Abstract
The majority of Shiga-toxigenic Escherichia coli (STEC) strains isolated from humans with gastrointestinal disease carry large (approximately 90-kb) plasmids. We have been analyzing the megaplasmid (designated pO113) from an O113:H21 STEC strain (98NK2). This strain lacks the locus for enterocyte effacement (LEE) and yet was responsible for an outbreak of hemolytic uremic syndrome. In the present study, we demonstrate that pO113 carries a novel type IV pilus biosynthesis locus (pil) related to those of the IncI plasmids R721, R64, and ColIb9. The pO113 pil locus consists of 11 closely linked genes (pilL through pilV) with an additional separately transcribed upstream gene (pilI). It directs the expression of long thin pili on the 98NK2 surface and the hemagglutination of guinea pig erythrocytes. We also demonstrate that pO113 can be transferred by conjugation. However, the type IV pilus encoded by pO113 does not appear to be involved in the adherence of 98NK2 to HEp-2 or Hct-8 cells in vitro. Homologues of the pO113 pil locus were present in several other LEE-negative STEC strains but not in LEE-positive STEC strains belonging to serogroup O26, O111, or O157.
Shiga-toxigenic Escherichia coli (STEC) are an important cause of gastrointestinal disease in humans, particularly since infections with these bacteria may result in life-threatening sequelae, such as hemolytic uremic syndrome (HUS) (14, 20, 25). The production of Shiga toxin (Stx) is associated with over 200 E. coli O:H serotypes (20), but epidemiological data indicate that not all of these are highly virulent for humans. Thus, although Stx is a sine qua non of virulence, additional STEC properties, including the capacity to adhere to the intestinal epithelium and colonize the gut, undoubtedly contribute to the pathogenic process. Indeed, STEC strains with the capacity to form attaching and effacing lesions on intestinal mucosae appear to be responsible for the majority of serious cases (those complicated by HUS) (14, 25). This property is encoded by a pathogenicity island termed the locus of enterocyte effacement (LEE) (26), which is present in several important STEC serogroups, most notably O157 and O111. These two serogroups have been responsible for the vast majority of recorded outbreaks of STEC disease complicated by HUS. However, the presence of LEE is not essential for pathogenesis, and a proportion of sporadic cases of HUS, as well as occasional outbreaks, are caused by LEE-negative STEC strains (23, 25).
The mechanism whereby LEE-positive strains adhere intimately and generate attaching and effacing lesions has been the subject of intensive study in recent years (for reviews, see references 8 and 20), but it seems likely that additional (non-LEE-encoded) adherence mechanisms also operate in STEC. For example, O157:H7 STEC have been reported to produce a chromosomally encoded 67-kDa homologue of Vibrio cholerae IrgA, termed Iha, which mediates the adherence of O157:H7 STEC to HeLa cells (33). Other putative STEC adhesins include fimbriae, which were initially thought to be encoded by pO157, the 93-kb virulence-related plasmid of O157:H7 STEC. Karch et al. (13) found that the presence of the plasmid correlated with the expression of fimbriae and adherence to Henle 407 cells but not to HEp-2 cells. However, recent genome sequence analyses of two O157:H7 STEC strains indicated that fimbrial loci are not present on pO157 (6, 17), although several such loci are located on the chromosome (11, 27).
Adherence phenotypes have been examined for several LEE-negative STEC strains (7, 9, 30), but virtually nothing is known of the actual mechanisms involved. A gene has recently been isolated from the megaplasmid (designated pO113) of a LEE-negative O113:H21 STEC strain responsible for an outbreak of HUS (23), which encodes an autoagglutinating adhesin designated Saa (STEC autoagglutinating adhesin) (22). The introduction of saa cloned in pBC resulted in a 9.7-fold increase in adherence of E. coli JM109 to HEp-2 cells and a semilocalized adherence pattern. Mutagenesis of saa, or curing the wild-type strain of pO113, resulted in a significant reduction in adherence. Homologues of saa were found in several unrelated LEE-negative STEC serotypes (including additional isolates from patients with HUS) but were not present in LEE-positive STEC strains (22). These findings underscore the fact that there are fundamental differences in the genetic compositions of the megaplasmids of LEE-positive and LEE-negative STEC. In the present study, we have continued our examination of pO113 and describe a novel type IV pilus biosynthesis locus encoded thereon.
MATERIALS AND METHODS
Bacterial strains and cloning vectors.
