATAGCTA | +821 to +840 | ||
| CtCYTb (cytochrome b) | AB044933 | CYTb F | TGTCAGCCATCCCATTCATT | +8 to +27 |
| CYTb R | TGCGTAGAATGGTAAGAGGT | +377 to +396 |
Nucleotide coordinates refer to the corresponding gene sequence in the GenBank database.
mRNA extraction and real-time reverse transcription-PCR.
Total RNA was extracted from exponential-phase YEPD broth cultures. Cells were collected by centrifugation for 5 min at 3000 × g and then washed in sterile distilled water and resuspended in 2 ml of 50 mM sodium acetate, 10 mM EDTA, 10% (wt/vol) sodium dodecyl sulfate. Two milliliters of phenol was added, and the suspension was vigorously shaken. The suspension was incubated at 65°C for 4 min and then quickly frozen at −80°C for 15 min, reincubated at 65°C for thawing, and finally centrifuged. The aqueous supernatant was collected, and RNA was recovered by phenolic extraction followed by ethanol precipitation and finally resuspended in 100 μl of diethyl pyrocarbonate-treated water.
Total RNA was used to perform real-time reverse transcription-PCR in order to quantify the expression of the CtERG11, CtMDR1, and CtCYTb genes. Reverse transcription was performed on 1 μg of total RNA incubated for 1 h at 37°C in a thermocycler in the presence of 2.65 μg random oligonucleotides (Amersham Biosciences), 2 μl 10 mM each deoxynucleoside triphosphate, 40 U RNasin RNase inhibitor, and 200 U Moloney murine leukemia virus reverse transcriptase (Promega, Charbonnières, France) in a final volume of 50 μl. The obtained cDNAs were purified with the Qiaquick PCR purification kit (QIAGEN) by following the manufacturer's recommendations and diluted 10-fold in distilled water. Quantitative PCR was performed on a capillary real-time thermal cycler (LightCycler; Roche Diagnostics), and the results were normalized to β-actin mRNA. Amplification was done in a 10-μl final volume containing 1 μl from the LightCycler Fastart DNA Master SYBR green kit (Roche Diagnostics), 1 μl of primer mix (5 nM each), 1.2 μl MgCl2 (4 mM final concentration), 1.8 μl H2O, and 5 μl cDNA. The following run protocol was used: denaturation at 95°C for 10 min, followed by 40 cycles consisting of 15 s at 95°C for denaturation, 11 s at 54°C for annealing, and 22 s at 72°C for elongation. For the CtCYTb gene, a real-time PCR was also performed on 1 μg of total genomic DNA, without a reverse transcription step, in order to quantify the number of gene copies. Primers used to perform real-time PCR and real-time reverse transcription-PCR experiments are described in Table 1.
Nucleotide sequence accession numbers.
The CtERG11 gene sequences of the four C. tropicalis isolates were deposited in the GenBank database under the accession numbers AY942643, AY942644, AY942645, and AY942646 for isolates 21232, 21233, 21234, and 10264, respectively.
RESULTS
Antifungal susceptibility.
The disk diffusion assay revealed for isolate 21234 a complete resistance to all azoles tested except ketoconazole on RPMI 1640-glucose agar and Casitone agar, whereas the three other control isolates were susceptible to azoles (Table 2). Indeed, on Casitone agar plates, no inhibition zones were seen for isolate 21234 with azoles, whereas well-defined inhibition zones were observed for isolate 21232. On RPMI 1640-glucose agar, isolate 21234 showed a markedly decreased susceptibility to ketoconazole, with an inhibition zone diameter of 12.7 mm versus 25.3 to 28.7 mm for the three susceptible isolates (Table 2). On Casitone agar plates this isolate also showed a greater susceptibility to polyenes than the other isolates, but this result was not confirmed on RMPI-glucose agar (Table 2).
TABLE 2.
