Abstract
Iturin A and its derivatives are lipopeptide antibiotics produced by Bacillus subtilis and several closely related bacteria. Three iturin group operons (i.e., iturin A, mycosubtilin, and bacillomycin D) of those antibiotic-producing strains have been cloned and sequenced thus far, strongly implying the horizontal transfer of these operons. To examine the nature of such horizontal transfer in terms of antibiotic production, a 42-kb region of the B. subtilis RB14 genome, which contains a complete 38-kb iturin A operon, was transferred via competent cell transformation to the genome of a non-iturin A producer, B. subtilis 168, using a method based on double-crossover homologous recombination with two short landing pad sequences (LPSs) in the genome. The recombinant was positively selected by confirming the elimination of the cI repressor gene, which was localized between the two LPSs and substituted by the transferred segment. The iturin A operon-transferred strain 168 was then converted into an iturin A producer by the introduction of an sfp gene, which encodes 4′-phosphopantetheinyl transferase and is mutated in strain 168. By inserting the pleiotropic regulator degQ, the productivity of iturin A increased sevenfold and was restored to about half that of the donor strain RB14, without the transfer of additional genes, such as regulatory or self-resistance genes.
Horizontal gene transfer is thought to be a ubiquitous event (18). In terms of antibiotic production genes, the β-lactam-class antibiotic genes are found in some organelles of fungi and in gram-positive and gram-negative bacteria and are supported to horizontally transfer from bacteria to fungi (1). In contrast, the syringomycin operon of Pseudomonas syringae, which encodes nonribosomal peptide synthetases, is postulated to be horizontally transferred from a eukaryote to a prokaryote (6). The number of possible examples of the transferred antibiotic production gene is increasing, and studies of successful artificial horizontal transfer of antibiotic production genes to a heterologous host have been reported (4, 22, 29). However, due to the limited numbers of artificial horizontal transfer, there is little knowledge of the genes required for the conversion from a heterologous host into an antibiotic producer.
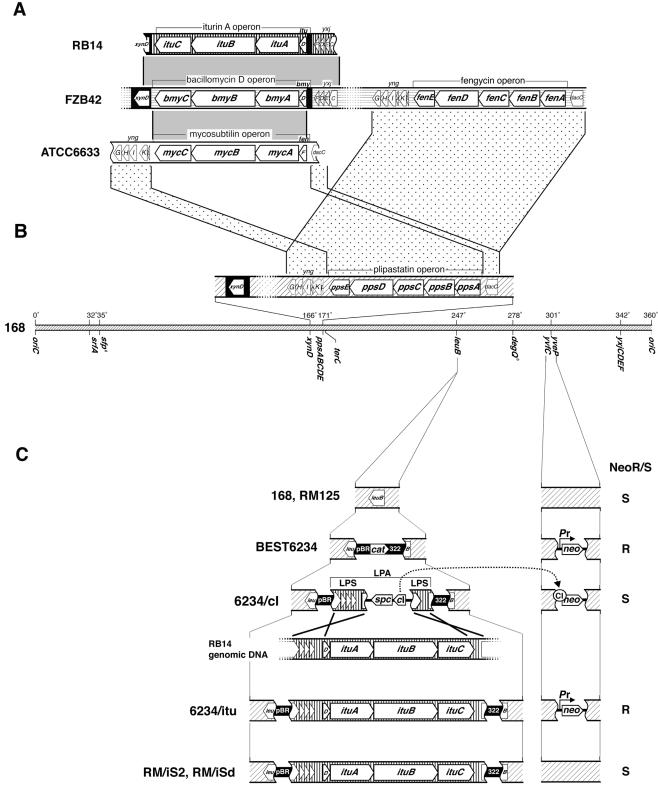
Many Bacillus subtilis strains simultaneously produce some lipopeptide antibiotics, whose peptide moiety is synthesized nonribosomally by large template enzyme complexes (e.g., B. subtilis RB14 [33]). Such lipopeptide antibiotics identified thus far have been divided into three groups according to their structure, as follows: the surfactin group (28), the plipastatin and fengycin group (34), and the iturin group (10, 24, 27). Iturin group lipopeptides are composed of seven α-amino acids and one β-amino acid with a long lipid moiety and are potent antifungal agents (24). Three distinct operons that belong to the iturin group have been cloned and sequenced thus far: the mycosubtilin operon of B. subtilis ATCC 6633 (3), the iturin A operon of B. subtilis RB14 (33), and the bacillomycin D operon of B. amyloliquefaciens FZB42 (19). All of these operons are composed of a putative transcriptional unit with four genes: one small gene that encodes malonyl-coenzyme A transferase and is probably responsible for β-amino acid synthesis and three large genes that encode large template enzymes for the synthesis of peptides with defined sequences and chiralities. Although the percent amino acid sequence identities between the operons range from approximately 75% to 85%, the flanking regions of the operons are quite different from each other, as shown in Fig. 1. The flanking region of the mycosubtilin operon is identical to that of the plipastatin operon of B. subtilis 168 (3). On the other hand, the iturin A and bacillomycin D operons are flanked by sequences homologous to xynD of strain 168 (19, 33) (Fig. 1). These findings strongly indicate the dynamic features of the iturin group operons.
