Abstract
A retrospective study was performed to identify methicillin-resistant Staphylococcus aureus (MRSA) isolates obtained from patients enrolled in phase 3 clinical trials for tigecycline that were genotypically similar to known community-associated MRSA (CA-MRSA) strains. The clinical trials were double-blind comparator studies for complicated skin and skin structure infections or complicated intra-abdominal infections. We obtained 85% of the MRSA isolates from patients with complicated skin and skin structure infections. Using ribotyping, MRSA isolates were compared with well-characterized North American CA-MRSA strains and negative-control hospital-associated (HA) MRSA strains by cluster analysis; 91 of the 173 isolates clustered with two groups of known CA-MRSA strains, 60% of which shared an indistinguishable ribotype. These isolates were subsequently tested for the presence of SCCmec type IV and the Panton-Valentine leukocidin (PVL)-encoding genes as well as susceptibility to clindamycin, characteristics that are typically associated with CA-MRSA; 89 of the 91 isolates carried the type IV SCCmec element and 76 were also positive for the PVL-encoding genes; 73 of these isolates were susceptible to clindamycin. A similar analysis performed on 26 nonclustering isolates identified only four with these characteristics; 89 of the 91 clustering isolates were inhibited by tigecycline at MICs of ≤0.5 μg/ml. On the basis of clustering information and preliminary genetic characterization, it appears that ribotyping is a useful tool in identifying potential CA-MRSA isolates and 76 MRSA isolates from patients enrolled in the tigecycline phase 3 trials have genetic markers typically associated with CA-MRSA.
Staphylococcus aureus is the causative agent for a wide variety of human diseases ranging from superficial skin infections to life-threatening conditions such as pneumonia, sepsis, and endocarditis (46). Infections caused by methicillin-resistant S. aureus (MRSA) have traditionally been confined to the healthcare setting. Treatment options for MRSA are often limited due to the resistance of this pathogen to multiple antibiotics, including most β-lactam antibiotics (44). However, the prevalence of community-associated MRSA (CA-MRSA) infections has increased in recent years and is a growing public health concern (41). CA-MRSA infections are most commonly associated with skin and soft tissue infections (12) and cases are often reported among institutionalized populations and sports participants (23, 38). Misdiagnosis of these infections can result in inappropriate therapy.
Clinically, CA-MRSA has been defined as an MRSA isolate obtained within 48 h of hospitalization from a patient with no known risk factors for MRSA infections (19, 33). Established risk factors include previous hospitalization or contact with the healthcare setting, dialysis, presence of an indwelling catheter or percutaneous device or previous isolation of MRSA (19, 33). Evidence from epidemiological and molecular typing studies suggests that CA-MRSA isolates are genetically distinct from hospital-associated MRSA (HA-MRSA) isolates (5, 8, 36, 45).
A number of other features have been identified that distinguish CA-MRSA from HA-MRSA isolates. Unlike HA-MRSA, CA-MRSA isolates are typically susceptible to most non-β-lactam antibiotics. For example, clindamycin susceptibility has been shown to correlate with CA-MRSA, whereas nosocomial isolates are typically resistant (17). CA-MRSA isolates carry the type IV staphylococcal chromosomal cassette (SCC)mec element encoding β-lactam resistance (10). This element differs from SCCmec types I to III that are commonly associated with HA-MRSA in its smaller size and lack of non-β-lactam resistance determinants (28).
The Panton-Valentine leukocidin (PVL)-encoding genes lukF and lukS are commonly found in CA-MRSA isolates and this toxin is associated with skin infections and severe necrotizing pneumonia (27). Vandenesch and colleagues described the PVL-encoding locus as a stable marker for CA-MRSA (45). However, a recent study suggests that while the prevalence of PVL is significantly increased in CA-MRSA isolates, its presence is not uniform (40). In addition to these genetic differences, CA-MRSA isolates grow significantly faster than HA-MRSA isolates (36). The presence of the PVL-encoding locus and SCCmec type IV and the higher growth rate of CA-MRSA isolates may confer a selective advantage for community-based MRSA pathogens.
In the current study, potential CA-MRSA were retrospectively identified among isolates obtained from patients enrolled in the phase 3 trials of tigecycline, a new broad-spectrum glycylcycline antibiotic recently approved for complicated skin and skin structure infections and complicated intra-abdominal infections (3, 14). The utility of ribotyping was evaluated as a screening tool to identify MRSA isolates that had similar genotypes to previously characterized CA-MRSA and to further characterize these isolates by testing for genetic markers that are commonly associated with CA-MRSA.
MATERIALS AND METHODS
Bacterial strains.
173 MRSA isolates were collected from 171 patients suffering from complicated skin and skin structure infections or complicated intra-abdominal infections enrolled in phase 3 double blind comparator trials for tigecycline (only one representative isolate for each ribotype was included for patients with multiple isolates). The control strains described in Table 1 were previously defined as CA-MRSA on the basis of clinical definitions or genetic characterization. Positive control strains included MW2 (2) (the prototype CA-MRSA strain) and its methicillin susceptible progenitor, MSSA-476 (22). The CA-MRSA control strains carry SCCmec type IV and have genetic backgrounds characterized by multilocus sequence typing (MLST), spa type, or pulsed-field gel electrophoresis (PFGE).
TABLE 1.