The O113:H21 STEC strain 98NK2 was isolated from a patient with HUS at the Women's and Children's Hospital, Adelaide, South Australia, Australia (23). A derivative of 98NK2 which had been cured of the megaplasmid (98NK2-Cu) and one in which a 1,402-bp internal portion of the saa gene carried by the megaplasmid had been deleted and replaced with a kanamycin resistance cartridge (98NK2-S) have also been described previously (22). All other STEC strains used in this study were also isolated at the Women's and Children's Hospital and have been described previously (24), except for the O157:H7 strain EDL933 (provided by R. Robins-Browne), the O91:H21 strain B2F1 (provided by A. Melton-Celsa), and the O113 strains 3848 and 1183 (provided by J. Bennet). E. coli K-12 strains DH1 and JM109 have been described previously (10, 35). The cosmid vector pHC79 has also been described previously (12). The phagemid pBC SK (encoding chloramphenicol resistance) was obtained from Stratagene (La Jolla, Calif.). All E. coli strains were routinely grown in Luria-Bertani (LB) medium (18) with or without l.5% Bacto Agar (Difco Laboratories, Detroit, Mich.). Where appropriate, ampicillin or chloramphenicol was added to growth media at a concentration of 50 or 40 μg/ml, respectively.
DNA manipulations.
Routine DNA manipulations (restriction digestion, agarose gel electrophoresis, ligation, transformation, Southern hybridization analysis, etc.) were carried out essentially as described by Maniatis et al. (18).
Construction of cosmid gene bank.
DNA from the megaplasmid (pO113) of 98NK2 was extracted by using a Qiagen (Hilden, Germany) plasmid mini kit and was digested partially with Sau3A1 to optimize the yield of fragments in the size range of 35 to 40 kb. This DNA was ligated with a fivefold molar excess of pHC79 DNA, which had been digested with BamHI. Ligated DNA was packaged into lambda heads by using a Packagene kit (Promega Biotec, Madison, Wis.) and transfected into E. coli DH1, which had been grown in LB medium plus 2% maltose. The cells were then plated onto LB agar supplemented with ampicillin, and after incubation, the clones were stored in LB medium plus ampicillin plus 15% glycerol in microtiter plates at −70°C.
DNA sequencing.
For DNA sequencing, the plasmid DNA template was purified by using a QIAPrep spin miniprep kit (Qiagen); alternatively, PCR products were purified by using an Ultraclean PCR clean-up kit (Mo Bio Laboratories, Solana Beach, Calif.). The sequences of both strands were then determined by dye terminator chemistry with either universal M13 sequencing primers or custom-made oligonucleotide primers on an Applied Biosystems model 3700 automated DNA sequencer.
Hemagglutination assay.
Bacteria were grown overnight in LB medium supplemented with appropriate antibiotics, diluted 1:20 in 10 ml of fresh LB medium, and grown with aeration to an A600 of approximately 0.5. Cells were pelleted by centrifugation, resuspended in 10 ml of Dulbecco modified Eagle medium (DMEM) (supplemented with 20 μg of l-proline/ml for E. coli BH101), and incubated for 3 h at 37°C in 5% CO2-95% air. Bacteria were then pelleted, washed once in phosphate-buffered saline (PBS), and resuspended in 100 μl of PBS containing 1% d-mannose. Guinea pig and chicken erythrocytes (obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia) or human type A, B, and O erythrocytes (obtained from the Red Cross Blood Transfusion Service, Adelaide, Australia) were washed three times in PBS (pH 7.4) and suspended at a density of 0.3% (vol/vol) in PBS. Hemagglutination assays were carried out in 96-well round-bottom microtiter plates containing 50 μl of serial 10-fold dilutions of each bacterial suspension in PBS (pH 7.4) containing 1% d-mannose. Fifty microliters of washed erythrocyte suspension was then added to the appropriate wells, and the plates were examined for macroscopic hemagglutination after 2 to 4 h at room temperature. Complete hemagglutination involving all of the erythrocytes was scored as “+++”; progressively lesser degrees of hemagglutination were scored as “++” and “+”. Trace hemagglutination was scored as “±”, whereas reactions in which the erythrocytes formed a tight button indistinguishable from that seen in the presence of PBS were scored as “−”.
Conjugation.
Conjugation was carried out with 98NK2-S as the donor (the megaplasmid of this strain has a kanamycin resistance cartridge in lieu of the saa gene). The recipient was a streptomycin-resistant derivative of 98NK2-Cu (the megaplasmid-cured derivative of 98NK2). Donor and recipient strains were grown overnight at 37°C with shaking in LB medium supplemented with 50 μg of kanamycin/ml or 30 μg of streptomycin/ml, respectively. Donor and recipient strains were washed twice in PBS to eliminate antibiotic and then combined at ratios of 1:5, 1:10, and 1:20. The cells were pelleted by centrifugation, gently resuspended in 200 μl of LB broth, spread onto cellulose acetate filters (0.45-μm pore size, type HA; Millipore Corp., Bedford, Mass.) on LB agar, and incubated for 3 h at 37°C. The cells were resuspended in 10 ml of saline, and aliquots were plated onto LB agar plates supplemented with both 50 μg of kanamycin/ml and 30 μg of streptomycin/ml. The transconjugants were counted after overnight incubation at 37°C.
