Abstract
While early transposon (ETn) endogenous retrovirus (ERV)-like elements are known to be active insertional mutagens in the mouse, little is known about their transcriptional regulation. ETns are transcribed during early mouse embryogenesis in embryonic stem (ES) and embryonic carcinoma (EC) cell lines. Despite their lack of coding potential, some ETns remain transposition competent through their use of reverse transcriptase encoded by a related group of ERVs—MusD elements. In this study, we have confirmed high expression levels of ETn and MusD elements in ES and EC cells and have demonstrated an increase in the copy number of ETnII elements in the EC P19 cell line. Using transient transfections, we have shown that ETnII and MusD LTRs are much more active as promoters in P19 cells than in NIH 3T3 cells, indicating that genomic context and methylation are not the only factors determining endogenous transcriptional activity of ETns. Three sites in the 5′ part of the long terminal repeat (LTR) were demonstrated to bind Sp1 and Sp3 transcription factors and were found to be important for high LTR promoter activity in P19 cells, suggesting that as yet unidentified Sp binding partners are involved in the regulation of ETn activity in undifferentiated cells. Finally, we found multiple transcription start sites within the ETn LTR and have shown that the LTR retains significant promoter activity in the absence of its noncanonical TATA box. These findings lend insight into the transcriptional regulation of this family of mobile mouse retrotransposons.
Endogenous retrovirus-like (ERV) sequences comprise approximately 10% of the mouse genome and 8% of human genomes (18, 19). While humans retain few, if any, ERVs capable of retrotransposition, mouse ERVs are highly active, accounting for 10 to 15% of all spontaneous insertional mutations (19, 22) and contributing to numerous cases of cancer (1, 10, 16).
One class of mouse ERVs is the early transposon (ETn) elements, first identified as a family of middle repetitive sequences transcribed during early mouse embryogenesis, with expression peaking between embryonic day 3.5 (E3.5) and E7.5 (8, 9). ETn elements were originally divided into two groups—ETnI and ETnII—which differ only in the 3′ one-third of the long terminal repeat (LTR) and the 5′ end of the adjacent internal region (20, 49, 51). The rest of the internal region contains mainly nonretroviral, noncoding sequences of unknown origin, with the exception of a short stretch of a retroviral pol gene (31). ETn RNA is highly transcribed in early (E3.5 to E7.5) mouse embryos (8, 9), in developing tissues of E7.5 to E13.5 embryos in a tissue- and stage-specific manner (27), and in embryonic stem (ES) cell lines (4), embryonic carcinoma (EC) lines (9), and several plasmocytoma cell lines (48), as well as in primary acute myeloid leukemia cells (56). However, primary cells of other tumor types, such as hepatoma and lymphoma, do not possess elevated amounts of ETn RNA (56). ETn expression is low or undetectable in the differentiated cell lines tested, including the embryonic fibroblast line 3T3 (9), and the level of ETn expression drops dramatically upon differentiation of the teratocarcinoma cell line F9 (17).
ETns are flanked by LTRs and do not contain retroviral open reading frames. However, some remain transposition competent, inducing novel insertional mutations (for a review, see reference 3). We have previously described a close relative of ETn in the mouse genome, a mouse ERV, MusD, possessing a full-length open reading frame, which has similarity to the gag, pro, and pol genes of primate type D retroviruses, or betaretroviruses (31). MusD and ETnII LTRs are highly similar (92 to 100% identical), as are the internal regions bordering the LTRs (4, 31) (see Fig. 1B). There are 80 to 100 MusD and about 40 ETnII copies in the C57BL/6 mouse genome (2, 4, 44), but both ETnII and MusD elements exhibit polymorphic sites of insertions (3), resulting in copy number differences among different mouse strains (4). Despite the lack of coding potential, ETnII elements, but not ETnI or MusD elements, account for most of the insertional mutations, with the ETnII-β subfamily being the most active (4). It was recently shown that ETnII elements utilize MusD-encoded proteins for their retrotransposition (44) (our unpublished data), clarifying the mechanism of integration of noncoding ETnII elements.