FIG. 1.
(A) Structure and flanking regions of the iturin A, mycosubtilin, and bacillomycin D operons. Shaded bridges between different strains connect regions homologous with the iturin A operon of RB14, while dotted bridges connect regions homologous with the plipastatin operon of 168. Solid black regions in the iturin A and bacillomycin D operons indicate regions homologous with the xynD gene of 168. Vertical lines, horizontal lines, and areas not linedrepresent B. subtilis RB14 DNA, B. amyloliquefaciens FZB42 DNA (19), and B. subtilis ATCC 6633 DNA (3), respectively. Fengycin is another name for plipastatin (19). (B) Whole-genome map of strain 168 (diagonal lines). The counterparts of the relevant genes in panel A are mapped. (C) Strain development. The positive-selection system is also indicated. The CI repressor in 6234/cI represses Pr-neo expression, resulting in neomycin susceptibility. However, once substitution of the cI gene by transformed DNA takes place, the transformant becomes neomycin resistant. Solid black regions show the tetracycline (labeled pBR) and ampicillin (labeled 322) resistance gene sides of the split pBR322 sequence. The genes cat and spc represent the chloramphenicol and spectinomycin resistance genes, respectively. The genes cI and Pr-neo indicate the CI repressor gene and the Pr promoter-driven neomycin resistance gene, respectively. BEST6234, 6234/cI, and 6234/itu also have a Pr-neo cassette in their genome; however, this cassette is absent in the RM/iS2 and RM/iSd series. The NeoR/S labels at the far right indicate neomycin resistance (R) or sensitivity (S). LPS, landing pad sequence; LPA, LPSs array.
Although 168 does not produce lipopeptides due to its mutation of the 4′-phosphopantetheine transferase gene sfp, which is responsible for the conversion of nascent antibiotic synthetase to its active form (21), it is a potential producer of two lipopeptide antibiotics, surfactin and plipastatin. We previously converted 168 into a coproducer of surfactin and plipastatin by introducing sfp and the pleiotropic regulator gene degQ, which is also mutated in 168 (32). However, on the basis of whole-genome sequence data, 168 does not have iturin group operons (20).
To investigate the nature of the horizontal transfer of an antibiotic producer gene in terms of antibiotic production, we transferred the iturin A operon of RB14 to the non-iturin-producing strain 168, using a positive-selection system that employs the cI repressor gene of lambda phage as a reverse marker (14, 15, 16). In this study, although sfp is necessary, we show that the iturin A operon is essentially the only operon required for conversion of strain 168 into an iturin A producer. We also demonstrate that, in the presence of degQ, an iturin A production almost comparable to that of the donor strain wasachieved.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and media.
The bacterial strains, phage, and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium was used for the cultivation of Escherichia coli and B. subtilis (30). When necessary, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 10 μg/ml; tetracycline, 20 μg/ml; and neomycin, 3 μg/ml. Number 3S (no. 3S) medium, containing (per liter) 10 g of Polypeptone S (Nihon Pharmaceutical Co.), 10 g of glucose, 1 g of KH2PO4, and 0.5 g of MgSO4 · 7H2O (pH 6.8), was used for lipopeptide production in liquid culture.
TABLE 1.