Electron microscopy.
Electron microscopic examination for pili was performed essentially as described previously (32). Bacteria were grown overnight in LB medium supplemented with appropriate antibiotics, diluted 1:20 in 10 ml of fresh LB medium, and grown with aeration to an A600 of approximately 0.5. Cells were pelleted by centrifugation, resuspended in 10 ml of DMEM (supplemented with 20 μg of l-proline/ml for E. coli BH101), and incubated for 3 h at 37°C in 5% CO2-95% air. Bacteria were then pelleted and resuspended in DMEM at a density of approximately 1010 CFU/ml. A 10-μl aliquot of each sample was spotted onto collodion-coated copper grids (300 mesh; TAAB). The grids were then washed twice in PBS and stained with 2% phosphotungstic acid (in PBS, pH 7.4). The grids were examined in a Philips CM 100 transmission electron microscope at an accelerating voltage of 80 kV.
Nucleotide sequence accession number.
The DNA sequence described in this paper has been deposited with GenBank under accession number AF399919.
RESULTS
Cloning of the pO113 pil locus.
In a previous study, it was demonstrated that a cosmid clone (designated pJCP561) derived from the megaplasmid pO113 of the O113:H21 STEC strain 98NK2 encoded a novel autoagglutinating adhesin, designated Saa (22). In the present study, we continued sequence analysis of this clone and found what appeared to be the 5′ portion of a novel type IV pilus biosynthesis locus approximately 11 kb downstream of saa. BLASTX analysis (1) indicated that the 3′ terminus of the pO113 insert in pJCP561 comprised open reading frames (ORFs) whose highest deduced amino acid similarities were with the PilL lipoproteins from two closely related IncI plasmids (ColIb-P9 from Shigella sonnei [GenBank accession number AB021078] and R64 from Salmonella enterica serovar Typhimurium [GenBank accession number AB027308]) (42% identity) and PilM from plasmid R721 (GenBank accession number AP002527) (36% identity). In order to isolate the complete locus, we constructed a cosmid gene bank of pO113 DNA (see Materials and Methods) and screened it by dot blot hybridization analysis for clones overlapping the 3′ end of pJCP561. One of these, designated pJCP575, was selected for further study. In order to determine the DNA sequence of the insert of pJCP575, a series of EcoRI and HindIII restriction fragments from pJCP575 were subcloned into pBC SK and transformed into E. coli JM109. The various subclones were then subjected to sequence analysis as described in Materials and Methods. Where subclones did not overlap, the sequence across the junction was determined with custom-designed oligonucleotide primers with pJCP575 DNA as the template.
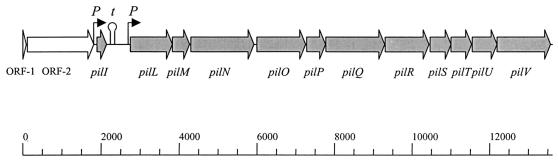
The positions of ORFs within the compiled 13,585-bp sequence of part of the 98NK2 DNA insert in pJCP575 are shown in Fig. 1. Examination of the DNA sequence indicated that it contained 13 complete ORFs and 1 partial ORF, and some features of the genes and their predicted protein products are listed in Table 1. The sequenced region commenced with the 3′ end of an ORF with a high degree of similarity to YqiJ, a 23.1-kDa putative oxidoreductase located in the GLGS-WAAE region of E. coli K-12. The following ORF was similar to YqiK, a 60.7-kDa putative membrane protein encoded by the same region. Homologues of these two proteins are also present in the genome of E. coli O157:H7, although the degree of amino acid identity is slightly lower. The first apparently pilus-related gene, designated pilI, was sufficient to encode an 80-amino-acid polypeptide with 32% identity over 62 amino acids with PilI from the S. enterica serovar Typhimurium plasmid R64. The pilI gene was preceded by a ribosome binding site, and the region immediately 5′ of this contained a putative promoter sequence, as predicted by the NNPP program (http://dot.imgen.bcm.tmc.edu) (28); transcription was predicted to start at T1878. Immediately downstream of pilI (nucleotides [nt] 2152 to 2181) is a potential stem-loop element (ΔG = −18.4 kcal/mol) which may function as a transcription terminator. There is a 610-nt noncoding region between pilI and the next gene, designated pilL. The pilL gene is also preceded by a weak ribosome-binding site, and a predicted promoter sequence is located approximately 150 nt upstream (the predicted transcription start site is nt 2602). The remainder of the pil locus comprises a further 10 genes (designated pilM through pilV), which appear to be linked. The intergenic gaps ranged from −11 to 75 nt (mean = 24 nt). Ribosome binding sites were present before each gene, with the exception of pilR, and there were no stem-loop elements within the pilLMNOPQRSTUV region, suggesting that the genes may form an operon. BLASTX analysis indicated significant similarity between each of the ORFs and the respective components of the pil operon from the IncI plasmid R721 (30 to 66% deduced amino acid sequence identity). Slightly lesser degrees of similarity were observed between the pO113 Pil proteins and their homologues from R64 and ColIb9 and those encoded by the pil locus on the large pathogenicity island of S. enterica serovar Typhi (38) (Table 1). Putative functions for some of the Pil proteins based on amino acid similarities also are also listed in Table 1. On this basis, pilS and pilV are predicted to encode the major and minor prepilin proteins, respectively.