Susceptibilities of C. tropicalis isolates to polyene and azole drugsa
| Antifungal | Diam (mm) of the inhibition zone of the indicated isolate on:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Casitone
|
RPMI 1640
|
|||||||
| 21234 | 10264 | 21233 | 21232 | 21234 | 10264 | 21233 | 21232 | |
| Polyenes | ||||||||
| Amphotericin B | 32 | 24 | 27.3 | 28.7 | 22.7 | 20.3 | 24.7 | 23 |
| Nystatin | 35.3 | 25.7 | 29.3 | 31.3 | 24 | 22.7 | 24.7 | 24 |
| Azoles | ||||||||
| Clotrimazole | TR | 22.3 | 30 | 31.3 | TR | 19.3 | 26 | 22 |
| Ketoconazole | TR | 34 | 41.3 | 45.3 | 12.7 | 25.3 | 28.7 | 26.3 |
| Fluconazole | TR | 24.7 | 30.7 | 11.7 | TR | 30 | 33.7 | 31.3 |
| Voriconazole | TR | 21 | 34 | 22 | TR | 27 | 33 | 32 |
In vitro susceptibility testing was performed by a disk diffusion method on Casitone agar and RPMI 1640-glucose agar plates using Neosensitabs tablets (containing 1 μg of drug for voriconazole, 10 μg for amphotericin B and clotrimazole, 15 μg for fluconazole and ketoconazole, and 50 μg for nystatin). Results, which are expressed as mean values of three independent determinations, correspond to the diameters of growth inhibition zones. Standard deviations of the means represented less than 10%. TR, total resistance (no inhibition zone). Data were analyzed with the Wilcoxon-Mann-Whitney test at the unilateral risk α of 0.05. Mean diameters of the inhibition zones were statistically different between isolate 21234 and the three other isolates (i.e., smaller for azoles and larger for polyenes), except for amphotericin B and nystatin on RMPI 1640-glucose agar.
These results were in agreement with MICs determined for amphotericin B, voriconazole, ketoconazole, and fluconazole using the Etest procedure on Casitone agar and RPMI 1640-glucose agar plates (Table 3). Indeed, the amphotericin B MIC for isolate 21234 was lower than for the other isolates on Casitone agar exclusively, and no inhibition zones were seen for isolate 21234 with fluconazole and voriconazole on both culture media. Moreover, this method confirmed the discrepancy observed with isolate 21234 for ketoconazole by the disk diffusion method on Casitone and RPMI 1640-glucose agar, since the ketoconazole MIC was 1.5 μg/ml on RPMI 1640-glucose agar, whereas it was >32 μg/ml on Casitone agar. Determination of the clotrimazole MIC90 performed by the broth microdilution method confirmed the decreased susceptibility of isolate 21234, as evidenced by the disk diffusion method (Table 3).
TABLE 3.
MICs of polyene and azole drugs for C. tropicalis isolatesa
| Antifungal | MIC (μg/ml) of indicated isolate on:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Casitone
|
RPMI 1640
|
|||||||
| 21234 | 10264 | 21233 | 21232 | 21234 | 10264 | 21233 | 21232 | |
| Amphotericin B | 0.037 | 0.064 | 0.058 | 0.042 | 0.029 | 0.064 | 0.024 | 0.094 |
| Clotrimazole | >250 | 1 | 1 | 1 | >250 | 4 | 8 | 1 |
| Ketoconazole | >32 | 0.02 | 0.024 | 0.02 | 1.5 | 0.052 | 0.021 | 0.074 |
| Fluconazole | >256 | 4 | 3 | 9 | >256 | 0.43 | 0.27 | 0.75 |
| Voriconazole | >32 | 0.6 | 0.025 | 0.046 | >32 | 0.104 | 0.115 | 0.147 |
Results are expressed as mean values of three independent determinations. Data were obtained by the Etest procedure using antifungal strips on Casitone agar and RPMI 1640-glucose agar plates, except for clotrimazole, which was not available as an Etest strip and which was therefore tested using the microbroth dilution method. Data were analyzed with the Wilcoxon-Mann-Whitney test at unilateral risk α of 0.05. MICs were statistically different between isolate 21234 and the three other isolates (i.e., higher for azoles and lower for amphotericin B), except for amphotericin B on RMPI 1640-glucose agar.
Respiratory activity.
The activity of the mitochondrial respiratory chain was analyzed by flow cytometry after staining of the cells with rhodamine 123. The azole-resistant isolate incorporated more fluorescent dye than the three susceptible isolates, with a mean fluorescence intensity of the cells of 2,150 arbitrary units, versus an average of 1,650 for the three others. Moreover, the respiratory activity of the resistant isolate was more sensitive to sodium azide, with a threefold decrease in mean fluorescence intensity of the cells after their preincubation with this inhibitor (down to 770 arbitrary units), whereas it remained almost unchanged for susceptible isolates (data not shown).