FIG. 1.
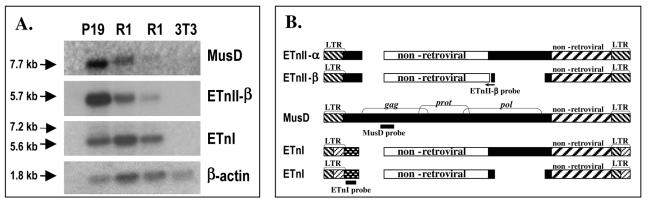
ETn and MusD transcript abundance in undifferentiated and differentiated cell lines. (A) Northern blot. Ten micrograms of RNA isolated from EC P19, ES R1, and fibroblast NIH 3T3 cells was hybridized with ETnI, ETnII-β, MusD, or β-actin probes. ETnII-β and MusD probes produced single bands, and the ETnI probe produced a single major band of 5.6 kb and a minor band of 7.2 kb. The blot was initially hybridized with an ETnII-β oligonucleotide probe, then stripped and rehybridized in turn with the other indicated probes. (B) Structures of ETn elements and locations of probes used in Northern blotting.
LTR sequences have a key role in controlling the expression of retroviral elements and the tissue tropism of retroviruses (28, 35, 43), employing a wide spectrum of cellular transcription factors. Despite the fact that ETn elements are active mouse mutagens, very little is known about the factors controlling their expression or that of their coding-competent MusD relatives. In this study, we have performed functional analysis of ETnII LTRs to gain insight into the transcriptional regulation of this intriguing family of retrotransposons.
MATERIALS AND METHODS
Cell culture, transient transfections, and luciferase assays.
Mouse embryonic teratocarcinoma cell line P19 and mouse embryonic fibroblast cell line NIH 3T3 (ATCC) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells were seeded 24 h prior to transfection into six-well plates at a density of 3 × 105 cells/well. Cells grown in monolayers were transfected with 1 μg of plasmid DNA and 100 ng of the Renilla luciferase vector pRL-TK using Lipofectamine-2000 (Invitrogen) according to the manufacturer's instructions. Cells were harvested into 1× passive lysis buffer (Promega) 24 h after transfection, and firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. The data were standardized to the internal Renilla luciferase control and expressed with regard to the promoter efficiency of the promoterless pGL3-Basic (pGL3B) vector. The results are means and standard deviations from three separate experiments performed in duplicate.
Plasmids and constructs.
ETnII and MusD LTRs were cloned into the KpnI-BglII cloning site of the pGL3B luciferase reporter vector (Promega). The 318-bp LTRs were amplified from C57BL/6 mouse genomic DNA by nested PCR, except for LTR#3, which was amplified from a bacterial artificial chromosome clone. All oligonucleotides used in plasmid construction are listed in Table S1 in the supplemental material. The following forward primers, located in the flanking sequence, were used for the first-round PCR: for LTR#1 (GenBank accession no. AC074208), primer wiz_2921as; for LTR#7 (AC079540), 540_1s; for LTR#6 (AC090879), 879_1s; for LTR#13 (AC079497), 497_1s; for LTR#10 (AL589871), 871_1s; for LTR#8 (AC087263), 263_1s; and for LTR#9 (AF132039), 039_1s. The reverse primer MusD/ETnII_361as was used in common for all first-round PCRs. Nested-PCR primers were identical for all LTRs: forward primer IM_LTR_1s and reverse primer IM_LTR_2as. Progressive 5′ deletion constructs of the ETnII LTR#6 were generated by PCR amplification with a common reverse primer, IM_LTR_2as, and forward primers IM_ETnII_18s, IM_ETnII_29s, IM_ETnII_43s, IM_ETnII_61s, IM_ETnII_61s-T, IM_ETnII_76s, IM_ETnII_140s, and IM_ETnII_162s. 3′ deletion constructs of the ETnII LTR#6 were generated by PCR amplification with a common forward primer, IM_LTR_1s, and reverse primers IM_ETnII_84-as, IM_ETnII_164-as, IM_ETnII_197-as, IM_ETnII_234-as, and IM_ETnII_277-as. The resulting LTR fragments were cloned into the KpnI-BglII site of the pGL3B reporter vector and sequenced.