FIG. 1.
Map of the portion of the 98NK2 DNA insert of pJCP575 subjected to sequence analysis, showing the location of ORFs (thick arrows) and putative promoter (P) and transcription termination (t) sites. The scale below the figure is in base pairs.
TABLE 1.
Properties of pO113 pil genes and their products
| Gene or ORF | Location in nucleotide sequence | % G+C | No. of amino acids | Similar protein(s); source (accession no.) | % Identical/% similar (no. of amino acids) | Putative function |
|---|---|---|---|---|---|---|
| ORF 1 | 1 to 93 | 50.6 | 29 | YqiJ; E. coli K-12 (P76657) | 62/82 (29) | Oxidoreductase |
| ORF 2 | 120 to 1814 | 47.3 | 564 | YqiK; E. coli K-12 (P77306) | 82/89 (541) | Membrane protein |
| pilI | 1895 to 2137 | 44.8 | 80 | PilI; plasmid R64 (AB027308) | 32/51 (62) | |
| PilI; plasmid ColIb-P9 (AB021078) | 27/51 (61) | |||||
| pilL | 2747 to 3820 | 41.5 | 357 | PilL; plasmid R64 | 43/57 (355) | Lipoprotein |
| PilL; plasmid ColIb-P9 | 42/56 (355) | |||||
| PilL; plasmid R721 (AP002527) | 28/44 (272) | |||||
| pilM | 3825 to 4262 | 46.1 | 145 | PilM; plasmid R721 | 36/65 (145) | |
| PilM; plasmids R64 and ColIb-P9 | 29/52 (145) | |||||
| PilM; S. enterica serovar Typhi (AF000001) | 31/46 (145) | |||||
| PilM; S. enterica serovar Dublin (AF247502) | 31/47 (142) | |||||
| pilN | 4293 to 5912 | 46.1 | 539 | PilN; plasmid R721 | 64/77 (539) | Outer membrane lipoprotein |
| PilN; plasmids R64 and ColIb-P9 | 39/58 (537) | |||||
| PilNa; S. enterica serovar Typhi and S. enterica serovar Dublin | 45/65 (253) | |||||
| BfpB; enteropathogenic E. coli EAF plasmid (U27184) | 26/45 (529) | |||||
| pilO | 5988 to 7235 | 47.3 | 415 | PilO; plasmid R721 | 40/52 (385) | |
| PilO; S. enterica serovar Typhi and S. enterica serovar Dublin | 25/37 (377) | |||||
| PilO; plasmids R64 and ColIb-P9 | 21/37 (328) | |||||
| pilP | 7225 to 7710 | 41.5 | 161 | PilP; plasmid R721 | 51/69 (61) | |
| pilQ | 7761 to 9269 | 48.3 | 502 | PilQ; plasmid R721 | 66/80 (495) | Nucleotide-binding protein |
| PilQ; S. enterica serovar Typhi and S. enterica serovar Dublin | 45/60 (438) | |||||
| pilR | 9271 to 10368 | 49.1 | 365 | PilR; plasmid R721 | 45/67 (361) | Integral membrane protein |
| PilR; S. enterica serovar Typhi and S. enterica serovar Dublin | 29/46 (344) | |||||
| BfpB; EPEC EAF plasmid | 29/49 (337) | |||||
| pilS | 10442 to 10978 | 43.3 | 178 | PilS; plasmid R721 | 43/57 (180) | Major pilin subunit |
| pilT | 10978 to 11505 | 46.6 | 175 | PilT; plasmid R721 | 61/76 (149) | Transglycosylase |
| PilT; plasmids R64 and ColIb-P9 | 54/67 (157) | |||||
| PilT; S. enterica serovar Typhi and S. enterica serovar Dublin | 54/70 (129) | |||||
| pilU | 11524 to 12159 | 53.2 | 211 | PilU; plasmid R721 | 30/51 (166) | Prepilin peptidase |
| HopD; E. coli (leader peptidase) | 28/43 (141) | |||||
| pilV | 12164 to 13531 | 49.0 | 456 | PilV (constant region); plasmid R721 | 52/65 (354) | Minor pilin subunit |
Electron microscopy.