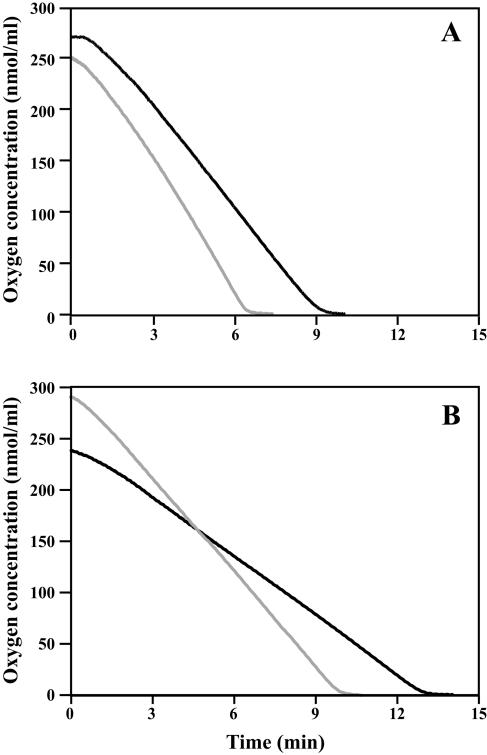
The oxygen consumption rate of the four isolates, determined by oxygraphy, confirmed the higher respiratory activity of isolate 21234 as suggested by flow cytometry. Indeed, its respiration rate was 44 nmol/ml/min versus an average of 25 nmol/ml/min for susceptible isolates (Fig. 1). These results led us to investigate the respiratory activity at a biomolecular level, by quantifying the CtCYTb gene mRNA level and copy number. However, no differences were seen between resistant and susceptible isolates (data not shown).
FIG. 1.
Oxygen consumption by C. tropicalis isolates 21234 (A, gray line), 21232 (A, black line), 10264 (B, gray line), and 21233 (B, black line). Comparison of the respiration rates showed an increase in oxygen consumption by resistant cells (42.9 nmol · ml−1 · min−1 versus 33, 18.9, and 30.6 nmol · ml−1 · min−1 for susceptible cells 21232, 10264, and 21233, respectively).
Sterol composition.
Analysis of the sterol content by gas chromatography did not reveal qualitative differences between resistant and susceptible isolates. The same molecular species were detected for all isolates, with ergosterol always being the prominent sterol, with a peak area ranging from 78.9% to 92.5% of the total sterols, depending on the isolates. However, spectrophotometric evaluation of the sterol content showed an accumulation of ergosterol in the cells of the azole-resistant isolate 21234, with an ergosterol content 1.5-fold higher than in cells of susceptible isolates (1.6 μg/mg [dry weight] for isolate 21234 versus 0.95, 1.09, and 0.94 μg/mg for isolates 21232, 21233, and 10264, respectively).
Uptake and efflux of rhodamine 6G.
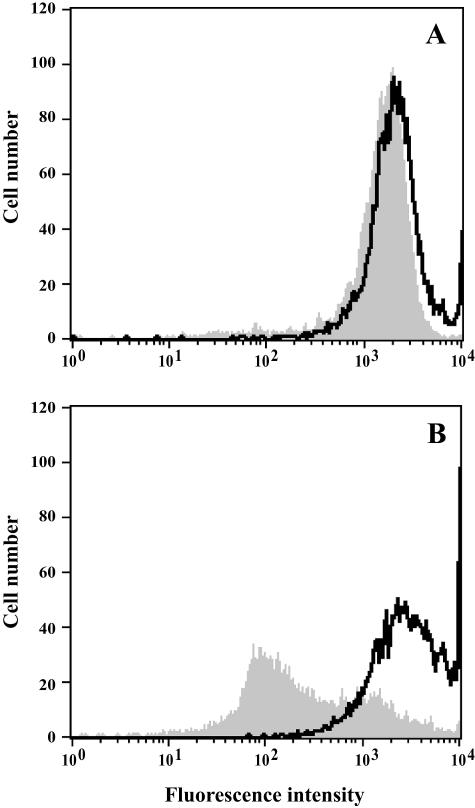
The accumulation and efflux of rhodamine 6G were quantified by flow cytometry. As illustrated in Fig. 2, the azole-resistant isolate incorporated less rhodamine 6G than susceptible isolates. After a 30-min incubation in YEPD broth with the fluorochrome, the mean fluorescence intensity of the cells was lower for isolate 21234 than for wild-type isolates (2,150 arbitrary units against an average of 2,850 for susceptible isolates). Moreover, the efflux of rhodamine 6G was very low for isolate 21234, since the residual fluorescence intensity after removal of the free dye and an additional 15-min incubation corresponded to 76% of the total quantity accumulated, whereas it was only 27%, 52%, and 44% for isolates 21232, 21233, and 10264, respectively.