Control strains used in this study
| Position in clustera | Group | Ribo group | Strain name | MLSTb | PFGE typeb | spa typeb | SCCmec type | PVL locus | Strain typec | Refer- ence(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | A4 | BK11235 | ST8 | 7; YHGCMBQBLO | IV | No | CA-MRSA | 40 | |
| 2 | 1 | A4 | BK10654 | ST8 | 7; YHGCMBQBLO | IV | No | CA-MRSA | 40 | |
| 3 | 1 | A4 | BK2443 | ST8 | 7; YHGCMBQBLO | IV | Yes | CA-MRSA | 40 | |
| 4 | 1 | A4 | BK11108 | ST8 | 7; YHGCMBQBLO | IV | No | CA-MRSA | 40 | |
| 5 | 1 | A4 | COL | ST250 | I | No | Archaic MRSA | 20 | ||
| 6 | 1 | A2 | BK11490 | ST8 | 1; YHGFMBQBLO | IV | Yes | CA-MRSA | 40 | |
| 7 | 1 | A2 | BK11472 | ST8 | 7; YHGCMBQBLO | IV | Yes | CA-MRSA | 40 | |
| 8 | 1 | A2 | BK11358 | ST8 | 1; YHGFMBQBLO | IV | Yes | CA-MRSA | 40 | |
| 9 | 1 | A5 | CDC 3 | ST8 | USA300 | IV | Yes | CA-MRSA | 13, 30 | |
| 10 | 1 | A5 | CDC 4 | ST8 | USA300 | IV | Yes | CA-MRSA | 13, 30 | |
| 11 | 1 | A5 | CDC 1 | ST8 | USA300 | IV | Yes | CA-MRSA | 13, 30 | |
| 12 | 1 | A5 | CDC 6 | ST8 | USA300 | IV | Yes | CA-MRSA | 13, 30 | |
| 13 | 2 | H5 | CDC 13 | ST1 | USA400 | IV | No | CA-MRSA | 13, 30 | |
| 14 | 2 | H5 | MSSA-476 | ST1 | MSSA | No | CA-MSSA | 22 | ||
| 15 | 2 | H5 | CDC 9 | ST1 | USA400 | IV | Yes | CA-MRSA | 13, 30 | |
| 16 | 2 | H5 | CDC 10 | ST1 | USA400 | IV | No | CA-MRSA | 13, 30 | |
| 17 | 2 | H5 | CDC 12 | ST1 | USA400 | IV | Yes | CA-MRSA | 13, 30 | |
| 18 | 2 | H5 | MW2 | ST1 | IV | Yes | CA-MRSA | 2, 22 | ||
| 19 | 2 | G7 | BK10370 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 20 | 2 | G7 | BK11514 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 21 | 2 | G7 | BK11632 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 22 | 2 | G2 | BK11580 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 23 | 2 | G2 | BK11118 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 24 | nc | J10 | BK2370 | ST8 | 7; YHGCMBQBLO | IV | No | CA-MRSA | 40 | |
| 25 | nc | M13 | BK9362 | ST8 | 7; YHGCMBQBLO | II | No | HA-MRSA | 40 | |
| 26 | nc | A9 | BK10484 | ST8 | 1; YHGFMBQBLO | IV | No | CA-MRSA | 40 | |
| 27 | nc | A15 | BK10474 | ST8 | 1; YHGFMBQBLO | IV | Yes | CA-MRSA | 40 | |
| 28 | nc | O7 | BK2620 | 35; UJFKBPE | II | No | HA-MRSA | 40 | ||
| 29 | nc | K16 | BK10398 | ST1 | 131; UJJFKBPE | IV | Yes | CA-MRSA | 40 | |
| 30 | nc | K11 | BK9360 | ST8 | 1; YHGFMBQBLO | II | No | HA-MRSA | 40 | |
| 31 | nc | P18 | CDC 16 | ST5 | USA800 | IV | No | HA-MRSA | 13, 30 | |
| 32 | nc | 112 | BK6909 | 17; ZDMDMNKB | IV | No | CA-MRSA | 40 | ||
| 33 | nc | Q19 | CDC 17 | ST5 | USA100 | II | No | HA-MRSA | 13, 30 | |
| 34 | nc | Q19 | Mu50 | II | No | HA-MRSA | 22, 24 | |||
| 35 | nc | Q22 | N315 | II | No | HA-MRSA | 22, 24 | |||
| 36 | nc | R20 | CDC 18 | ST36 | USA200 | II | No | HA-MRSA | 13, 30 | |
| 37 | nc | S21 | MRSA-252 | ST36 | USA200 | II | No | HA-MRSA | 22 | |
| 38 | nc | N14 | BK10488 | 267; TAAMBMDMGMK | IV | No | CA-MRSA | 40 |
Strains belong to group 1 or group 2 or were nonclustering (nc) strains as illustrated in Fig. 1.
Information regarding pulsed-field type (U.S. designations) or spa type is provided where available.
CA-MSSA, community-associated methicillin-susceptible S. aureus. These designations were made according to criteria outlined in the relevant references.
The BK positive-control strains belong to a collection of 121 geographically diverse North American isolates, representatives of which were described previously (40). The Centers for Disease Control and Prevention positive control strains are community onset isolates belonging to PFGE types USA300 and USA400 (30). Negative control HA-MRSA strains used in this study included 3 isolates from the Centers for Disease Control and Prevention (CDC) with pulsed-field types USA100, 200, and 800, which are predominantly associated with the US healthcare setting and HA-MRSA strains with SCCmec type II, including the sequenced strains N315 (24), Mu50 (24), and MRSA-252 (22). The archaic MRSA strain COL (20) with ST250 was also included.