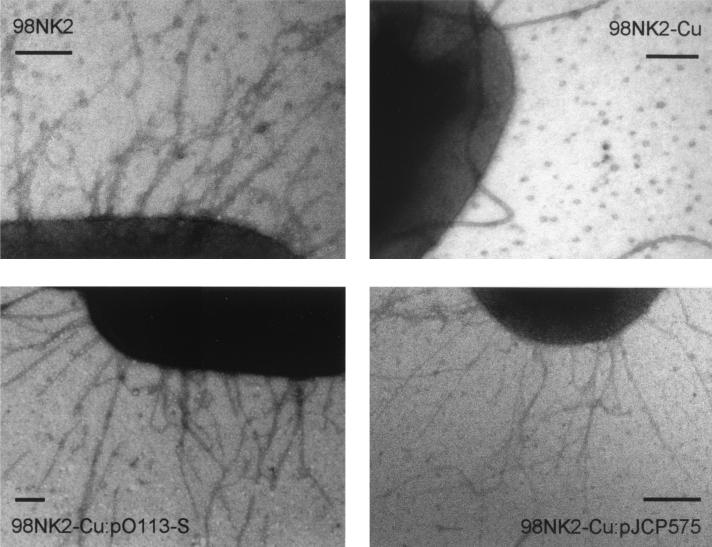
To examine whether the pil locus carried by pO113 is expressed under in vitro conditions, 98NK2 and various derivatives thereof were examined by transmission electron microscopy after being stained with phosphotungstic acid (see Materials and Methods). Long thin pili were observed on the surface of 98NK2, but these structures were absent in 98NK2-Cu, a derivative of 98NK2 which had been cured of pO113 (Fig. 2). Thin pili were, however, observed on the surface of 98NK2-Cu after transformation with either a derivative of pO113 containing a kanamycin resistance cartridge inserted into the saa gene or the cosmid pJCP575 (Fig. 2). Collectively, these data indicate that the pili observed on the surface of 98NK2 are indeed encoded by the pO113 pil locus.
FIG. 2.
Electron micrographs of STEC strains after staining with phosphotungstic acid. Bars, 0.2 μm.
Hemagglutination.
The capacity of the pO113 pil locus to mediate hemagglutination was examined by using guinea pig, chicken, and human group A, B, AB, and O erythrocytes, as described in Materials and Methods. For 98NK2, the strongest hemagglutination activity was observed with guinea pig cells. Maximal hemagglutination was observed at a bacterial cell density of 108 CFU/ml, with weak hemagglutination also detectable at 107 CFU/ml (Table 2). 98NK2 also agglutinated chicken erythrocytes at bacterial cell densities greater than 109 CFU/ml, but there was no agglutination with any of the human erythrocyte suspensions, even when undiluted bacterial suspensions were used (data not shown). Hemagglutination of guinea pig erythrocytes mediated by various 98NK2 derivatives is also shown in Table 2. Only trace hemagglutination was observed with undiluted suspensions of the megaplasmid-cured derivative 98NK2-Cu. However, hemagglutination activity was largely reconstituted in this strain after transformation with the kan-tagged megaplasmid pO113-S or the pil-carrying cosmid pJCP575. Thus, hemagglutination of guinea pig erythrocytes by 98NK2 appears to be mediated by the pO113 pil locus.
TABLE 2.
Hemagglutination of guinea pig erythrocytes
| Strain | Hemagglutination scorea at indicated bacterial cell density (CFU/ml)
|
|||||
|---|---|---|---|---|---|---|
| 1011 | 1010 | 109 | 108 | 107 | 106 | |
| 98NK2 | +++ | +++ | +++ | +++ | + | − |
| 98NK2-Cu | ± | − | − | − | − | − |
| 98NK2-Cu:pO113-S | +++ | +++ | +++ | + | − | − |
| 98NK2-Cu:pJCP575 | +++ | +++ | +++ | − | − | − |
Hemagglutination was scored after incubation of the various dilutions of bacteria with guinea pig erythrocytes for 2 to 4 h at room temperature (see Materials and Methods, “Hemagglutination assay”).
In vitro adherence to epithelial cells.