To generate a mutation in the first Sp1 binding site, in vitro mutagenesis was performed by amplification of ETnII LTR#6 with primers IM_LTR_mSp1-s and IM_LTR_2as. For the other two Sp1 binding site mutation constructs, the 3′ part of the ETnII LTR#6 was first amplified with a common primer, IM_LTR_1s, and the following mutating oligonucleotides: IM_mutSp1/32-as to generate a mutation in the second Sp1 binding site and IM_mutSp1/61-as for mutation in the third Sp1 binding site. Products of these first reactions were used as forward primers for amplification of ETnII LTR#6 with a common reverse primer, IM_LTR_2as. All PCRs were carried out under standard conditions using Pfu DNA polymerase (Invitrogen). All insertions cloned into the pGL3B vector were sequenced.
Preparation of RNA, Northern blotting, and 5′ rapid amplification of cDNA ends (5′-RACE).
P19 and NIH 3T3 cells grown to 90% confluence were lysed in 1 ml TRIzol (Invitrogen) per 10 cm2, and total RNA was extracted according to the TRIzol protocol. Two samples of RNA from the ES cell line R1 were generously provided by Lynn Mar and Laura Sly.
For Northern blotting, 10 μg of RNA for each lane was denatured, electrophoresed in a 1.2% agarose-3.7% formaldehyde gel in 1× morpholinepropanesulfonic acid (MOPS) buffer, transferred overnight to a Zeta-probe nylon membrane (Bio-Rad), and UV cross-linked. Probes specific for ETnI and MusD elements were synthesized by PCR using the following primers: for ETnI, forward primer STD1 and reverse primer STD2; for MusD, forward primer MusD_823s and reverse primer MusD_1192as. Amplified DNA fragments were α-32P labeled using a Random Primers DNA Labeling system (Invitrogen). α-32P-end-labeled antisense oligonucleotide probes were used as probes for ETnII-β elements (ETnIIgr1_3636as) (4) and β-actin (3′actin). Membranes were prehybridized in ExpressHyb (BD Biosciences) for 2 to 4 h at 68°C for fragment probes and 54°C for oligonucleotide probes, hybridized overnight at the same temperatures in fresh ExpressHyb, washed according to the manufacturer's instructions, and exposed to film.
5′-RACE was carried out using the FirstChoice RLM-RACE kit (Ambion) with 10 μg of total RNA from the untreated cell line P19 or from P19 cells transfected with expression constructs according to the manufacturer's instructions. Two rounds of nested PCR were carried out with forward primers specific for the 5′ RNA adapter and reverse primers specific for an ETnII element. Endogenously expressed ETnII elements were cloned using ETnI/II_665as as the reverse primer for the first-round PCR and IM_3as as the reverse primer for the second-round PCR. Both primers were located downstream of the 5′ LTR, prohibiting amplification of solitary LTRs. Transfected full-length and 3′-deleted constructs were cloned using primers specific for the luciferase gene: IM_Luc_208as as the reverse primer for the first-round PCR and GL-2 as the reverse primer for the second-round PCR. Specific amplification products were cloned into the pGEM-T vector (Promega). Several independent clones derived from each PCR product were sequenced.
Preparation of genomic DNA and Southern blotting.