Ribotyping.
MRSA isolates were ribotyped using the RiboPrinter microbial characterization system (Qualicon, Wilmington, DE) according to the manufacturer's instructions. Each isolate was analyzed with two restriction enzymes, EcoRI and PvuII and ribotypes were assigned using a letter for unique EcoRI patterns and a number for unique PvuII patterns. Clustering analysis was performed using Bionumerics software (Applied Maths, Austin, TX).
Susceptibility testing.
Tigecycline MICs were determined using the broth microdilution method with fresh (<12 h old) Mueller Hinton II broth using methods outlined by the Clinical Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (9, 34). Susceptibility to tigecycline was defined using the Food and Drug Administration-approved breakpoint for S. aureus (47) of ≤0.5 μg/ml. Methicillin-resistance was defined as an oxacillin MIC of ≥4 μg/ml. The susceptibility of isolates to clindamycin and erythromycin was determined by the disk diffusion method (35). An inducible clindamycin resistant phenotype was detected by placing erythromycin and clindamycin disks 15 to 26 mm apart on Mueller-Hinton Agar plates as detailed by the CLSI (35). A flattened (D-shaped) zone of inhibition around the clindamycin disk on the side facing the erythromycin disk was indicative of an inducible clindamycin resistant phenotype (25).
PCR analysis.
All PCR amplifications were carried out using the FailSafe system (Epicenter, Madison, WI) with buffer C. The SCCmec type was determined by the method of Oliveira and de Lencastre (37) using a 59°C annealing temperature with a 30-s extension time. Further confirmation of SCCmec type IV was achieved by testing for the presence of ccrA type 2 using the method described by Okuma et al. (36). The PVL-encoding genes were detected using the primers and conditions described by Lina et al. (27). The following S. aureus strains were used as controls: COL (SCCmec type I, PVL negative), N315 (SCCmec type II, PVL negative), ATCC BAA-39 (SCCmec type III, PVL negative) (11), and MW2 (SCCmec type IV, PVL positive).
RESULTS
Comparison of test MRSA isolates with control CA-MRSA strains by cluster analysis.
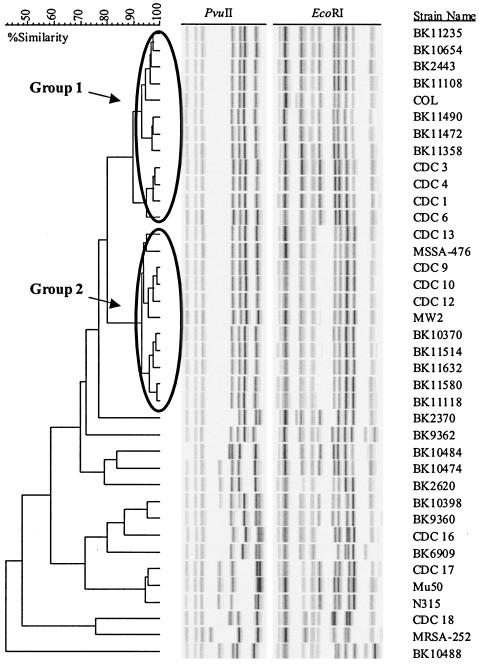
A series of positive-control CA-MRSA strains and negative-control HA-MRSA strains were analyzed by clustering on the basis of their EcoRI and PvuII riboprint patterns (Fig. 1). The majority of positive control strains fell into two tightly clustered groups with >90% similarity. The strains in group 1 were characterized by the ST8 genotype, all carried SCCmec type IV and 8/12 strains (67%) in this group carried the PVL-encoding genes (Table 1). ST information for the BK strains of spa type 1, 7 or 131 was derived from McDougal et al. (30). The archaic MRSA strain COL with ST250 also clustered with this group. The strains in group 2 had ST1 genotypes, carried SCCmec type IV and 8/10 (80%) of the MRSA strains were positive for the PVL genes. The prototype CA-MRSA strain, MW2, and its methicillin-susceptible parent strain, MSSA-476, belonged to this group. The remaining strains, including the negative control hospital associated strains had <70% similarity to these two groups (Fig. 1); 7/15 (47%) of the nonclustering strains carried SCCmec type IV but only 2/15 (13%) were positive for the PVL-encoding genes (Table 1).
FIG. 1.
Clustering of control CA-MRSA and HA-MRSA strains on the basis of ribotype. A series of positive-control CA-MRSA strains and negative-control HA-MRSA strains were compared by cluster analysis on the basis of riboprint patterns generated by digestion with EcoRI and PvuII. Groups 1 and 2 represent clusters of positive control strains with 90% or greater similarity.
