Abstract
Homologous recombination between closely related gene cassettes, such as aadA1 and aadA2, which are 89% identical, can create hybrid cassettes and hybrids of existing cassette arrays. A new cassette array, dfrA12-orfF-aadA8b, which was created by such a recombination event occurring within the aadA2 cassette in the dfrA12-orfF-aadA2 array, has been identified.
Gene cassettes are a major source of the resistance genes found in clinical, commensal, and environmental isolates of bacteria that are resistant to antibiotics. Most commonly, they are found in association with class 1 or class 2 integrons (3, 13). One growing group of gene cassettes encodes aminoglycoside (3′) (9) adenylyltransferases that confer resistance to both streptomycin and spectinomycin (9). The genes and the cassettes, which are named after the genes, are designated aadA with an Arabic numeral to distinguish distinct genes, namely, those that differ by at least 2% in both the DNA and protein sequences. Two of these cassettes, aadA1 and aadA2, were present in two of the earliest-known plasmids that confer resistance to multiple antibiotics, namely, NR1, also called R100 (7), and pSa (1, 14), and remain very common in modern-day isolates. Though they have both been found in various contexts, they recur in a few specific cassette arrays, e.g., aadA1 or aadA2 alone, oxa1-aadA1, dfrA1-aadA1, and dfrA12-orfF-aadA2, which appear to have become globally disseminated. These two cassettes have now been sequenced many times, leading to the identification of several variant sequences for each of them, each containing one or a few single base changes (Fig. 1; also Fig. 3 in reference 9). The high level of similarity between the aadA1 and aadA2 cassette sequences (89.3% DNA identity; cassette length, 856 bp) means that they share many stretches of sequence identity that allow homologous recombination between them to occur. Several hybrids between aadA1 and aadA2 that presumably arose by homologous recombination have already been reported (Table 1). By combining existing knowledge of variant sequences and of cassette arrays, it is potentially possible to track the movement of specific sets of gene cassettes within bacterial populations and identify events that have been involved in their creation and dissemination.
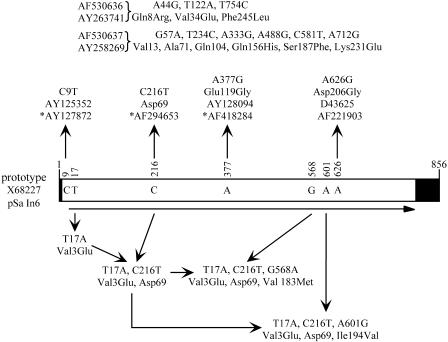
FIG. 1.
Variants of the aadA2 gene cassette. The linear form of the 856-bp aadA2 cassette is shown, with the 59-base element (59-be) represented by a filled box and an arrow below indicating the extent of the aadA2 gene. The bases found in the prototype aadA2 sequence (GenBank accession no. X68227) at positions that can vary are shown in the box, with their positions in the cassette sequence given above. Position 1 is the first T residue of the 1R site (GTTAGAC) of the 59-be. For simplicity, only changes found in at least two of the available aadA2 sequences are included and variations in the 59-be are not shown. Variant sequences are grouped according to base changes, with any corresponding amino acid changes indicated. Variants shown below the box are found in the dfrA12-orfF-aadA2 cassette array (accession numbers in Table 2). Variants found in other arrays are listed above the box, with accession numbers shown, except for T17A C216T variants found in GenBank accession no. AY263740, *AF458082, *AF486817, and *AY681136 (an asterisk indicates that the sequence contains additional unique changes). Seventeen further sequences are identical to the prototype, and seven more have additional differences found only in these entries.
TABLE 1.