P19 and NIH 3T3 cells grown to 90% confluence were lysed in 750 μl DNAzol (Invitrogen) per 10 cm2, and genomic DNA was extracted according to the DNAzol protocol. Genomic DNA from ES cell lines R1, EK.ECC, AB-1, and 671 was a gift from Lynn Mar; genomic DNA from mouse strains 129S1/Sv-+p +Tyr-c KitlSl-J/+ (stock 000090), 129X1/SvJ (stock 000691), and C3H/HeJ (stock 000659) was obtained from the Jackson Laboratory. One microgram of genomic DNA for each lane was digested overnight, run overnight in a 0.8% 1× Tris-acetate-EDTA agarose gel, and transferred overnight in 0.4 M NaOH to a Zeta-probe nylon membrane (Bio-Rad). A probe specific for both ETnII and MusD elements was amplified by PCR from the plasmid containing an ETnII clone using forward primer IM_ETnII/MusD_329s and reverse primer IM_ETnII/MusD_624as and was α-32P labeled using the Random Primers DNA Labeling system (Invitrogen). The membrane was prehybridized in ExpressHyb (BD Biosciences) at 60°C for 3 to 4 h, hybridized overnight at the same temperature in fresh ExpressHyb, washed according to the manufacturer's instructions, and exposed to film.
Nuclear extraction and electrophoretic mobility shift assays (EMSA).
Nuclear extracts were prepared from P19 and NIH 3T3 cells as follows. Cells were washed in phosphate-buffered saline, pelleted at 5,000 rpm for 5 min at 4°C, and resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, protease inhibitors, 0.1% NP-40). After incubation on ice for 5 min, the lysates were spun at 2,500 rpm at 4°C for 4 min. The pelleted nuclei were washed in lysis buffer without NP-40 and pelleted at 2,500 rpm for 4 min at 4°C. The nuclear pellet was resuspended in nuclear extraction buffer (20 mM Tris-HCl [pH 8.0], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 25% glycerol). After incubation on ice for 20 min, the nuclei were briefly vortexed and spun at 13,000 rpm for 6 min at 4°C. The supernatant was removed and used as a nuclear extract. Protein concentrations were determined using the Bio-Rad protein assay.
EMSA were performed with γ-32P-labeled double-stranded oligonucleotides identical to the three fragments of the ETnII LTR#6 containing putative Sp1/Sp3 binding sites (I, II, and III). Nuclear extracts (5 μg) were preincubated with 20 μl of a reaction mixture containing 0.5 μg of poly(dI · dC) in 10 mM HEPES, 4 mM dithiothreitol, 0.2 mM EDTA, 0.1 mM NaCl, 0.1 mg/ml bovine serum albumin, and 4% glycerol. Samples were incubated for 20 min on ice with either a 200-fold excess of unlabeled competitor oligonucleotide or an antibody specific for Sp1 or Sp3 (sc-59 X and sc-644 X; Santa Cruz Biotechnology) for supershift assays. Double-stranded oligonucleotides were labeled using [γ-32P]ATP and T4 polynucleotide kinase according to the manufacturer's protocols (Invitrogen) and purified on MicroSpin G-25 columns (Amersham Pharmacia Biotech). A γ-32P-labeled oligonucleotide probe was added to the reaction mixture, and the incubation was continued on ice for another 30 min. Reaction products were fractionated on a 5% polyacrylamide gel and run at 4°C in 0.5× Tris-borate-EDTA buffer at 8 to 15 mA for 3 h. Gels were transferred onto a Whatman paper, dried, and autoradiographed.
RESULTS AND DISCUSSION
Transcription of endogenous ETn and MusD in ES and EC cell lines.