FIG. 2.
Flow cytometric analysis of rhodamine 6G uptake and efflux. Uptake of fluorochrome was quantified by incubating cells of C. tropicalis isolates 21234 (A) and 21232 (B) in YEPD broth for 30 min with 100 μM rhodamine 6G (black line). Then, efflux was evaluated by quantifying the residual fluorescence of the cells after removal of the free dye and an additional incubation of 15 min in YEPD broth (gray area).
In addition, prior incubation of the cells for 2 h in the presence of potassium cyanide in order to deplete them of their endogenous ATP resulted in a 30% increase in the mean fluorescence intensity of the cells, whatever the isolate. This suggested that the lower accumulation of the dye in a resistant isolate than in susceptible ones was not related to a high intrinsic efflux capacity.
Analysis of CtERG11 gene sequences.
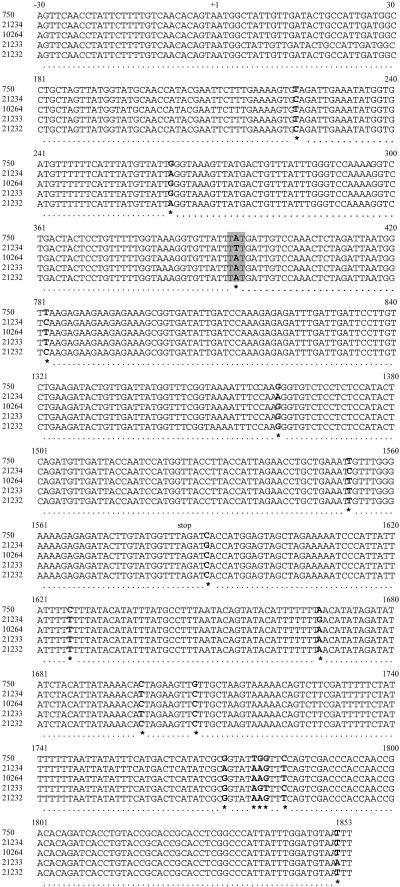
Comparison of the CtERG11 gene sequences of the four isolates studied with the available corresponding sequence in the GenBank database (accession number M23673) revealed several point mutations (Fig. 3). Indeed, some nucleotide substitutions located outside the coding sequence were detected: four in isolate 10264, five in isolates 21232 and 21233, and seven in isolate 21234. In addition, isolates 21232 and 21234 shared two point mutations in their coding sequences (T224C and G263A) that did not affect the deduced amino acid sequence for CtERG11p. The CtERG11 sequence of isolate 21234 presented three other silent mutations, T781C, G1360A, and T1553C. A missense mutation (A393T) was also found for isolate 21234, leading to the substitution of tyrosine by phenylalanine at position 132 in the deduced lanosterol 14α-demethylase sequence. In addition, analysis of chromatograms revealed a heterozygoty for some of the silent mutations (T224C and G1361A for the resistant isolate) or of the mutations located outside the coding sequence (A1668G for the resistant isolate and C1698T, T1777A, and C1782T for the susceptible isolate 21233). All other mutations, including the missense mutation A393T detected for the resistant isolate 21234, were shared by the two alleles.
FIG. 3.
CtERG11 gene sequence alignment of five C. tropicalis isolates (10264, 21232, 21233, and 21234 [this study] and GenBank strain 750). Only parts of DNA sequences showing differences between isolates are presented, and the number of the corresponding isolate is indicated on the left of each line. Numbers on the top of each five-line group indicate the position relative to the open reading frame. Start (+1) and stop codons are also indicated. Each mutation is positioned below the alignment by an asterisk, and corresponding nucleotides are in bold characters. The point mutation (A393T) leading to an amino acid substitution in the deduced protein sequence of the CtERG11 gene of the azole-resistant isolate 21234 is highlighted by a gray background.