Hybrids of aadA1 and aadA2 gene cassettes
| GenBank accession no. | Namea | Original name | Order | Crossover positionb | Change(s) from standard cassettec
|
Cassette array | Species | Reference or source | |
|---|---|---|---|---|---|---|---|---|---|
| aadA2d | aadA1d | ||||||||
| AF047479 | aadA3 | aadA3 | 2/1 | 758-760 | C759T | aadA3-aacA1/orfG-orfsH to M-catB3 | |||
| AF326210e | aadA8 | aadA8 | 2/1 | 550-600 | C759T + 4f | Unknown | Klebsiella pneumoniae | 12 | |
| AY139603 | aadA8b | aadA8 | 2/1 | 602-647 | A402Gg | C759T | aadA8b | Uncultured | 15 |
| AY852272 | aadA8b | 2/1 | 602-647 | T17A C216T | dfrA12-orfF-aadA8b | Mixed culture | This work | ||
| AY232671 | aadA1 | aadA1 | 2/1 | 88-101 | T17A | C759T | aadA2/1-blaP1 | Pasteurella multocida | 16 |
| AY171244e | aadA21 | aadA21 | 2/1 | 148-159 | GC634-635CGg | aadA21 | Salmonella enterica serovar Typhimurium | 2 | |
| AY550883 | aadA21 | aadA22 | 2/1 | 148-159 | G790Ag | aadA21 | Salmonella enterica serovar Choleraesuis | ||
| AJ809407 | aadA21 | aadA23 | 2/1 | 148-159 | G444Ag | aadA21 | Salmonella enterica serovar Agona | ||
| AB189176 | aadA21 | aadA23b | 2/1 | 148-159 | A529Gg | aadA21 | Escherichia coli | ||
| G790Ag | |||||||||
| AY665771h | aadA12 | aadA12 | 2/1/2 | 148-159 | Unknown | Escherichia coli | |||
| 550-600 | |||||||||
| AB107663 | aadA1 | aadA1 | 1/2 | 783-786 | G732C C759T | aadA1/2 | Vibrio cholerae | ||
Cassettes with high identity (>98%) to aadA1 or aadA21 are named aadA1 or aadA21, respectively.
For boundary x-y, the last base identifiable as matching only the “front” cassette is x − 1, and the first base identifiable as matching only the “back” cassette is y + 1.
Unless otherwise indicated, these differences are found in a number of other variants of the relevant cassette.
The standard cassettes used for comparison are found in GenBank accession no. X12870 for aadA1, variants of which commonly have C759T, and X68227 for aadA2.
The first 9 nucleotides of the cassette sequence are missing from AF326210, and the last 28 bp of the cassette sequence are missing from AY171244.
The four additional changes are C609A, A612G, G773A (also found in the aadA1 cassette variant in AY139597), and the unique change C599T.
The changes indicated have not been seen in any other variant of the relevant cassette.
The sequence of the aadA12 gene only is present in AY665771.
We used primers located in the 5′- and 3′-conserved segments (CS) of class 1 integrons (HS458, 5′-GTTTGATGTTATGGAGCAGCAACG-3′, and HS459, 5′-GCAAAAAGGCAGCAATTATGAGCC-3′ [5]) to screen DNA isolated from mixed bacterial samples recovered from the feces of human volunteers with no recent exposure to antibiotics. Mixed cultures were grown in L broth at 37°C under aerobic conditions, and the organisms recovered were predominantly Escherichia coli. DNA was recovered using alkaline lysis, and PCR conditions were as follows: denaturation at 94°C for 3.0 min; 35 cycles at 94°C for 30 s, 65°C for 1.0 min, and 72°C for 1.5 min or 4.0 min in the final cycle. A significant proportion of the samples screened yielded an amplicon of 2.2 kb corresponding to a cassette array of 1.7 to 1.8 kb when corrected for the amplified portions of the 5′-CS and 3′-CS (465 bp). Partial sequencing of seven of the PCR products revealed that the cassette array was either dfrA12-orfF-aadA2 (in one case) or a derivative in which the end of the aadA2 cassette is replaced by the corresponding part of the aadA1 cassette (in six cases) (Fig. 2). The 2.2-kb amplicon from one of the latter samples was cloned into pGEM-T Easy (Promega) by following the manufacturer's instructions, with selection on LB plates containing streptomycin (25 μg ml−1) and trimethoprim (50 μg ml−1), and the resultant plasmid (pMAQ697) also conferred resistance to spectinomycin (25 μg ml−1). The insert was sequenced using procedures described previously (11). The crossover in the hybrid aadA2/aadA1 cassette was located between positions 602 and 647 in the cassette (numbered from the conserved TT at beginning of the cassette) by comparison with the reference aadA1 (GenBank accession no. X12870) and aadA2 (X68227) cassette sequences. A similar hybrid with the same crossover position has recently been reported as aadA8 (AY139603 [15]). However, the original aadA8 cassette (AF326210 [12]), which is also an aadA2/1 hybrid, has a different crossover position, between 550 and 600 (Table 1). As the three sequences exhibit high levels (>98%) of DNA and protein identity, we have named the aadA2/1 hybrid cassettes with crossover between positions 602 and 647 aadA8b (Table 1) to indicate the difference in the crossover position.