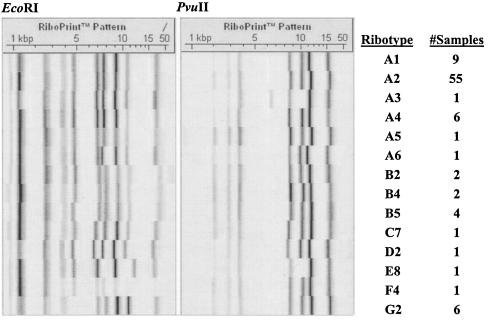
We subsequently compared 173 MRSA isolates collected during the phase 3 clinical trials for tigecycline to this group of control strains by cluster analysis using ribotyping data for these isolates; 63% (106/173) of these MRSA isolates were obtained from patients in North America and the remainder came from Europe, South Africa, Australia, India or Latin America; 85% of MRSA isolates were associated with complicated skin and skin structure infections; 85 isolates clustered with the group 1 control strains and six isolates were found to cluster with group 2 (data not shown). These 91 isolates were characterized by 14 unique riboprint patterns (Fig. 2). One particular ribotype, A2, was common to 65% of the isolates within group 1. All six MRSA isolates that clustered with the group 2 positive control strains had ribotype G2. A further 12 isolates were found to cluster with one of the CA-MRSA control strains (BK10474, BK10398 or BK2370) that did not cluster with either group 1 or group 2 (data not shown).
FIG. 2.
Riboprint patterns for MRSA isolates that cluster with positive-control CA-MRSA strains in groups 1 or 2. Fourteen representative EcoRI and PvuII riboprint patterns for the 91 MRSA isolates that cluster with positive-control CA-MRSA strains are displayed. The assigned ribotype and the number of isolates defined by this ribotype are indicated.
Determination of SCCmec type and distribution of the PVL-encoding locus among MRSA isolates that clustered with positive control CA-MRSA strains.
The 91 MRSA isolates that clustered with positive-control CA-MRSA strains in groups 1 and 2 (Fig. 1) were analyzed further to determine SCCmec type; 98% (89/91) were found to carry SCCmec type IV and 84% (76/91) were positive for the genes encoding PVL (Table 2). All PVL positive isolates were also SCCmec type IV. Of the 12 isolates that clustered with the positive control strains in the nonclustering group, only one carried SCCmec type IV and none were positive for the PVL-encoding genes; 26 MRSA isolates from the tigecycline studies that did not cluster with any of the CA-MRSA positive control strains were used as negative controls for this sample set and were tested for the presence of the PVL-encoding locus and SCCmec type IV; 31% (8/26) carried the SCCmec type IV element and only 15% (4/26) were positive for the genes encoding PVL (data not shown).
TABLE 2.
Molecular, susceptibility, and patient information for 91 MRSA isolates that cluster with positive-control CA-MRSA strains
| Strain no.a | Region | Country/state | Ribotype | Susceptibilityb
|
Inducible CLI resistance | PVL locus | SCCmec type | TGC MICc (μg/ml) | CA-MRSA characteristicsd | |
|---|---|---|---|---|---|---|---|---|---|---|
| CLI | ERY | |||||||||
| 1075 | Europe | Lithuania | A1 | S | S | No | No | IV | 0.12 | ++ |
| 1448 | USA | Ohio | A1 | S | R | No | Yes | IV | 0.25 | +++ |
| 3110 | Europe | Lithuania | A1 | S | S | No | No | Unclear | 0.06 | + |
| 3976 | Europe | France | A1 | S | S | No | No | IV | 0.25 | ++ |
| 4911 | USA | California | A1 | S | R | No | Yes | IV | 0.12 | +++ |
| 5278 | Europe | Russia | A1 | R | R | No | No | IV | 0.12 | +++ |
| 5385 | Europe | Russia | A1 | R | R | No | Yes | IV | 0.12 | +++ |
| 8104 | Europe | Bulgaria | A1 | R | R | No | No | III | 0.5 | + |
| 9375 | Europe | Lithuania | A1 | S | S | No | No | IV | 0.25 | ++ |
| 1180 | USA | Louisiana | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 1689 | USA | Louisiana | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 1854 | USA | Louisiana | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 1977 | USA | New Jersey | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2102 | USA | California | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 2227 | USA | California | A2 | R | R | No | Yes | IV | 0.12 | +++ |
| 2230 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2245 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2248 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2275 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2276 | USA | California | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 2637 | USA | Hawaii | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2644 | USA | Hawaii | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 2648 | USA | Hawaii | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 2654 | USA | Hawaii | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2668 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2670 | USA | California | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 2672 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2968 | USA | Florida | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2970 | USA | Florida | A2 | R | R | No | Yes | IV | 0.12 | +++ |
| 2974 | USA | Oklahoma | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 2986 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 2993 | USA | Louisiana | A2 | S | R | No | Yes | IV | 1 | +++ |
| 3024 | USA | New Jersey | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3044 | USA | Hawaii | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 3161 | USA | Florida | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3166 | USA | Florida | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3167 | USA | Florida | A2 | S | R | Yes | Yes | IV | 0.25 | +++ |
| 3221 | USA | California | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 3320 | USA | Hawaii | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3346 | USA | Ohio | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 3412 | USA | Florida | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3462 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3490 | USA | Indiana | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3497 | USA | Florida | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 3523 | USA | New Jersey | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3527 | USA | New Jersey | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3764 | Europe | Poland | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3877 | USA | Florida | A2 | S | S | No | Yes | IV | 0.12 | +++ |