It has previously been shown that 98NK2-Cu has a significantly reduced level of in vitro adherence to HEp-2 cells compared with that of wild-type 98NK2 (22). To determine whether the pil locus carried by pO113 contributes to this adherence, 98NK2, 98NK2-Cu, and 98NK2-Cu:pJCP575 were tested in the HEp-2 adherence assay as previously described (22). However, the presence of pJCP575 did not significantly increase the adherence of 98NK2-Cu, which remained roughly 50% of that of the wild-type strain (data not shown). The introduction of pJCP575 also had no effect on the adherence of 98NK2-Cu to a human colonic cell line (Hct-8) (data not shown).
Conjugation.
The similarity of the pO113 pil locus to those of R721, R64, and ColIb9 raised the possibility that this plasmid could also be mobilized by conjugation. To examine this, a streptomycin-resistant derivative of 98NK2-Cu was mated with 98NK2-S, which carries pO113 with a kanamycin resistance cassette inserted into the saa gene (see Materials and Methods). Transconjugants resistant to both streptomycin and kanamycin were obtained at all donor-to-recipient ratios tested (1:5, 1:10, and 1:20). However, there was no growth in the presence of streptomycin plus kanamycin when either donor or recipient cells were omitted from the mating mix. The transconjugants were all positive by PCR for a variety of genes present in the donor plasmid but absent in 98NK2-Cu, including the saa::kan locus, pilS, espP, iha, and ehxA (data not shown).
Presence of the pO113 pil locus in other E. coli strains.
The presence of related pil loci in other E. coli strains was examined by Southern hybridization analysis with a digoxigenin-labeled pilS-specific probe as well as by PCR with primers specific for pilS (Table 3). None of the LEE-positive STEC strains tested (including representatives of serogroups O157, O111, and O26) contained pilS-related sequences. Among the LEE-negative STEC strains, pilS-related sequences were detected in 5 of 5 O113 strains as well as in 11 of 14 strains belonging to other serogroups. Interestingly, the presence of pilS did not necessarily correlate with the presence of the other putative virulence genes carried by the megaplasmid, saa and ehxA, a result which further underscores the heterogeneity of these plasmids in STEC.
TABLE 3.
Presence of pilS homologues in other STEC strains
| E. coli strain | Serotypea | Sourceb | Presence of:
|
|||
|---|---|---|---|---|---|---|
| pilSc | saad | LEEe | ehxAe | |||
| 98NK2 | O113:H21 | HUS | + | + | − | + |
| 97MW1 | O113:H21 | BD, MHA, T | + | + | − | + |
| 3848 | O113:H21 | HUS | + | + | − | + |
| 1183 | O113:H21 | HUS | + | + | − | + |
| MW10 | O113:H21 | Food | + | + | − | + |
| 94CR | O48:H21 | HUS | + | + | − | + |
| B2F1 | O91:H21 | HUS | − | + | − | + |
| MW13 | O98 | Food | + | + | − | + |
| MW2 | Ont | Food | + | + | − | + |
| MW14 | ND | Food | + | + | − | + |
| MW8 | Ont | Food | + | + | − | + |
| MW6 | OR | Food | + | + | − | + |
| MW3 | O82:H8 | Food | + | + | − | + |
| MW7 | Ont:H11 | Food | + | + | − | + |
| 95HE4 | O91 | D | + | + | − | + |
| MW15 | O141 | Food | + | − | − | − |
| MW4 | Ont | Food | + | − | − | − |
| MW11 | ND | Food | − | − | − | − |
| MW12 | O159 | Food | − | − | − | − |
| EDL933 | O157:H7 | BD | − | − | + | + |
| 96GR1 | O157:H− | HUS | − | − | + | + |
| 95ZG1 | O26 | BD | − | − | + | + |
| 96RO1 | O111:H− | HUS | − | − | + | + |
| 95NR1 | O111:H− | HUS | − | − | + | + |
Ont, O nontypeable; OR, O rough; ND, serotype not determined.
STEC strains were isolated either from foods or from the feces of patients with uncomplicated diarrhea (D), bloody diarrhea (BD), microangiopathic hemolytic anemia (MHA), thrombocytopenia (T), or hemolytic uremic syndrome (HUS).
Determined by Southern hybridization analysis with a digoxigenin-labeled PCR product corresponding to the complete pilS open reading frame as probe and by PCR.
Determined by Southern hybridization analysis (22).