Expression levels of the CtMDR1 and CtERG11 genes.
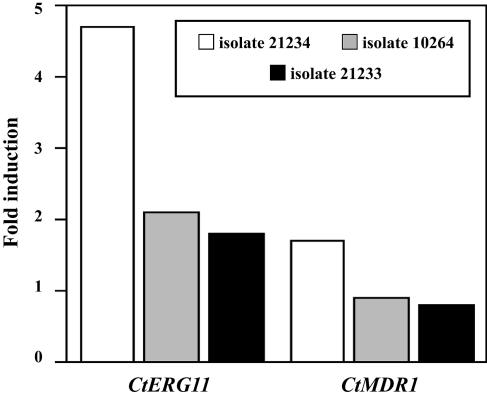
Quantification of the expression of the CtMDR1 and CtERG11 genes was performed by real-time reverse transcription-PCR. The MFS transporter and lanosterol 14α-demethylase mRNA values were normalized to that for β-actin mRNA. The specificity of the amplification reaction was verified by determination of the lengths of the PCR products by agarose gel electrophoresis (data not shown). Figure 4 shows the induction factors calculated from three independent experiments for isolates 21234, 10264, and 21233 compared to that for isolate 21232. No significant difference was found between the CtMDR1 mRNA levels of the four isolates tested, as the induction factors did not exceed 1.7 compared to that for isolate 21232. In contrast, the CtERG11 mRNA level was approximately fivefold higher in the azole-resistant isolate 21234 than in isolate 21232, whereas it was increased approximatively twofold for the two other isolates, 10264 and 21233.
FIG. 4.
Quantification of the relative transcript levels of CtERG11 and CtMDR1 genes from C. tropicalis isolates 21234, 10264, and 21233 compared to those of isolate 21232. Measured quantities of each mRNA were normalized using the expression level of the CtACT gene. Results, which are mean values from triplicate experiments, represent the numbers of times the genes are expressed compared to that in isolate 21232.
DISCUSSION
Little information is available concerning the molecular mechanisms leading to azole resistance in the pathogenic yeast C. tropicalis. However, due to the extensive use of antifungals in the prophylaxis or treatment of candidiasis, azole-resistant clinical isolates are increasingly reported for this species (18, 33, 38). Here we studied a clinical isolate of C. tropicalis presenting a resistance to azole antifungals and determined its resistance mechanisms.
Determination of the susceptibility of C. tropicalis to azoles by a disk diffusion method on Casitone and RPMI 1640-glucose agar showed a complete resistance (or a markedly decreased susceptibility) to all azoles tested. In contrast, this isolate showed an increased susceptibility to polyene drugs compared to three azole-susceptible isolates used as controls, but only on Casitone agar. Interestingly, determination of antifungal MICs by the Etest procedure or the broth microdilution method confirmed the markedly decreased susceptibility of isolate 21234 to ketoconazole and its complete resistance to the other azoles tested. Such differences in antifungal activity between ketoconazole and the other azole antifungals, including triazoles, have already been reported for azole-resistant clinical isolates of C. glabrata (35). Moreover, a greater antifungal activity of ketoconazole versus fluconazole has been shown in vitro against C. albicans, although fluconazole presented the higher efficiency in vivo (24).
As suggested by previous studies performed in our hospital laboratory or by other groups on petite mutants of Saccharomyces cerevisiae (9) and C. glabrata (2, 3, 6), cross-resistance to azoles may be linked to a loss of respiratory activity. Relationships between mitochondrial functions and azole susceptibility are not surprising considering the extensive cross talk that exists between the nucleus and mitochondria (20, 34). Thus, we analyzed the respiratory status of our azole-resistant clinical isolate. Strikingly, we did not find a loss of respiratory activity in the resistant isolate 21234 but in contrast found an increased respiration. Indeed, cytometric analysis of the cell fluorescence showed an increased uptake of rhodamine 123 after incubation of the cells with the fluorochrome and a more pronounced reduction when incubation with the fluorescent dye was performed in the presence of the mitochondrial respiratory chain inhibitor sodium azide. This increased respiratory activity, which was confirmed by the evaluation of oxygen consumption by oxygraphy, was not, however, linked to a modification of mitochondrial genome expression. Indeed, neither a multiplication of CtCYTb gene copy number nor an increased expression of this mitochondrial gene could explain the higher respiratory activity of this clinical isolate, and other mechanisms must be considered. Indeed, as suggested by Lupetti et al. (13), as well as by studies using petite mutants (2, 27), a link between respiration and susceptibility to azoles could exist in Candida yeasts.