Bacteria, phage, and plasmids used in this study
| Strain, phage, or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strain | ||
| JA221 | F−hsdR hsdM+trp leu lacY recA1 | 11 |
| B. subtilis strains | ||
| RB14 | IT+ SF+ PL+ | 33 |
| 168 | trpC2 sfp0degQ0, IT− SF− PL− | 20 |
| RM125 | leuB8 arg-15 ΔSPβ R(hsdR-cotA+-purB+)202-5 sfp0degQ0, IT− SF− PL− | 13 |
| BEST6225 | RM125 Δ(yvfC-yveP)::Pr-neo, IT− SF− PL− | This study |
| BEST6234 | RM125 Δ(yvfC-yveP)::Pr-neo leuB::pBR322::cat, Cmr Nmr IT− SF− PL− | This study |
| 6234/cI | BEST6234; leuB::pBR322::[(yxjCDEFRB14-ituD)-(cI-spc)-(ituC-xynDRB14)], Spr Cms Nms IT− SF− PL− | This study |
| 6234/itu | BEST6234 (host) × RB14 (DNA); leuB::pBR322::(yxjCDEFRB14-ituD+A+B+C+-xynDRB14), Nmr Sps IT− SF− PL− | This study |
| RM/iS2 | 6234/itu::pMMN6(sfp+cat) R[Δ(yvfC-yveP)::Pr-neo]RM125, Cmr Nms IT+ SF+ PL+ | This study |
| RM/iSd4, -12, -14, and -16 | RM/iS2::pUC19HP1NmrF(degQYB8+neo), Cmr Nmr IT+ SF+ PL+, clone numbers 4, 12, 14, and 16 | This study |
| RM/Sp6 | RM125::pMMN6(sfp+, cat), Cmr IT− SF+ PL+ | This study |
| Phage | ||
| λ69 | Lambda DASH II cloned downstream edge of iturin A operon | 33 |
| Plasmids | ||
| pBR322 | Cloning vector, Apr Tcr | 30 |
| pNEXT4PN-2 | Pr-neo cassette introduction plasmid, Apr Nmr | This study |
| RSF2124.B.leuB | EcoRI fragment of B. subtilis leuB gene cloned into ColiE1 | 25 |
| pBR322Cm | cat cassette cloned into EcoRI site of pBR322, Apr Tcr Cmr | 12 |
| pBMAP103-322CA | RSF2124.B.leuB-inserted pBRCm in the leuB gene | This study |
| pCISP303B#6 | cI-spc cassette cloned into pBR322, Apr Tcr Spr | 15 |
| pBRE9k | 9-kb EcoRI fragment from λ69 containing downstream edge of iturin A operon (i.e., ituC-xynDRB14) cloned in EcoRI site of pBR322, Apr Tcr | This study |
| pBRHd8k | 8-kb HindIII fragment from strain 1006 containing promoter region of iturin A operon (i.e., yxjCDEFRB14-ituDA) inserted into HindIII site of pBR322, Apr Cmr | 33 |
| pBRE4H8 | pBRE9k-inserted 8-kb HindIII fragment from pBRHd8k into EcoRV site by replacing small EcoRV fragments, Apr Cmr | This study |
| pBRE2H4cI (LPA) | pBRE4H8-inserted 2.5-kb SmaI fragment containing cI-spc cassette from pCISP303B#6 into AgeI-CpoI site by replacing a small fragment, Apr Spr | This study |
| pMMN6 | E. coli plasmid containing functional sfp, Apr Cmr | 26 |
| pUC19HPINmrF | degQ of strain YB8 cloned into E. coli plasmid, Apr Nmr | 32 |
IT, iturin A production; SF, surfactin production; PL, plipastatin production; Apr, ampicillin resistant; Tcr, tetracycline resistant; Cmr or Cms, chloramphenicol resistant or sensitive; Nmr or Nms, neomycin resistant or sensitive; Spr or Sps, spectinomycin resistant or sensitive; cat, chloramphenicol resistance gene; neo, neomycin resistance gene; spc, spectinomycin resistance gene.
Transformation and DNA manipulation.
The B. subtilis strain was transformed by the method of Anagnostopoulos and Spizizen (2), as described previously (31). Routine DNA manipulation, E. coli transformation, contour-clamped homogeneous electric-field (CHEF) pulsed-field gel electrophoresis, and Southern hybridization were performed as described previously (32). The CHEF conditions were as follows: field strength, 3 V/cm; pulse time, 45 s; and running time, 44 h.
Construction of LPA.
A landing pad sequence array (LPA), in which the cI repressor gene and spectinomycin resistance gene are flanked by two landing pad sequences (LPSs), was constructed in plasmid pBR322 in E. coli as follows. A 9-kb EcoRI fragment with a downstream edge of the iturin A operon was obtained from recombinant phage λ69, derived from a lambda DASH II phage library of the RB14 chromosome (33), and cloned into the EcoRI site of pBR322 to obtain plasmid pBRE9k. An 8-kb HindIII fragment, with an upstream edge of the iturin A operon and a transposon, was obtained from pBRHd8k (33) and ligated to the largest fragment of the products of the EcoRV digestion of pBRE9k, generating the plasmid pBRE4H8. pBRE4H8 was digested with both AgeI and CpoI, and then the largest fragment (10 kb) obtained was purified by electrophoresis. The obtained fragment was dephosphorylated by alkaline phosphatase from E. coli (Toyobo, Inc., Japan), blunted with a DNA blunting kit (Takara Shuzo Co., Japan), and then ligated to the SmaI fragment of the cI-spc cassette from pCISP303B#6 (15), resulting in plasmid pBRE2H4cI, containing the LPA.