DISCUSSION
The majority of STEC strains isolated from humans with gastrointestinal disease carry large (approximately 90-kb) plasmids (25). These encode a number of putative accessory virulence proteins, such as the enterohemolysin EhxA (29) and the serine protease EspP (5). The complete DNA sequences of pO157 plasmids from two distinct O157:H7 STEC strains have been determined and found to be very similar to each other (6, 17). However, PCR, Southern hybridization, and partial sequence analyses of the megaplasmids from STEC strains belonging to different serotypes suggest that there are major differences in both the genes present and their organization (4). We have been analyzing the megaplasmid pO113 from an O113:H21 STEC strain (98NK2), which lacks the LEE locus and yet was responsible for an outbreak of hemolytic uremic syndrome. It has previously been demonstrated that this plasmid encodes a novel adhesin (Saa) which is unique to LEE-negative STEC strains (22). In the present study, we have shown that pO113 also carries a novel type IV pilus biosynthesis locus comprising 11 closely linked genes (pilL through pilV) with an additional, separately transcribed gene (pilI) upstream. This pilus locus is most closely related to those of the IncI plasmids R721, R64, and ColIb9, but there are several differences in organization. In R721, pilL and pilM are separated from pilNOPQRSTUV by a 10.6-kb region containing 12 genes (ygeA, traB through traK, and yfdA) and there is also no homologue of pilI. On the other hand, the R64 and ColIb9 loci contain two additional genes (pilJ and pilK) between pilI and pilL, and both loci appear to form a single 14-gene transcriptional unit. However, in R64, pilI and pilJ have been shown not to be essential for pilus biogenesis (37). The organization of the pil loci from the major pathogenicity islands of S. enterica serovar Typhi (38) and S. enterica serovar Dublin (AF247502) is essentially the same as that of the pilL-through-pilV region of pO113. The pO113 pilI and pilL gene products are more similar to their respective R64 homologues than they are to those of R721, whereas the reverse is true for the products of pilM through pilV; this also suggests the possibility of recombination events during the evolution of the pO113 pil locus.
Type IV pili have been shown to be involved in the adherence of a number of pathogenic bacteria to host epithelial cells. Among the better characterized of these are the bundle-forming pilus of enteropathogenic E. coli, which has been implicated in human virulence, autoaggregation, and localized adherence (2, 31, 32), and the toxin-coregulated pilus of V. cholerae, which is essential for gut colonization and virulence in an infant mouse model (19). The type IV pilus encoded on the major pathogenicity island of S. enterica serovar Typhi, which is even more closely related to the pO113 pilus, has also been shown to be important for adherence to and invasion of intestinal epithelial cells (38). In the present study, we demonstrated that 98NK2, but not the plasmid-cured derivative 98NK2-Cu, expressed long thin pili visible by electron microscopy after negative staining. Pili were also present on the surfaces of derivatives of 98NK2-Cu after electroporation with pO113-S or the cosmid pJCP575, both of which carry complete pil loci. Both of these 98NK2-Cu derivatives were also capable of hemagglutinating guinea pig erythrocytes. However, the introduction of pJCP575 into 98NK2-Cu did not increase the in vitro adherence to HEp-2 cells. It has previously been reported that 98NK2-Cu exhibits a level of adherence to HEp-2 cells similar to that seen for 98NK2 carrying pO113-S, which lacks the saa gene (in both cases, roughly 50% of that for wild-type 98NK2) (22). Thus, the HEp-2 cell adherence conferred by pO113 appears to be largely attributable to saa. It is possible, of course, that the pO113-encoded pili may mediate adherence to receptors on human intestinal epithelial cells that are not present on HEp-2 cells, although in the present study, the introduction of pJCP575 did not increase the adherence of 98NK2-Cu to Hct-8, a colonic cell line of human origin.
The R64 pilus was the first member of the type IV family to be implicated in bacterial conjugation, and several genes within this locus have been shown to be essential for liquid mating, presumably by facilitating the attachment of donor cells to recipient bacteria (16, 37). The minor pilin subunit protein PilV appears to play a key role in recipient cell recognition. Interestingly, the C-terminal portion of this protein and its homologues encoded on ColIb9 and R721 undergo structural rearrangements under the control of shufflons (15, 36, 37). The shufflons comprise a pilV constant region followed by several DNA segments flanked and separated by inverted repeat sequences. These are followed by the rci gene, the product of which promotes site-specific recombination between any two inverted repeat sequences, resulting in the in-frame fusion of one of the alternative segments to the constant pilV ORF. In R64, the resultant variation in the C-terminal portion of PilV determines the recipient cell specificity in liquid mating (16). However, pO113 pilV does not appear to contain such repeat elements. In the present study, we have demonstrated that like R721, R64, and ColIb9, the kan-tagged megaplasmid derivative pO113-S can be transferred by conjugation into 98NK2-Cu. Confirmation that the pil locus is essential for this will require construction of pO113 derivatives with defined pil knockout mutations. Nevertheless, the fact that pO113 is transferable distinguishes it from pO157, the megaplasmid of O157:H7 STEC strains, which lacks pil loci or tra genes and is presumed to be nonconjugable (6, 17).