| 3891 | USA | Florida | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 3912 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 3984 | USA | Louisiana | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 4190 | USA | Louisiana | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 4666 | USA | Texas | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 4667 | USA | Texas | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 4912 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 4914 | USA | California | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 5156 | USA | Washington | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 5416 | USA | Ohio | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 5455 | USA | Washington | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 5732 | USA | Illinois | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 5857 | USA | Texas | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 7130 | USA | Louisiana | A2 | S | R | No | Yes | IV | 1 | +++ |
| 7448 | USA | California | A2 | S | R | No | Yes | IV | 0.12 | +++ |
| 9271 | USA | Florida | A2 | S | R | No | Yes | IV | 0.25 | +++ |
| 7947 | USA | Florida | A3 | S | R | No | Yes | IV | 0.25 | +++ |
| 1303 | USA | Louisiana | A4 | S | R | No | Yes | IV | 0.25 | +++ |
| 1720 | Europe | Ukraine | A4 | S | S | No | No | IV | 0.12 | ++ |
| 1874 | Europe | Lithuania | A4 | S | S | No | No | IV | 0.25 | ++ |
| 3252 | Europe | Russia | A4 | R | R | No | No | IV | 0.12 | ++ |
| 4372 | Europe | Belgium | A4 | R | R | No | No | IV | 0.12 | ++ |
| 4742 | Europe | Ukraine | A4 | S | S | No | No | IV | 0.12 | ++ |
| 2237 | USA | Louisiana | A5 | S | R | No | Yes | IV | 0.12 | +++ |
| 9336 | USA | Illinois | A6 | S | R | No | Yes | IV | 0.12 | +++ |
| 2243 | USA | California | B2 | S | R | No | Yes | IV | 0.25 | +++ |
| 2249 | USA | California | B2 | S | S | No | Yes | IV | 0.25 | +++ |
| 1497 | Europe | Lithuania | B4 | S | S | No | No | IV | 0.12 | ++ |
| 2233 | USA | California | B4 | S | S | No | Yes | IV | 0.25 | +++ |
| 1334 | USA | Louisiana | B5 | S | R | No | Yes | IV | 0.25 | +++ |
| 1568 | USA | Louisiana | B5 | S | R | No | Yes | IV | 0.12 | +++ |
| 2207 | USA | California | B5 | S | R | No | Yes | IV | 0.12 | +++ |
| 2269 | USA | Ohio | B5 | S | R | No | Yes | IV | 0.12 | +++ |
| 5913 | USA | Louisiana | C7 | S | R | No | Yes | IV | 0.12 | +++ |
| 4269 | USA | California | D2 | S | R | Yes | Yes | IV | 0.12 | +++ |
| 7051 | Africa | South Africa | E8 | S | R | Yes | Yes | IV | 0.12 | +++ |
| 3547 | Europe | Russia | F4 | R | R | No | No | IV | 0.12 | ++ |
| 3267 | USA | Washington | G2 | S | R | Yes | Yes | IV | 0.25 | +++ |
| 4946 | USA | Washington | G2 | S | R | Yes | Yes | IV | 0.06 | +++ |
| 5806 | Europe | Romania | G2 | S | S | No | Yes | IV | 0.12 | +++ |
| 6031 | Europe | Romania | G2 | S | R | Yes | No | IV | 0.12 | ++ |
| 6072 | USA | Louisiana | G2 | S | R | Yes | Yes | IV | 0.25 | +++ |
| 9167 | Europe | Romania | G2 | S | R | Yes | No | IV | 0.12 | ++ |
Strain numbers in bold cluster with the group 2 positive-control CA-MRSA strains (see Fig. 1). Strain numbers 1334 and 7130 were isolated from the same patient.
Clindamycin (CLI) and erythromycin (ERY) susceptibility and inducible clindamycin resistance were tested using the disk diffusion method. R, resistant; S, susceptible.
Tigecycline (TGC) MICs were determined using the broth microdilution method.
Each + indicates that this isolate has one of the following markers that are typically associated with CA-MRSA; +, genotypically similar to CA-MRSA; ++, PVL-encoding genes; +++, SCCmec type IV.
On the basis of genotype as determined by ribotyping, and the presence of both the PVL-encoding locus and SCCmec type IV, it appears that 76 MRSA isolates identified in this study had characteristics consistent with known CA-MRSA strains (Table 2). The degree of similarity to known CA-MRSA strains on the basis of the criteria described above is indicated. Isolates fulfilling all three criteria are subsequently referred to as putative CA-MRSA. However, it is possible that isolates fulfilling only two of these criteria could still be considered CA-MRSA if more detailed clinical information were available; 95% (72/76) of the putative CA-MRSA isolates detected in this study were North American in origin. Two of these isolates (1334 and 7130) were unique isolates from the same patient and the remainder represented unique isolates from different patients; 74 of the 75 patients infected with putative CA-MRSA isolates were diagnosed with complicated skin and skin structure infections (data not shown).
Antimicrobial susceptibility of putative CA-MRSA isolates.
The 91 isolates that clustered with the positive control CA-MRSA strains in groups 1 or 2 and the 26 nonclustering isolates were tested for susceptibility to clindamycin and erythromycin as well as inducible clindamycin resistance in the presence of erythromycin (Table 2 and data not shown); 12% (9/76) of the putative CA-MRSA isolates (PVL+, SCCmec type IV+) and 27% (7/26) of the nonclustering isolates were susceptible to erythromycin; 96% (73/76) of putative CA-MRSA isolates were susceptible to clindamycin compared to 50% (13/26) of nonclustering isolates; 8% (6/73) of the putative CA-MRSA isolates that were clindamycin susceptible and 46% (6/13) of the clindamycin-susceptible nonclustering isolates demonstrated inducible clindamycin resistance. Five of the six MRSA isolates that clustered with the group 2 positive-control strains exhibited an inducible clindamycin resistance phenotype; the sixth was susceptible to erythromycin and clindamycin.