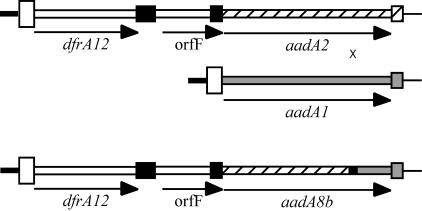
FIG. 2.
Schematic of the original and hybrid cassette arrays. The aadA8b cassette has arisen by homologous recombination between the aadA2 cassette (hatched) in a dfrA12-orfF-aadA2 cassette array (e.g., GenBank accession no. AF284063) and an aadA1 cassette (shaded) in another integron. Cassettes are represented as narrow open boxes with a larger box at one end representing the 59-base element and are shown to scale. × indicates the position of the crossover. The region (between positions 602 and 647) where recombination took place is indicated by a small filled box. The attI1 site is shown as a tall open box and defines the end of the 5′-CS (thick black line). The thin black line beyond the last cassette represents the 3′-CS sequence.
To examine if the recombination event could have occurred between the dfrA12-orfF-aadA2 cassette array in one integron and an aadA1 cassette located in a second integron, the sequences were examined in detail. The dfrA12-orfF-aadA2 cassette array was first isolated from an E. coli strain from Finland (4), but the sequence of the aadA2 cassette was not completed (GenBank accession no. Z21672). However, this array has since been found in many bacterial species (Table 2) isolated in many different countries from clinical and animal-associated sources and from wastewater. Examination of the sequences of this array recorded in the relevant GenBank entries revealed that five are identical to one another (reference sequence AF284063) and to the incomplete Z21672 sequence. The remainder differ by only a few single base changes (Table 2; Fig. 1), and some of these variations may represent either sequence errors or errors arising during PCR amplification. This suggests that this array arose on a single occasion and has become globally disseminated but subsequently acquired occasional base substitutions.
TABLE 2.
dfrA12-orfF-aadA2 cassette arrays
| GenBank accession no. | Sequence difference(s) in cassettea
|
Organism | Reference or source | ||
|---|---|---|---|---|---|
| dfrA12 | orfF | aadA2 | |||
| Z21672b | —c | Escherichia coli | 4 | ||
| AF284063d | Serratia marcescens | ||||
| AB154407 | Escherichia coli | ||||
| AB191048 | Staphylococcus aureus | ||||
| AF550415 | Citrobacter freundii | ||||
| AY748453 | Klebsiella pneumoniae | ||||
| AY852272 | —e | Mixed culture | This work | ||
| AB191047 | T178C | Pseudomonas aeruginosa | |||
| AF175203 | AAAA528-31AAAf | GG302-303GGGf | Citrobacter freundii | ||
| AY522923 | G568A | Aeromonas hydrophila | |||
| AF188331 | GGG101-103GGg | G568A | Shigella flexneri | ||
| AB196348 | G300A G360A | G302Tf | A601G | Enterococcus faecalis | |
| AY115474 | C567Gf | C50G G75A T133C C140G T248C T267Gf | A601G | Uncultured | 15 |
| AY139605 | C567Gf | C50G G75A T133C C140G T248C T267Gf | A601G | Uncultured | 15 |
| AY551331 | T115C C179T | A732G | Salmonella enterica serovar Choleraesuis | 6 | |
| AF335108 | T216Ch CCG826-828Δf | Escherichia coli | 8 | ||
| AF180731 | GC209-210CG | A34G G642T C826Δf G830Δf | Klebsiella pneumoniae | ||
Unless indicated otherwise, these changes are seen only in GenBank entries included in this table.
Standard sequence for the dfrA12 and orfF cassettes.
Sequence is only available to position 125 of the aadA2 cassette.