Several mechanisms have been described for azole resistance in clinical isolates of C. albicans and C. glabrata, the most frequent being an increased efflux of the drug. However, analysis of the uptake and efflux of rhodamine 6G, which uses the same transporters as azoles in yeasts (14), suggested that overexpression of efflux pumps was not involved in the azole resistance of our isolate. Indeed, the efflux of rhodamine 6G was very low in this isolate compared to that of the azole-susceptible isolates. Likewise, real-time reverse transcription-PCR analysis of the expression of the CtMDR1 gene, which encodes an MFS transporter, did not reveal an overexpression of this gene. This was not surprising since MFS transporters are known for their relative specificity for fluconazole among azole drugs, whereas overexpression of genes encoding ABC proteins are usually responsible for a cross-resistance to all azole antifungals (29). So an increased expression of CtMDR1 is unlikely to be responsible for the susceptibility pattern of our clinical isolate. No ABC transporters in C. tropicalis have been characterized as yet. However, Barchiesi et al. (1) have demonstrated by Northern blotting, using C. albicans CDR1 as a probe, the presence of a homologous gene in C. tropicalis. Although flow cytometry experiments performed on native cells, as well as on cells previously depleted from their endogenous ATP, did not show increased activity of the efflux pumps and therefore favor the prediction of a modest contribution—or even no contribution—of azole efflux as a potential resistance mechanism, an upregulation of as-yet-unknown C. tropicalis ABC transporter genes cannot be ruled out.
Sterols were also analyzed qualitatively and quantitatively in order to determine if changes in the ergosterol biosynthesis pathway could be responsible for azole resistance in this clinical isolate. The major change consisted in an increased ergosterol content in cells of the resistant isolate 21234. This may be related to its greater susceptibility to polyene drugs on Casitone agar, as revealed by the disk diffusion method and confirmed by determination of the amphotericin B MIC using the Etest procedure. Induction of ergosterol biosynthesis also suggested an increased expression of the azole target, lanosterol 14α-demethylase, which could be responsible for the azole resistance of this isolate. Accordingly, an increase in the mRNA level of the CtERG11 gene, which encodes the lanosterol 14α-demethylase, was found by real-time reverse transcription-PCR in the resistant isolate 21234 compared to the level in control isolates. Overexpression of ERG11 may arise from an augmentation of the mRNA half-life by modification of their 3′-untranslated region. Likewise, a modification in the ERG11 gene promoter region, as well as a chromosome duplication leading to multiplication of the ERG11 gene, may also lead to an increase in mRNA level. This last mechanism has already been described for an azole-resistant isolate of C. glabrata, in which Marichal et al. (16) showed an overexpression of ERG11 due to the duplication of the entire chromosome bearing the ERG11 gene. Further investigations are needed to determine the precise mechanism of increased CtERG11 mRNA expression in isolate 21234.
Although less frequent than increased efflux of azole drugs, point mutations in the ERG11 gene may lead to a decrease in the affinity of azoles for their target. Besides silent nucleotide mutations, an adenine mutation to thymine at position 393 was found in the CtERG11 sequence of isolate 21234, leading to a mutation from tyrosine to phenylalanine at position 132 in the enzyme sequence. Such a modification in the amino-acid sequence could lead to structural changes sufficient to induce a decreased affinity of azoles but still permitting the enzyme activity. Several point mutations in the ERG11 gene sequence, some of them leading to a decreased affinity of azoles for their target, have been described to occur in other yeast species (15). At least 12 have been reported to occur in azole-resistant clinical isolates of C. albicans and 7 in C. glabrata, and the mutations most frequently associated with azole resistance were Y132H, D278E, S405F, G464S, and R467K. In C. albicans, the tyrosine 132 substitution of ERG11p has already been described by N. S. Ryder and B. Favre (Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-13, 1997), and Sanglard et al. (28) demonstrated its involvement in azole resistance, since this mutation induced a decrease in the affinity of azoles for lanosterol 14α-demethylase. In azole-resistant clinical isolates of C. tropicalis, only Loeffler et al. (11) have investigated the CtERG11 gene sequence as a possible resistance mechanism. A point mutation (T1554C), recovered eight times in a set of 21 isolates, was identified, but this mutation was silent and therefore it was not linked to azole resistance.