We performed reverse transcription-PCR (data not shown) and Northern blot analysis to estimate the amounts of endogenous ETn and MusD transcripts in the EC cell line P19, two samples of the ES cell line R1, and the differentiated embryonic fibroblast cell line NIH 3T3 (Fig. 1A). For ETnII, an oligonucleotide probe specific for the most abundant subtype, ETnII-β, was used. This probe spans a small deletion found only in ETnII-β elements (Fig. 1B) (4). ETn and MusD RNAs were abundant in P19 and R1 cells, with both groups of elements expressed at higher levels in P19 cells. The NIH 3T3 cell line, as expected, did not express ETn or MusD elements. Single major bands depicted by Northern blotting clearly indicate a preference for transcription of a group of elements of a specific size, suggesting that other subtypes either are transcriptionally inert or are transcribed at far lower levels. Similar results were previously demonstrated for ETnI and ETnII elements combined (56). Thus, our analysis has confirmed previously reported high levels of ETn and MusD expression in undifferentiated EC and ES cell lines (4, 9) and their absence in the differentiated cell line NIH 3T3 (9).
Promoter activities of ETnII and MusD LTRs in the P19 and NIH 3T3 cell lines.
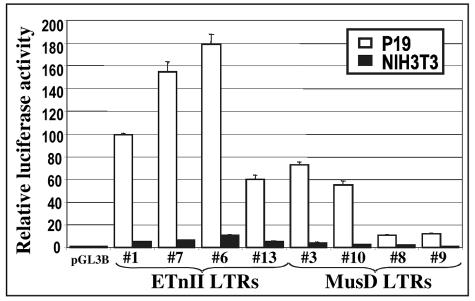
Since ETnII elements are most frequently found at the sites of recent insertions and are more highly transcribed than MusD elements (3, 4), we analyzed the promoter efficiencies of ETnII and MusD LTRs in differentiated versus undifferentiated cell lines. The 5′ LTRs of four ETnII and four MusD elements belonging to different subfamilies were chosen for the study. These elements possess various degrees of nucleotide identity between their 3′ and 5′ LTRs, with evolutionarily younger elements displaying 100% identity (Table 1). The LTRs were cloned into the pGL3B luciferase reporter vector, transfected into P19 and NIH 3T3 cells, and analyzed for their promoter efficiencies by measuring luciferase gene expression (Fig. 2). While the sample size is small, we observed that the LTRs of younger elements had higher promoter activities, suggesting degeneration of promoter-competent LTRs with time due to accumulation of mutations either by random genetic drift or by negative selection aimed at the elimination of deleterious sequences.
TABLE 1.
Description of the LTRs used in this study
| Element no. | % Identity of 5′- and 3′ LTRsa | Accession no. |
|---|---|---|
| ETnII | ||
| LTR#1 | 100 (317/317) | AC074208 |
| LTR#7 | 100 (317/317) | AC079540 |
| LTR#6 | 100 (318/318) | AC090879 |
| LTR#13 | 94 (301/318) | AC079497 (13-bp deletion in 5′ LTR) |
| MusD | ||
| LTR#3 | 100 (319/319) | AC084696 |
| LTR#10 | 99 (316/319) | AL589871 |
| LTR#8 | 96 (308/319) | AC087263 |
| LTR#9 | 97 (310/319) | AF132039 |
Numbers in parentheses are number of identical nucleotides/total number of nucleotides.
FIG. 2.
Promoter activities of ETnII and MusD LTRs in P19 and NIH 3T3 cells. Four ETnII and four MusD LTRs (see Table 1 for accession numbers) cloned into a pGL3B luciferase reporter vector were assayed for promoter efficiency by transient transfections into P19 and NIH 3T3 cells.