Standard sequence for the complete dfrA12-orfF-aadA2 array.
The aadA8b hybrid cassette is identical to the aadA2 cassette in AF284063 to position 602 and to the standard aadA1 cassette in X12870 from positions 647 to 856.
In the 59-base element.
This change disrupts the dfrA12 reading frame, and there are likely errors elsewhere in this sequence (10).
Found in the prototype aadA2 sequence in X68227.
There are several variant sequences for the aadA2 cassette found among those deposited in GenBank (Fig. 1), but aadA2 in the dfrA12-orfF-aadA2 arrays (Fig. 1) usually differs at two positions (T17A and C216T) from the reference aadA2 cassette found in In6 from plasmid pSa (X68227). In the dfrA12-orfF-aadA2 and dfrA12-orfF-aadA8b arrays recovered here, the aadA2 gene cassette and the aadA2 portion of the aadA8b cassette are identical to the aadA2 variant found in the standard dfrA12-orfF-aadA2 array (AF284063), indicating that the recombination event is likely to have occurred within this array. Beyond the position of the switch to aadA1, the sequence of the aadA8b cassette is identical to the prototype for aadA1 (X12870) and to single aadA1 cassettes recovered from other samples in our commensal mixed-culture collection. In contrast, the variant of aadA8b found as a single cassette in AY139603 (15) differs from the version reported here at three positions in the aadA2 portion and one in the aadA1 portion (Table 1), indicating that the two variants are likely to have arisen via independent recombination events occurring within the same span (positions 602 to 647).
It is clear that recombination events involving the aadA1 and aadA2 cassettes have occurred on several occasions (Table 1) and thus contribute significantly to the creation of new cassette arrays. However, the fact that the dfrA12-orfF-aadA8b array was recovered from the fecal flora of a number of different individuals indicates that it has already become widely distributed, at least in Australia.
Nucleotide sequence accession number.
The sequence of the dfrA12-orfF-aadA8b PCR amplicon has been deposited in GenBank under accession no. AY852272.
Acknowledgments
S.R.P. was supported by grant no. 192108 from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Bito, A., and M. Susani. 1994. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother. 38:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faldynova, M., M. Pravcova, F. Sisak, H. Havlickova, I. Kolackova, A. Cizek, R. Karpiskova, and I. Rychlik. 2003. Evolution of antibiotic resistance in Salmonella enterica serovar Typhimurium strains isolated in the Czech Republic between 1984 and 2002. Antimicrob. Agents Chemother. 47:2002-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 4.Heikkilä, E., M. Skurnik, L. Sundström, and P. Huovinen. 1993. A novel dihydrofolate reductase cassette inserted in an integron borne on a Tn21-like element. Antimicrob. Agents Chemother. 37:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes, A. J., M. R. Gillings, B. S. Nield, B. C. Mabbutt, K. M. H. Nevalainen, and H. W. Stokes. 2003. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5:383-394. [DOI] [PubMed] [Google Scholar]
- 6.Huang, T.-M., Y.-F. Chang, and C.-F. Chang. 2004. Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan. Vet. Microbiol. 100:247-254. [DOI] [PubMed] [Google Scholar]
- 7.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morabito, S., R. Tozzoli, A. Caprioli, H. Karch, and A. Carattoli. 2002. Detection and characterization of class 1 integrons in enterohemorrhagic Escherichia coli. Microb. Drug Resist. 8:85-91. [DOI] [PubMed] [Google Scholar]
- 9.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partridge, S. R., and R. M. Hall. 2004. Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob. Agents Chemother. 48:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters, E. D., M. A. Leverstein-van Hall, A. T. Box, J. Verhoef, and A. C. Fluit. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 14.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 15.Tennstedt, T., R. Szczepanowski, S. Braun, A. Pühler, and A. Schlüter. 2003. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45:239-252. [DOI] [PubMed] [Google Scholar]
- 16.Wu, J.-R., H. K. Shieh, J.-H. Shien, S.-R. Gong, and P.-C. Chang. 2003. Molecular characterization of plasmids with antimicrobial resistant genes in avian isolates of Pasteurella multocida. Avian Dis. 47:1384-1392. [DOI] [PubMed] [Google Scholar]