To our knowledge, this study constitutes the first exhaustive analysis of the mechanisms responsible for azole resistance in a clinical isolate of C. tropicalis. This isolate presented a resistance or markedly decreased susceptibility to all azole antifungals tested. Although increased efflux is the most frequent mechanism involved in azole resistance, the decreased susceptibility to azoles of this isolate seemed to be due to an overexpression of CtERG11, associated with a missense mutation of this gene. However, due to the absence of matched susceptible isolates, one cannot disregard the possibility that the difference observed in CtERG11 expression between resistant and control isolates may be due to strain variations. Moreover, an increased respiratory activity was seen for this isolate by flow cytometry and oxygraphy. Further experiments are needed to determine if it plays a role in the azole resistance of this isolate. In addition, it would be interesting to investigate the respiratory activities of other azole-resistant clinical yeast isolates.
Acknowledgments
We thank Pascal Reynier (EMI-U00.18, Centre Hospitalier Universitaire, Angers, France) and Alain Morel (INSERM U564, Centre Hospitalier Universitaire, Angers, France) for their help in sequence analysis of the CtERG11 gene and quantitative PCR analysis of CtERG11, CtMDR1, and CtCYTb gene expression and Dorian McIlroy (CNRS UMR 6204, Nantes, France) for reading the manuscript.
REFERENCES
- 1.Barchiesi, F., D. Calabrese, D. Sanglard, L. Falconi Di Francesco, F. Caselli, D. Giannini, A. Giacometti, S. Gavaudan, and G. Scalise. 2000. Experimental induction of fluconazole resistance in Candida tropicalis ATCC 750. Antimicrob. Agents Chemother. 44:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brun, S., C. Aubry, O. Lima, R. Filmon, T. Bergès, D. Chabasse, and J.-P. Bouchara. 2003. Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun, S., T. Bergès, P. Poupard, C. Vauzelle-Moreau, G. Renier, D. Chabasse, and J.-P. Bouchara. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casalinuovo, I. A., P. Di Francesco, and E. Garaci. 2004. Fluconazole resistance in Candida albicans: a review of mechanisms. Eur. Rev. Med. Pharmacol. Sci. 8:69-77. [PubMed] [Google Scholar]
- 5.Colombo, A. L., and T. Guimaraes. 2003. Epidemiology of hematogenous infections due to Candida spp. Rev. Soc. Bras. Med. Trop. 36:599-607. [DOI] [PubMed] [Google Scholar]
- 6.Defontaine, A., J.-P. Bouchara, P. Declerk, C. Planchenault, D. Chabasse, and J.-N. Hallet. 1999. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48:663-670. [DOI] [PubMed] [Google Scholar]
- 7.Hsueh, P.-R., Y.-J. Lau, Y.-C. Chuang, J.-H. Wan, W.-K. Huang, J.-M. Shyr, J.-J. Yan, K.-W. Yu, J.-J. Wu, W.-C. Ko, Y.-C. Yang, Y.-C. Liu, L.-J. Teng, C.-Y. Liu, and K.-T. Luh. 2005. Antifungal susceptibilities of clinical isolates of Candida species, Cryptococcus neoformans, and Aspergillus species from Taiwan: surveillance of multicenter antimicrobial resistance in Taiwan program data from 2003. Antimicrob. Agents Chemother. 49:512-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, M. D., and J. R. Perfect. 2003. Caspofungin: first approved agent in a new class of antifungals. Expert Opin. Pharmacother. 4:807-823. [DOI] [PubMed] [Google Scholar]
- 9.Kontoyiannis, D. P. 2000. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:191-197. [DOI] [PubMed] [Google Scholar]
- 10.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50:243-260. [DOI] [PubMed] [Google Scholar]
- 11.Loeffler, J., L. Hagmeyer, H. Hebart, N. Henke, U. Schumacher, and H. Einsele. 2000. Rapid detection of point mutations by fluorescence resonance energy transfer and probe melting curves in Candida species. Clin. Chem. 46:631-635. [PubMed] [Google Scholar]
- 12.Loeffler, J., and D. A. Stevens. 2003. Antifungal drug resistance. Clin. Infect. Dis. 36:31-41. [DOI] [PubMed] [Google Scholar]