Preparation of a recipient strain containing the LPA.
The Pr-neo cassette introduction plasmid was constructed with a substitutional insertion of an EcoRI fragment of Pr-neo from pBEST515C (14) between two NotI sites in pNEXT4 (11). The resulting plasmid, pNEXT4PN-2, was used to transform B. subtilis RM125, a strain 168 derivative that lacks a restriction/modification system, generating BEST6225. The LPA carried by pBRE2H4cI was inserted into the leuB region in the BEST6225 genome as follows. Plasmid pBR322Cm (12), which harbors a chloramphenicol resistance gene (cat) cassette in the EcoRI site of pBR322, was linearized at a unique PvuII site and then inserted into a unique BamHI site in the leuB gene of plasmid RSF2124.B.leuB (25) by using a T4 DNA polymerase-based blunting kit. The resulting plasmid, pBMAP103-322CA, was used to transform BEST6225, generating BEST6234, which has pBR322Cm inserted in the genomic leuB region (Fig. 1C). BEST6234 was transformed with pBRE2H4cI and selected using spectinomycin. Transformants were assayed for chloramphenicol sensitivity by plate replication, and then a chloramphenicol-sensitive strain, which resulted from the double-crossover recombination between pBR322Cm and pBRE2H4cI (Fig. 1C), was selected. The obtained strain was designated 6234/cI and used as the recipient of the iturin A operon.
Iturin A operon transfer.
High-molecular-weight whole DNA of strain RB14 was prepared according to a method reported previously (11). One hundred microliters of competent cell culture of 6234/cI was mixed with 10 μl of 1 μg/μl RB14 chromosome. Following incubation at 37°C for 30 min, 300 μl of LB medium was added to the culture, which was incubated with gentle agitation at 37°C for 3 h to allow the expression of neomycin resistance. The culture was then plated on LB plates containing neomycin and incubated at 30°C overnight. Colonies that appeared on the plates were streaked on two LB plates, one containing spectinomycin and the other containing neomycin, for the screening of spectinomycin-sensitive colonies. Selected colonies were then picked up with a toothpick and inoculated in 25 μl of PCR solution (5 U of TaKaRa Ex Taq DNA polymerase [Takara Shuzo, Kyoto, Japan], 10 μl of Ex Taq buffer, and 8 μl of deoxynucleoside triphosphate solution [2.5 mM each]) with the primers ITUP1-F (5′-AGCTTAGGGAACAATTGTCATCGGGGCTTC-3′, positioned from nucleotide 15353 to 15383 of the iturin A operon sequence [DDBJ/EMBL/GenBank accession no. AB050629]) and ITUP2-R (5′-TCAGATAGGCCGCCATATCGGAATGATTCG-3′, complementary sequence positioned from nucleotide 17326 to 17355 of AB050629), which are able to detect a 2-kb region that includes the intergenic sequence between ituA and ituB. The colony PCR conditions were as follows: 96°C for 5 min; 30 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 150 s.
Introduction of sfp and degQ.
The sfp-harboring E. coli plasmid pMMN6 (26) was inserted into the genome of 6234/itu by Campbell-type insertion. In this transformation, genomic DNA of RM125 was simultaneously transferred to remove Pr-neo from the yvfC-yveP region for the following experiment. Thus, a chloramphenicol-resistant, neomycin-sensitive colony was selected and designated RM/iS2. This strain harbors pMMN6 in the sfp0 region. We did not determine in which site the actual insertion in RM/iS2 occurred. The degQYB8-containing E. coli plasmid pUC19HP1NmrF (32) was transformed into the RM/iS2 strain and selected for neomycin. Since pUC19HP1NmrF has three potential sites for Campbell-type insertion in the RM/iS2 genome (one is degQ0 and the others are ampicillin resistance genes in genomic pBR322 and pMMN6), several transformants were selected and designated the RM/iSd series.
Quantitative analysis of iturin A, plipastatin, and surfactin.
The culture (40 ml of no. 3S medium, 30°C) of the B. subtilis strain was acidified to pH 2.0 with 12 N HCl. Then, the precipitate formed was collected by centrifugation and extracted with methanol. Iturin A, plipastatin, and surfactin in the extracted solution were quantified by reversed-phase high-performance liquid chromatography (HPLC) using a two-eluent gradient as described previously (32, 33). For the detailed composition analysis of the fatty moiety of the β-amino acid of iturin A, the methanol extract was subjected to another reversed-phase HPLC using one eluent as described previously (8).
RESULTS
Horizontal transfer of iturin A synthetase operon to strain 168.