CpG methylation is an important factor in suppression of LTR-driven transcription, as shown for intracisternal A particle elements and other classes of mouse ERVs (6, 15, 37, 57). A wave of global mammalian CpG methylation is known to occur during cellular differentiation around the time of implantation (58), correlating with the reduction in ETn transcription. However, despite the lack of CpG methylation of the expression plasmids, transiently transfected LTR promoter constructs revealed 15-times-lower promoter activity in NIH 3T3 cells than in P19 cells (Fig. 2), suggesting an important role for cell type-specific transcription factors. We propose that, in addition to possible suppressing effects of methylation, ETn transcription in NIH 3T3 cells may be restricted by the lack of certain activating transcription factors present in the P19 cell line or by the presence of suppressor transcription factors in the NIH 3T3 cell line. This is further supported by the recently published evidence that CpG demethylation increases the level of transcription initiating from human endogenous retrovirus-K LTRs only in the context of the specific transcription factor pool present in germ cell tumors (24). However, since the absolute difference in LTR promoter activities between P19 and NIH 3T3 cells is difficult to ascertain, our subsequent experiments focus on the relative promoter activities within each cell line of partially deleted or mutated LTRs.
Regions in the ETnII LTR crucial for high promoter ability in P19 cells.
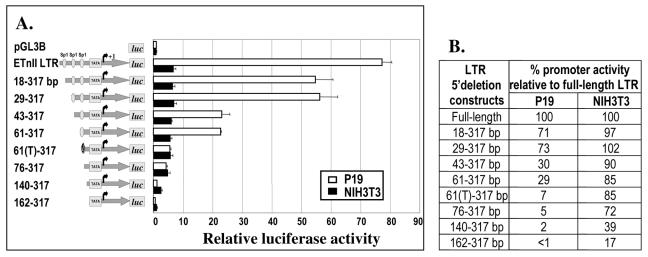
To identify regulatory regions in the ETn LTRs responsible for their exceptionally high promoter activities in P19 cells, we performed functional analysis of the ETnII LTR#6 (GenBank accession no. AC090879), which displayed the highest promoter activity among the LTRs tested (see Fig. 2). The promoter activities of the 5′ LTR deletion constructs were analyzed in P19 and NIH 3T3 cells. We identified three regions in the ETnII LTR essential for high LTR promoter efficiency in P19 cells. All were positioned within the first 76 bp of the LTR, 190 to 120 bp upstream of the previously mapped transcription start site (20) (Fig. 3A and B). Regions overlapping nucleotides 1 to 18, 29 to 43, and 61 to 76 are clearly responsible for P19-specific expression, since these deletions lead to a severe reduction of activity in P19 cells (down to 5% of the wild-type level) but not in NIH 3T3 cells. All of the three critical regions contain predicted recognition sequences for Sp1 and Sp3 transcription factors (Fig. 4). Notably, a single-nucleotide C→T mutation positioned 72 bp from the beginning of the LTR, abrogating the third Sp1 binding site, is found in ETnII LTR#13 and MusD LTR#8 and LTR#9 and may account for their lower promoter activities (see Fig. 2). A 5′ deletion construct incorporating this mutation [61(T)-317] demonstrates significantly reduced promoter activity in P19 cells in comparison to the wild-type deletion construct (61-317). We hypothesize that the second and third regions described above are necessary for high P19-specific, and possibly embryo- specific, expression of ETn elements.
FIG. 3.
Promoter activities of 5′-deleted ETnII LTRs in P19 and NIH 3T3 cells. (A) Graphical representation of the promoter efficiencies of 5′-deleted LTR#6 constructs. Numbers indicate the span of the construct, with base pairs corresponding to those in LTR#6. The putative TATA box and the transcription start site identified previously (20) are shown. Ovals represent possible Sp1 binding sites. The Sp1 binding site mutated in the 61(T)-317 construct is crossed. (B) Promoter activities of the LTR deletion constructs in P19 and NIH 3T3 cells as percentages of the promoter activities of full-length LTR#6.
FIG. 4.