- 13.Lupetti, A., A. Paulusma-Annema, M. M. Welling, H. Dogterom-Ballering, C. P. J. M. Brouwer, S. Senesi, J. T. van Dissel, and P. H. Nibbering. 2003. Synergistic activity of the N-terminal peptide of human lactoferrin and fluconazole against Candida species. Antimicrob. Agents Chemother. 47:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maesaki, S., P. Marichal, H. Vanden Bossche, D. Sanglard, and S. Kohno. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27-31. [DOI] [PubMed] [Google Scholar]
- 15.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 16.Marichal, P., H. Vanden Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. Van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morschhäuser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 18.Myoken, Y., T. Kyo, M. Fujihara, T. Sugata, and Y. Mikami. 2004. Clinical significance of breakthrough fungemia caused by azole-resistant Candida tropicalis in patients with hematologic malignancies. Haematologica 89:378-380. [PubMed] [Google Scholar]
- 19.Nissapatorn, V., C. Lee, Q. K. Fatt, and K. A. Abdullah. 2003. AIDS-related opportunistic infections in Hospital Kuala Lumpur. Jpn. J. Infect. Dis. 56:187-192. [PubMed] [Google Scholar]
- 20.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genome can influence nuclear gene expression in yeast. Science 235:576-580. [DOI] [PubMed] [Google Scholar]
- 21.Perfect, J. R. 2004. Antifungal resistance: the clinical front. Oncology 18:15-22. [PubMed] [Google Scholar]
- 22.Polak, A. 2003. Antifungal therapy—state of the art at the beginning of the 21st century. Prog. Drug Res. Spec. No. 59-190. [DOI] [PubMed]
- 23.Price, M. F., M. T. LaRocco, and L. O. Gentry. 1994. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob. Agents Chemother. 38:1422-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers, T. E., and J. N. Galgiani. 1986. Activity of fluconazole (UK 49,858) and ketoconazole against Candida albicans in vitro and in vivo. Antimicrob. Agents Chemother. 30:418-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronot, X., L. Benel, M. Adolphe, and J. C. Mounolou. 1986. Mitochondrial analysis in living cells: the use of rhodamine 123 and flow cytometry. Biol. Cell 57:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Sanglard, D. 2002. Clinical relevance of mechanisms of antifungal drug resistance in yeasts. Enferm. Infecc. Microbiol. Clin. 20:462-469. [DOI] [PubMed] [Google Scholar]
- 27.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 30.Sanguinetti, M., B. Posteraro, B. Fiori, S. Ranno, R. Torelli, and G. Fadda. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 49:668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Servouse, M., and F. Karst. 1986. Regulation of early enzymes of ergosterol biosynthesis in Saccharomyces cerevisiae. Biochem. J. 240:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, A., I. Bairy, and P. G. Shivananda. 2003. Spectrum of opportunistic infections in AIDS cases. Indian J. Med. Sci. 57:16-21. [PubMed] [Google Scholar]
- 33.Tortorano, A. M., A. L. Rigoni, E. Biraghi, A. Prigitano, M. A. Viviani, and the FIMUA-ECMM Candidaemia Study Group. 2003. The European Confederation of Medical Mycology (ECMM) survey of candidaemia in Italy: antifungal susceptibility patterns of 261 non-albicans Candida isolates from blood. J. Antimicrob. Chemother. 52:679-682. [DOI] [PubMed] [Google Scholar]
- 34.Traven, A., J. M. S. Wong, D. Xu, M. Sopta, and C. J. Ingles. 2001. Interorganellar communication: altered nuclear gene expression profiles in a yeast mitochondrial mutant. J. Biol. Chem. 276:4020-4027. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Bossche, H., P. Marichal, F. C. Odds, L. Le Jeune, and M. C. Coene. 1992. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 36:2602-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermitsky, J. P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wingard, J. R. 1995. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin. Infect. Dis. 20:115-125. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Y. L., Y. A. Ho, H. H. Cheng, M. Ho, and H. J. Lo. 2004. Susceptibilities of Candida species to amphotericin B and fluconazole: the emergence of fluconazole resistance in Candida tropicalis. Infect. Control Hosp. Epidemiol. 25:60-64. [DOI] [PubMed] [Google Scholar]