CA-MRSA isolates exhibit less antibiotic resistance compared to HA-MRSA isolates.
Historical MIC data for the 76 putative CA-MRSA isolates carrying the PVL-encoding genes and SCCmec type IV and the 26 nonclustering isolates was examined (data not shown). No intermediate or high-level vancomycin resistant isolates were detected in either group; 100% of putative CA-MRSA isolates were susceptible to minocycline and imipenem compared to 88% and 69%, respectively, for the nonclustering group. Furthermore, whereas 95% and 66% of putative CA-MRSA isolates were susceptible to tobramycin and levofloxacin, respectively, only 35% and 19% of the nonclustering isolates were susceptible to the same antibiotics (data not shown); 89/91 MRSA isolates that clustered with the positive-control CA-MRSA strains were found to be susceptible to tigecycline (MICs ≤0.5 μg/ml) (Table 2). The two nonsusceptible isolates had tigecycline MICs of 1.0 μg/ml. All of the nonclustering MRSA isolates were also susceptible to tigecycline (data not shown).
DISCUSSION
Tigecycline, a novel broad-spectrum glycylcycline antibiotic, has activity against multidrug-resistant S. aureus isolates (4, 39); 171 patients enrolled in the recently completed phase 3 clinical trials for tigecycline had infections caused by MRSA. 85% of these patients were diagnosed with complicated skin and skin structure infections. MRSA is most commonly associated with the hospital environment but recently MRSA outbreaks have been described in the community (CA-MRSA) and have caused serious, sometimes fatal, infections that include necrotizing fasciitis (6, 31). The tigecycline trials were not designed to capture clinical information that could be used to identify potential CA-MRSA isolates. Furthermore, only limited information regarding health care-related risks for MRSA infection was available. In the absence of adequate clinical information, this study assessed the utility of ribotyping to retrospectively identify MRSA isolates collected during the tigecycline trials that were genotypically related to known CA-MRSA strains. Subsequently, these isolates were tested for genetic markers and antimicrobial susceptibility phenotypes that are commonly associated with CA-MRSA.
It has been shown using MLST or PFGE that CA-MRSA isolates are genotypically distinct from the MRSA isolates commonly found in health care institutions (33, 45). Okuma et al. found that a set of CA-MRSA isolates studied belonged to five clonal complexes (36). Said-Salim et al. also describe five distinct CA-MRSA groups based upon spa typing (40). Therefore, CA-MRSA isolates appear to be derived from a number of genetic backgrounds rather than the worldwide dissemination of a single clone. A number of studies have reported CA-MRSA isolates with ST1 (36, 45), ST8 (7, 12, 18, 21, 23, 32, 41), or ST30 (12, 36) genetic backgrounds.
The CA-MRSA strains used as positive controls in this study were analyzed by ribotyping and most fell into two groups on the basis of cluster analysis. These groups had ST8 or ST1. The archaic MRSA strain COL (ST250) clustered with the ST8 CA-MRSA controls. This strain was shown to have evolved from an MSSA strain of ST8 (15); 91 of the 173 MRSA isolates examined in this study clustered with the two groups of positive control strains, 85 with group 1 (ST8) and 6 with group 2 (ST1); 14 distinct ribotypes were associated with these 91 isolates. Interestingly, 55/85 (65%) of group 1 isolates shared a common ribotype (A2, Fig. 2) and all group 2 (6/6) isolates were characterized by the G2 ribotype. Recent reports have suggested that the group 1 CA-MRSA clone designated USA300 by the CDC (30) has become the predominant CA-MRSA clone in certain parts of the United States (7). This may explain why the majority of isolates examined here clustered with this group.
The presence of the PVL-encoding genes and SCCmec type IV are considered genetic markers for CA-MRSA (45) but a recent study suggests that while the PVL-encoding locus is more prevalent in CA-MRSA isolates this is not uniform (40) In this study however, we included the PVL-encoding locus as one of the genetic markers used to characterize putative CA-MRSA isolates; 76 of the 91 MRSA isolates that clustered with the positive control strains in this study carried the PVL locus and SCCmec type IV and were considered to be putative CA-MRSA isolates. An additional 12 MRSA isolates clustered with three positive control strains that did not cluster in either group 1 or 2. However, only one of these isolates carried SCCmec type IV and none were positive for the PVL-encoding genes. Furthermore, the prevalence of these genetic markers among isolates that did not cluster with any of the positive control strains was much lower than in the clustering group which suggests that ribotyping is a reasonable predictor for genotypes that carry the CA-MRSA-associated markers PVL and SCCmec type IV. Surprisingly, the HA-MRSA strain BK9360 (ST8, SCCmec type II) and the CA-MRSA strain BK10398 (ST1, SCCmec type IV) were found to cluster together on the basis of ribotype (Fig. 1). Both share an indistinguishable EcoRI riboprint pattern (as indicated by the letter K, Table 1).