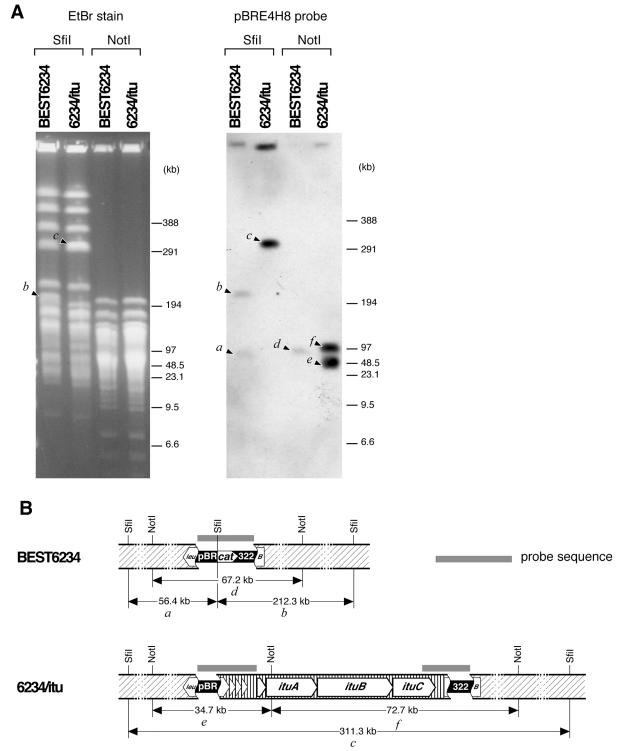
To horizontally transfer the complete iturin A operon (42 kb) in one step, B. subtilis BEST6234, which was constructed from RM125 (13), a derivative of 168, was used. Briefly, strain BEST6234 has two functional units in its genome. One is a Pr-neo cassette, which is a neomycin resistance gene (neo) whose original promoter was replaced with the Pr promoter from lambda phage to control neo expression by a cI gene of lambda phage. This cassette was substitutionally inserted between the NotI sites of yvfC and yveP. The other is pBR322Cm, containing a chloramphenicol resistance gene, inserted into the BamHI site in the leuB coding sequence (Fig. 1). Prior to the horizontal transfer of the iturin A operon, two landing pad sequences (2 kb and 4 kb), corresponding to the two edges of the region to be transferred and bracketing a cassette of the cI repressor gene with the spectinomycin resistance gene (spc) between them, were assembled in pBR322 in E. coli and then localized in the genomic pBR322 sequence in the leuB gene of BEST6234, as shown in Fig. 1. The resulting strain, 6234/cI, was cultivated to develop competent cells, supplemented with whole chromosomal DNA of RB14, and then plated onto neomycin-containing LB plates. About 100 neomycin-resistant transformants were then screened for sensitivity to spectinomycin, and 20 transformants showing a neomycin-resistant and spectinomycin-susceptible phenotype were selected. These transformants were subjected to a colony PCR assay using specific primers for the internal region of the iturin A operon. Two of them yielded a 2-kb PCR product, and one of these was chosen for further experiments and named 6234/itu. To confirm the transfer of the iturin A operon to the leuB region of the recipient strain, the whole chromosome of 6234/itu was digested with NotI or SfiI, fractionated by pulsed-field gel electrophoresis, and then subjected to Southern hybridization analysis using pBRE4H8 as the probe. As shown in Fig. 2, strong signals from 6234/itu were observed because pBRE4H8 has 12-kb-long iturin A operon sequences plus the pBR322 sequence. On the other hand, BEST6234 showed weak bands, which are due to hybridization between the genomic copy of pBR322 and the pBR322 sequences of pBRE4H8. The sizes of the observed bands are consistent with the expected values from the whole-genome sequence (Fig. 2). 6234/itu was thus confirmed to have integrated the 42-kb region containing the iturin A operon into the leuB region.
FIG. 2.
Confirmation of iturin A operon transfer. (A) Southern hybridization analysis. SfiI and NotI digestion of BEST6234 and 6234/itu genomic DNA prepared in an agarose gel block were fractionated by CHEF pulsed-field gel electrophoresis and subjected to Southern hybridization analysis using pBRE4H8 as the probe. Letters correspond to labeled portions of the map in panel B. EtBr, ethidium bromide. (B) Physical map around leuB of iturin A operon-transferred strain. Lettered fragments on the map were actually observed as bands on the photographs in panel A. This figure is drawn on the basis of the whole-genome sequence of strain 168 (DDBJ/EMBL/GenBank accession no. AL009126). Diagonal and vertical lines represent B. subtilis 168 DNA and B. subtilis RB14 DNA, respectively.
Conversion of iturin A operon-transferred strain into an iturin A producer.
In a previous study, we demonstrated that the 4′-phosphopantetheinyl transferase sfp gene is essential for iturin A production as well as for surfactin production in the RB14 strain (7, 9).