High-stringency homologues of pO113 pilS were found in a wide range of LEE-negative, but not in any LEE-positive, STEC strains. The presence of pilS sequences did not always correlate with the presence of other presumptive megaplasmid genes. For example, the O91:H21 strain B2F1 contains both saa and ehxA but lacks pilS, underscoring the likelihood of further differences in the genetic composition of LEE-negative STEC megaplasmids. Conversely, two other STEC strains tested contained a pilS homologue but lacked ehxA or saa. It is not yet known whether these two strains actually harbor large virulence plasmids, but Brunder et al. (4) have reported that some STEC megaplasmids lack ehxA as well other genes carried by pO157, such as espP and katP. Interestingly, although pO157 from O157:H7 STEC lacks pil loci, Brunder et al. (3) have recently described a six-gene operon on the megaplasmid of sorbitol-fermenting O157:H− STEC strains which encodes the synthesis of pili distantly related to uropathogenic E. coli P-pili. This operon was present in all sorbitol-fermenting O157:H− strains tested but not in sorbitol-negative O157 or any of the other STEC or other pathogenic E. coli strains tested. However, although this operon directed the expression of pili when cloned in E. coli K-12, the expression appeared to be repressed in wild-type sorbitol-fermenting O157:H− strains in vitro.
The conjugal transfer of STEC megaplasmids may help to explain the broad range of STEC strains in which they are found. However, it does not account for the fact that ehxA, which has been commonly used as a marker for the presence of an STEC megaplasmid, is rarely found in non-STEC strains (29). Since Shiga toxins are almost invariably phage encoded, this suggests that there is a tight association between the presence of a megaplasmid and the susceptibility to lysogeny by Stx-converting phages. Other closely related members of the type IV pilus family provide ample precedent for this possibility. TcpA, the major toxin-coregulated pilus subunit of V. cholerae, is known to be the receptor for Ctxφ, the phage which encodes cholera toxin (34). Similarly, phages Iα and PR64FS specifically adsorb to E. coli via the shaft (PilS) and tip (PilV), respectively, of the thin pili encoded by plasmid R64 (37). Thus, the very presence of the pil locus may have contributed to the diversity of E. coli strains carrying stx genes and capable of causing disease in humans.
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council of Australia.
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 3.Brunder, W., A. S. Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(−). Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 5.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 6.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Siofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Azavedo, J., E. McWhirter, M. Louie, and J. Brunton. 1994. Eae-negative verotoxin-producing Escherichia coli associated with haemolytic uremic syndrome and hemorrhagic colitis, p. 265-268. In M. A. Karmali and A. G. Goglio (ed.), Recent advances in verocytotoxin-producing Escherichia coli infections. Elsevier Science B.V., Amsterdam, The Netherlands.
- 8.Donnenberg, M. S., J. B. Kaper, and B. B. Finlay. 1997. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 5:109-114. [DOI] [PubMed] [Google Scholar]
- 9.Dytoc, M. T., A. Ismaili, D. J. Philpott, R. Soni, J. L. Brunton, and P. M. Sherman. 1994. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect. Immun. 62:3494-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 13.Karch, H., J. Heesemann, R. Laufs, A. D. O'Brien, C. O. Tacket, and M. M. Levine. 1987. A plasmid of enterohemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen and for adhesion to epithelial cells. Infect. Immun. 55:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S. R., and T. Komano. 1992. Nucleotide sequence of the R721 shufflon. J. Bacteriol. 174:7053-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, S. R., and T. Komano. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 179:3594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, H. C. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Manning, P. A. 1997. The tcp gene cluster of Vibrio cholerae. Gene 192:63-70. [DOI] [PubMed] [Google Scholar]
- 20.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stxf1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga-toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. 1997. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect. Immun. 65:3799-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perna, N. T., G. F. Mayhew, G. Posfai, S. J. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition. In L. Hunter and T. E. Klein (ed.), Biocomputing: proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co., Singapore.
- 29.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scotland, S. M., S. Knutton, B. Said, and B. Rowe. 1994. Adherence to Caco-2 cells of Vero cytotoxin-producing strains of Escherichia coli belonging to serogroups other than O157, p. 257-260. In M. A. Karmali and A. G. Goglio (ed.), Recent advances in verocytotoxin-producing Escherichia coli infections. Elsevier Science B.V., Amsterdam, The Netherlands.
- 31.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C.-Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone, K. D., H.-Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 33.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, T., N. Furuya, M. Ishikura, T. Isobe, K. Haino-Fukushima, T. Ogawa, and T. Komano. 1998. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J. Bacteriol. 180:2842-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, T., S. Kim, and T. Komano. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]