Susceptibility to clindamycin has also been associated with CA-MRSA (16, 17) but this trait is not sufficiently specific to use as a marker for CA-MRSA (43). Clindamycin is an attractive treatment option for CA-MRSA infections in the outpatient setting but the possibility of inducible clindamycin resistance is a concern where the erythromycin resistance determinants encoded by the erm genes are present (26, 29, 42); 96% of the putative CA-MRSA isolates (PVL, SCCmec type IV) were susceptible to clindamycin compared to only 50% of the nonclustering isolates. Inducible clindamycin resistance was more common in the negative control group (46% of susceptible isolates demonstrated inducible clindamycin resistance in the presence of erythromycin) compared to 8% of the putative CA-MRSA group. Unlike HA-MRSA isolates, CA-MRSA isolates are generally not multidrug resistant and increased susceptibility to non-β-lactam antibiotics, other than clindamycin, is typical (1, 33). The 76 putative CA-MRSA isolates identified during this study exhibited increased susceptibility to imipenem, minocycline, tobramycin and levofloxacin compared to the negative control group.
Tigecycline was active against all but two of the MRSA isolates examined in this study with MICs of ≤0.5 μg/ml. Tigecycline has demonstrated clinical efficacy against MRSA (including the putative CA-MRSA isolates described here) in complicated skin and skin structure infections (14). Despite the fact that 76 MRSA isolates were identified that were genotypically similar to CA-MRSA and positive for PVL and SCCmec type IV, a number of the patients infected with these isolates were exposed to previous antibiotic therapy or had underlying conditions such as intravenous drug abuse, diabetes, dermatitis, or human immunodeficiency virus infection, which are considered risk factors for MRSA infection (5, 19).
This study is limited by the fact that all of the positive control CA-MRSA strains were North American in origin thus leading to a geographical bias in the identification of putative CA-MRSA isolates based on genotypic comparison with this control group. Vandenesch and colleagues demonstrated that the genetic background of CA-MRSA isolates varies depending on geographic origin (45). The phase 3 clinical trials for tigecycline involved enrollment of patients at multiple sites worldwide. However, 63% of MRSA isolates were obtained from patients in North America. Of the 91 MRSA isolates that clustered with the positive control strains, only 19 were isolated from sources outside of the USA (18 from Europe and 1 from South Africa) and only 4 of the 76 putative CA-MRSA isolates (PVL+, SCCmec IV+) were not from the United States in origin. Further putative CA-MRSA isolates may have been identified if positive control CA-MRSA strains representative of geographically diverse locations had been included.
In the absence of adequate clinical information to identify CA-MRSA isolates, a genotyping approach based upon ribotyping information was used to identify MRSA isolates that clustered with previously characterized CA-MRSA strains and subsequently these isolates were tested for genetic markers that are typically associated with CA-MRSA. It was shown that two ribotypes in particular, A2 and G2, were strongly associated with the ST8 and ST1 genetic background, respectively, to which most of the positive-control CA-MRSA strains belonged. Ribotyping may represent a useful predictive tool to identify genotypes that are commonly associated with CA-MRSA. Tigecycline exhibited good activity against the majority of putative CA-MRSA isolates identified in this study and may represent a therapeutic option for patients hospitalized as a result of CA-MRSA infections.
Acknowledgments
We thank Frank Ritacco and Jeffrey Janso for assistance with BioNumerics software and Fred Tenover for the CDC control strains.
REFERENCES
- 1.Almer, L. S., V. D. Shortridge, A. M. Nilius, J. M. Beyer, N. B. Soni, M. H. Bui, G. G. Stone, and R. K. Flamm. 2002. Antimicrobial susceptibility and molecular characterization of community-acquired methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 43:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Babinchak, T., E. Ellis-Grosse, N. Dartois, G. M. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline for the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41(Suppl. 5):S354-S367. [DOI] [PubMed] [Google Scholar]
- 4.Bozdogan, B., D. Esel, C. Whitener, F. A. Browne, and P. C. Appelbaum. 2003. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52:864-868. [DOI] [PubMed] [Google Scholar]
- 5.Carleton, H. A., B. A. Diep, E. D. Charlebois, G. F. Sensabaugh, and F. Perdreau-Remington. 2004. Community-adapted methicillin-resistant Staphylococcus aureus (MRSA): population dynamics of an expanding community reservoir of MRSA. J. Infect. Dis. 190:1730-1738. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus-Minnesota and North Dakota, 1997-1999. JAMA 282:1123-1125. [PubMed] [Google Scholar]
- 7.Chambers, H. F. 2005. Community-associated MRSA-resistance and virulence converge. N. Engl. J. Med. 352:1485-1487. [DOI] [PubMed] [Google Scholar]
- 8.Charlebois, E. D., F. Perdreau-Remington, B. Kreiswirth, D. R. Bangsberg, D. Ciccarone, B. A. Diep, V. L. Ng, K. Chansky, B. R. Edlin, and H. F. Chambers. 2004. Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 39:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; informational supplement M100-S15, vol. 15. Clinical Laboratory Standards Institute, Wayne, Pa.