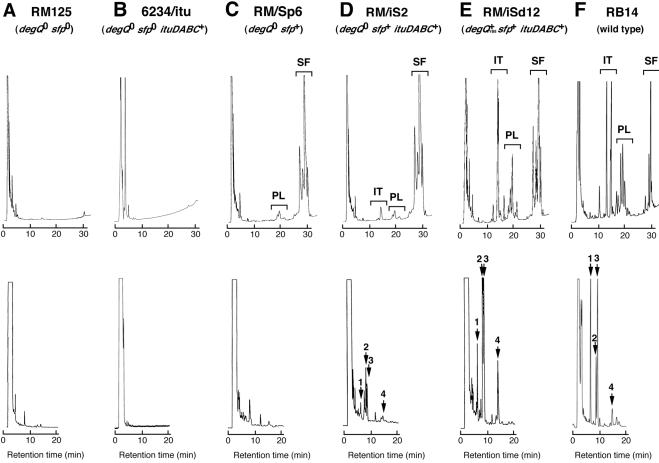
Since strain 168 lacks a functional 4′-phosphopantetheinyl transferase gene (sfp), a functional sfp gene is introduced by a Campbell-like insertion of the sfp-containing plasmid pMMN6 (26). When sfp was transferred to 6234/itu, the resulting strain, RM/iS2, produced iturin A at a concentration of 8 μg/ml in no. 3S medium at 30°C for 120 h, while the control strain, 6234/itu, and RM/Sp6 did not produce iturin A (Fig. 3). However, the production of iturin A by RM/iS2 was 13-fold less than that by RB14 (105 μg/ml).
FIG. 3.
HPLC peak patterns of three lipopeptides (iturin A [IT], surfactin [SF], and plipastatin [PL]) (top) and those focused on iturin A (bottom) produced by RM125 (A), 6234/itu (B), RM/Sp6 (C), RM/iS2 (D), RM/iSd12 as represented by four RM/iSd strains (E), and RB14 (F) in no. 3S medium at 30°C for 120 h. Two distinct HPLC conditions were used to analyze one sample. The top chromatographs were obtained with a two-eluent gradient, while bottom chromatographs were obtained with one eluent. Peaks 1, 2, 3, and 4 correspond to iturin A whose β-amino acids are n-C14-β-amino acid, anteiso-C15-β-amino acid, iso-C15-β-amino acid, and n-C16-β-amino acid, respectively.
In our previous study, we also demonstrated that the introduction of the degQYB8 gene into the sfp+ strain 168 derivative causes plipastatin hyperproduction (32). Analogous with our previous study, we examined the effect of degQYB8 on iturin A production by the sfp+ strain RM/iS2. By transformation with plasmid pUC19HP1NmrF, which carries degQYB8, 28 transformants (RM/iSd series) were obtained, and 4 of them, randomly selected (RM/iSd4, -12, -14, and -16), were subjected to further investigation. Southern hybridization analysis was performed to analyze the integration sites of pUC19HP1NmrF in these transformants. In each of them, the integration site of the plasmid was confirmed to be into one of the two ampicillin resistance genes that are in the genomic copies of pBR322 (RM/iSd4, -12, and -14) or pMMN6 (RM/iSd16) and not into the degQ0 genomic allele (data not shown). This occurred probably because the length of degQYB8 in pUC19HP1NmrF is 0.5 kb, shorter than the 1-kb-long ampicillin resistance gene present in the vector portion of pUC19HP1NmrF. In consideration of the two integration sites of pUC19HP1NmrF in RM/iS2, all of the four transformants of the RM/iSd series were assayed for iturin A production. The production levels by the four strains ranged from 51 to 64 μg/ml, which was six- to eightfold higher than that of RM/iS2 (not containing degQYB8) and one-half that of RB14. The time course of iturin A production, as well as those of surfactin and plipastatin production in the RM/iSd series strains, was similar to that of iturin A production in RB14 (data not shown).
A slight difference was observed in the detailed peak compositions of the iturin A produced, which was caused by a structural difference of the β-amino acids in iturin A (Fig. 3). Generally, the highest peak of iturin A produced by RB14 was peak 1, which corresponds to the n-C14-β-amino acid, while that of the RM/iSd series strains was peak 2, which corresponds to the anteiso-C15-β-amino acid. As well as the differences in iturin A production, the fatty acid compositions of surfactin and plipastatin produced by RB14 were different from those seen for the 168 derivatives.
DISCUSSION
Using a positive-selection system, we transferred a 42-kb region that contains a 38-kb complete iturin A operon that had no selectable markers. In the previous artificial transfer of the subtilin operon or the bacitracin operon into strain 168, transformants were obtained from a donor DNA which had a selection marker inserted into the relevant operon at the original host level for the direct selection of the transferred recombinant (4, 22). It is implied that the donor of this type of horizontal transfer is limited to the organisms that have established genetic transformation to introduce the selectable marker in the donor genome prior to horizontal transfer. In our study, we performed the positive-selection system, which employs the CI repressor and a Pr promoter-driven neomycin resistance gene (Pr-neo) to detect transfer. In this system, the CI repressor in the host strain usually represses Pr-neo expression, causing a neomycin-susceptible phenotype. However, once the transferred DNA is substituted for the cI gene, the host strain is no longer able to repress the Pr-neo expression and becomes neomycin resistant. Since this system does not require any selection marker in the donor DNA, it is suitable for the study of general horizontal transfer (14, 15, 16).
Of the utmost relevance in this study was to identify which genes are required for the conversion of a heterologous host into an antibiotic producer. Several genes are thought to be involved in antibiotic production: for example, regulator genes that control the conditional expression of antibiotic synthetase genes; antibiotic synthetase or structural genes; modification genes; and self-resistance, or efflux pump, genes. In general, these antibiotic production-related genes would form a cluster in the genome. In particular, synthetase genes are usually in one transcriptional unit that is expressed from one promoter in a polycistronic manner. This centered regulation appears to reflect the need for a strict expression control of synthetases. However, Guenzi et al. demonstrated that by inserting a secondary promoter into the surfactin operon, coordinate transcription of surfactin synthesis is not necessary for surfactin production (5). From the viewpoint of horizontal transfer, the cluster feature is reasonable, due to the very low frequency of transformation of multiple-donor regions. If these genes are separated into several parts, the simultaneous transfer of several genes necessary for conversion into a producer may be quite rare. When the cluster feature is a consequence of horizontal transfer, it is probable that the transfer of one cluster may be sufficient for conversion into a producer. Indeed, in a previous horizontal transfer of a bacitracin operon to strain 168, the transferred genes contained self-resistance genes and their regulator genes as well as synthetase genes, all of which are members of the cluster (4). In the transfer of the antibiotic subtilin, the transferred segment has not only the structural gene of the antibiotic but also the regulator genes of the structural gene, modification gene, immunity gene, and efflux gene, which are also of the same cluster (17, 22, 23). Thus, we postulated that the iturin A operon, composed of one transcriptional unit encoding four synthetase genes, has the ability to completely convert a heterologous host into an iturin A producer upon transfer of this operon.
The 6234/itu transformant carrying the iturin A operon was not able to produce iturin A due to the presence of a null sfp allele in this strain. We previously demonstrated that the sfp homologue gene lpa-14 is essential for iturin A production in RB14 (7, 9). When sfp was introduced into the 6234/itu strain, the strain produced iturin A, although at a very low level (Fig. 3). However, the integration of the additional pleiotropic regulator, degQ, which is also mutated in strain 168, resulted in about half of the iturin A production as that of RB14. Therefore, we concluded that the iturin A operon is the only cluster required for conversion of the non-iturin A producer B. subtilis into an iturin producer.
Even without the introduction of a self-resistance, or efflux pump, gene, the transferred strain produced iturin A in the heterologous host, indicating that it has an intrinsic resistance to or secretion mechanism for iturin A. We are currently investigating the resistance, or efflux, mechanism in the transferred strain.