- 10.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 11.de Lencastre, H., E. P. Severina, H. Milch, M. K. Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 12.Diep, B. A., G. F. Sensabaugh, N. S. Somboona, H. A. Carleton, and F. Perdreau-Remington. 2004. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J. Clin. Microbiol. 42:2080-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., W. Mounts, F. McAleese, F. Immermann, D. Macapagal, E. Marsilio, L. McDougal, F. C. Tenover, P. A. Bradford, P. J. Petersen, S. J. Projan, and E. Murphy. 2004. Uses of Staphylococcus aureus GeneChips in genotyping and genetic composition analysis. J. Clin. Microbiol. 42:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis-Grosse, E. J., T. Babinchak, N. Dartois, G. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of skin and skin-structure infections: results of 2 double-blind phase 3 comparison studies with vancomycin-aztreonam. Clin. Infect. Dis. 41(Suppl. 5):S341-S353. [DOI] [PubMed] [Google Scholar]
- 15.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Community-acquired and clindamycin-susceptible methicillin-resistant Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 18:993-1000. [DOI] [PubMed] [Google Scholar]
- 17.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Increase in community-acquired methicillin-resistant Staphylococcus aureus in children. Clin. Infect. Dis. 29:935-936. [DOI] [PubMed] [Google Scholar]
- 18.Frazee, B. W., J. Lynn, E. D. Charlebois, L. Lambert, D. Lowery, and F. Perdreau-Remington. 2005. High prevalence of methicillin-resistant Staphylococcus aureus in emergency department skin and soft tissue infections. Ann. Emerg. Med. 45:311-320. [DOI] [PubMed] [Google Scholar]
- 19.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, B. E., G. Martinez-Aguilar, K. G. Hulten, W. A. Hammerman, J. Coss-Bu, A. Avalos-Mishaan, E. O. Mason, Jr., and S. L. Kaplan. 2005. Severe Staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. Pediatrics 115:642-648. [DOI] [PubMed] [Google Scholar]
- 22.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468-475. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 25.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, J. S., II, and J. H. Jorgensen. 2005. Inducible clindamycin resistance in Staphylococci: should clinicians and microbiologists be concerned? Clin. Infect. Dis. 40:280-285. [DOI] [PubMed] [Google Scholar]
- 27.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 28.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Aguilar, G., W. A. Hammerman, E. O. Mason, Jr., and S. L. Kaplan. 2003. Clindamycin treatment of invasive infections caused by community-acquired, methicillin-resistant and methicillin-susceptible Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 22:593-598. [DOI] [PubMed] [Google Scholar]
- 30.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, L. G., F. Perdreau-Remington, G. Rieg, S. Mehdi, J. Perlroth, A. S. Bayer, A. W. Tang, T. O. Phung, and B. Spellberg. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 352:1445-1453. [DOI] [PubMed] [Google Scholar]
- 32.Mishaan, A. M., E. O. Mason, Jr., G. Martinez-Aguilar, W. Hammerman, J. J. Propst, J. R. Lupski, P. Stankiewicz, S. L. Kaplan, and K. Hulten. 2005. Emergence of a predominant clone of community-acquired Staphylococcus aureus among children in Houston, Texas. Pediatr. Infect. Dis. J. 24:201-206. [DOI] [PubMed] [Google Scholar]
- 33.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A5, 5th ed., vol. 17. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 35.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests: approved standard M2-A8, 8th ed., vol. 23. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 36.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, E. S., B. A. Diep, H. A. Carleton, E. D. Charlebois, G. F. Sensabaugh, B. L. Haller, and F. Perdreau-Remington. 2003. Increasing prevalence of methicillin-resistant Staphylococcus aureus infection in California jails. Clin. Infect. Dis. 37:1384-1388. [DOI] [PubMed] [Google Scholar]
- 39.Petersen, P. J., P. A. Bradford, W. J. Weiss, T. M. Murphy, P. E. Sum, and S. J. Projan. 2002. In vitro and in vivo activities of tigecycline (GAR-936), daptomycin, and comparative antimicrobial agents against glycopeptide-intermediate Staphylococcus aureus and other resistant gram-positive pathogens. Antimicrob. Agents Chemother. 46:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Said-Salim, B., B. Mathema, K. Braughton, S. Davis, D. Sinsimer, W. Eisner, Y. Likhoshvay, F. R. DeLeo, and B. N. Kreiswirth. 2005. Differential distribution and expression of Panton-Valentine leukocidin among community-acquired methicillin resistant Staphylococcus aureus. J. Clin. Microbiol. In press. [DOI] [PMC free article] [PubMed]
- 41.Said-Salim, B., B. Mathema, and B. N. Kreiswirth. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect. Control Hosp. Epidemiol. 24:451-455. [DOI] [PubMed] [Google Scholar]
- 42.Schreckenberger, P. C., E. Ilendo, and K. L. Ristow. 2004. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J. Clin. Microbiol. 42:2777-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suntharam, N., D. Hacek, and L. R. Peterson. 2001. Low prevalence of community-acquired Methicillin-resistant Staphylococcus aureus in adults at a university hospital in the central United States. J. Clin. Microbiol. 39:1669-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenover, F. C., and R. P. Gaynes. 2000. The epidemiology of Staphylococcus aureus infection, p. 414-421. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 45.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldvogel, F. A. 2000. Staphylococcus aureus, p. 2069-2092. In G. L. Mandel, J. E. Bennett, and R. Dolin (ed.), Principle and practice of infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 47. Wyeth Pharmaceuticals Inc. 20. June 2005. Tygacil package insert. Available at http://www.fda.gov/cder/foi/label/2005/021821lbl.pdf.