REFERENCES
- 1.Aharonowitz, Y., G. Cohen, and J. F. Martin. 1992. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation, and evolution. Annu. Rev. Microbiol. 46:461-495. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eppelmann, K., S. Doekel, and M. A. Marahiel. 2001. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 276:34824-34831. [DOI] [PubMed] [Google Scholar]
- 5.Guenzi, E., G. Galli, I. Grgurina, E. Pace, P. Ferranti, and G. Grandi. 1998. Coordinate transcription and physical linkage of domains in surfactin synthetase are not essential for proper assembly and activity of the multienzyme complex. J. Biol. Chem. 273:14403-14410. [DOI] [PubMed] [Google Scholar]
- 6.Guenzi, E., G. Galli, I. Grgurina, D. C. Gross, and G. Grandi. 1998. Characterization of the syringomycin synthetase gene cluster. J. Biol. Chem. 273:32857-32863. [DOI] [PubMed] [Google Scholar]
- 7.Hiraoka, H., T. Ano, and M. Shoda. 1992. Molecular cloning of a gene responsible for the biosynthesis of the lipopeptide antibiotics iturin and surfactin. J. Ferment. Bioeng. 74:323-326. [Google Scholar]
- 8.Hiraoka, H., O. Asaka, T. Ano, and M. Shoda. 1992. Characterization of Bacillus subtilis RB14, coproducer of peptide antibiotics iturin A and surfactin. J. Gen. Appl. Microbiol. 38:635-640. [Google Scholar]
- 9.Huang, C. C., T. Ano, and M. Shoda. 1993. Nucleotide sequence and characteristics of the gene, lpa-14, responsible for biosynthesis of the lipopeptide antibiotics iturin A and surfactin from Bacillus subtilis RB14. J. Ferment. Bioeng. 76:445-450. [Google Scholar]
- 10.Isogai, I., S. Takayama, S. Murakoshi, and A. Suzuki. 1982. Structures of β-amino acids in antibiotics iturin A. Tetrahedron Lett. 23:3065-3068. [Google Scholar]
- 11.Itaya, M., and T. Tanaka. 1991. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J. Mol. Biol. 220:631-648. [DOI] [PubMed] [Google Scholar]
- 12.Itaya, M. 1993. Integration of repeated sequences (pBR322) in the Bacillus subtilis 168 chromosome without affecting the genome structure. Mol. Gen. Genet. 241:287-297. [DOI] [PubMed] [Google Scholar]
- 13.Itaya, M., and T. Tanaka. 1997. Predicted and unsuspected alterations of the genome structure of genetically defined Bacillus subtilis 168 strains. Biosci. Biotechnol. Biochem. 61:56-64. [Google Scholar]
- 14.Itaya, M. 1999. Effective cloning of unmarked DNA fragments in the Bacillus subtilis genome. Biosci. Biotechnol. Biochem. 63:602-604. [DOI] [PubMed] [Google Scholar]
- 15.Itaya, M., T. Nagata, T. T. Shiroishi, K. Fujita, and K. Tsuge. 2000. Efficient cloning and engineering of giant DNAs in a novel Bacillus subtilis genome vector. J. Biochem. (Tokyo) 128:869-875. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, M., K. Fujita, M. Ikeuchi, M. Koizumi, and K. Tsuge. 2003. Stable positional cloning of long continuous DNA in the Bacillus subtilis genome vector. J. Biochem. (Tokyo) 134:513-519. [DOI] [PubMed] [Google Scholar]
- 17.Izaguirre, G., and J. N. Hansen. 1997. Use of alkaline phosphatase as a reporter polypeptide to study the role of the subtilin leader segment and the SpaT transporter in the posttranslational modifications and secretion of subtilin in Bacillus subtilis 168. Appl. Environ. Microbiol. 63:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koumoutsi, A., X.-H. Chen, A. Henne, H. Liesegang, G. Hitzeroth, P. Franke, J. Vater, and R. Borriss. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst, F., et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 22.Liu, W., and J. N. Hansen. 1991. Conversion of Bacillus subtilis 168 to a subtilin producer by competence transformation. J. Bacteriol. 173:7387-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, W., and J. N. Hansen. 1992. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J. Biol. Chem. 267:25078-25085. [PubMed] [Google Scholar]
- 24.Maget-Dana, R., and F. Peypoux. 1994. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151-174. [DOI] [PubMed] [Google Scholar]
- 25.Nagahari, K., and K. Sakaguchi. 1978. Cloning of Bacillus subtilis leucine A, B, and C genes with Escherichia coli plasmids and expression of the leuC gene in E. coli. Mol. Gen. Genet. 158:263-270. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M. M., N. Corbell, J. Besson, and P. Zuber. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol. Gen. Genet. 232:313-321. [DOI] [PubMed] [Google Scholar]
- 27.Peypoux, F., M. Guinand, G. Michel, L. Delcambe, B. C. Das, and E. Lederer. 1978. Structure of iturine A, a peptidolipid antibiotic from Bacillus subtilis. Biochemistry 17:3992-3996. [DOI] [PubMed] [Google Scholar]
- 28.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer, B. A., S. J. Admiraal, H. Gramajo, D. E. Cane, and C. Khosla. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790-1792. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, N.Y.
- 31.Tsuge, K., T. Ano, and M. Shoda. 1996. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotics plipastatin B1 and surfactin in Bacillus subtilis YB8. Arch. Microbiol. 165:243-251. [DOI] [PubMed] [Google Scholar]
- 32.Tsuge, K., T. Ano, M. Hirai, Y. Nakamura, and M. Shoda. 1999. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 43:2183-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umezawa, H., T. Aoyagi, T. Nishikiori, A. Okuyama, Y. Yamagishi, M. Hamada, and T. Takeuchi. 1986. Plipastatins: new inhibitors of phospholipase A2, produced by Bacillus cereus BMG202-fF67. I. Taxonomy, production, isolation and preliminary characterization. J. Antibiot. 39: 737-744. [DOI] [PubMed] [Google Scholar]