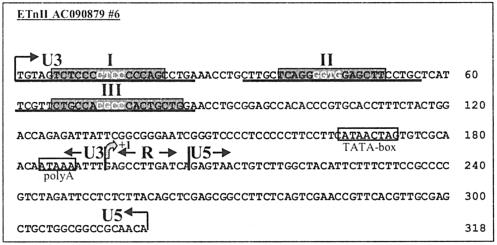
Sequence of the ETnII LTR#6, with positions of deletion constructs and EMSA oligonucleotides shown. The transcription start site, as identified in reference 20, is indicated as +1. The predicted TATA-like sequence and polyadenylation signals (20) are boxed, and U3, R, and U5 regions are indicated. Oligonucleotides used as probes in EMSA (I, II, and III) are underlined. Sp1/Sp3 recognition sequences are highlighted in gray; core regions are specified in white letters on a gray ground.
The three ETnII LTR critical regions bind Sp1 and Sp3 transcription factors.
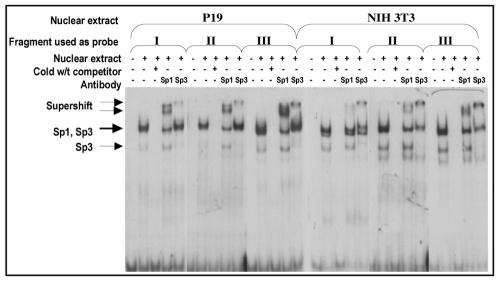
To determine whether the three regions critical for high ETnII LTR promoter activity bind Sp transcription factors, we performed EMSA with double-stranded DNA oligonucleotides corresponding to each of the three regions (Fig. 4). Supershift results demonstrate efficient binding of Sp1 and Sp3 transcription factors from both P19 and NIH 3T3 nuclear extracts to all of the three candidate regions, resulting in extremely similar band patterns. However, incubation of the first segment with an NIH 3T3 nuclear extract produced an extra band, indicating the presence of additional complexes (Fig. 5), suggestive of repression. However, based on the deletion analysis results described above and the mutational analysis described in the next section, this hypothesis is unlikely. The binding of Sp1 and Sp3 to these regions in both P19 and NIH 3T3 cells suggests that Sp1-dependent transcription may be mediated by cofactors that bind to Sp proteins. This hypothesis is supported by numerous recent reports of heterodimerization of various transcription factors with Sp1, resulting in transcriptional activation through Sp1 binding sites (21, 40, 46).
FIG. 5.
Binding of Sp1 and Sp3 transcription factors to the regions crucial for high LTR promoter activity. γ-32P-labeled oligonucleotides identical in sequence to the regions essential for high LTR promoter ability in P19 cells (I, II, and III) were incubated with nuclear extracts from P19 and NIH 3T3 cells. Competition assays were performed with a 200-fold excess of wild-type unlabeled oligonucleotide. Antibodies against Sp1 and Sp3 supershifted specific bands.
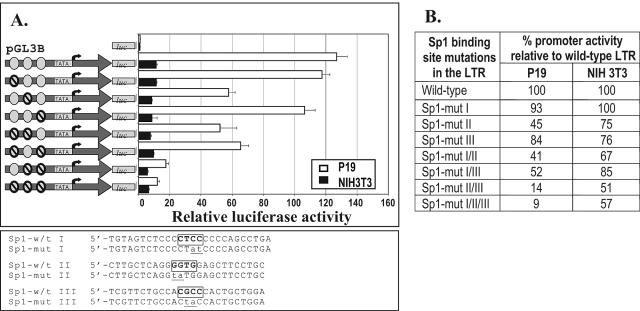
Mutational analysis of the ETnII LTR.
To assess the importance of the Sp1/Sp3 binding sites for the P19 cell-specific promoter activity of ETnII LTRs, we introduced mutations abolishing Sp1/Sp3 binding sites into the ETnII LTR#6 and assayed the promoter efficiencies of these constructs. Prior to transfection experiments, we employed EMSA to demonstrate that mutations identical to those introduced into the LTR completely abolished the binding of proteins to these regions (data not shown). Mutation of either the first or the third Sp binding site did not lead to a significant reduction in promoter activity, whereas mutation of the second Sp binding site led to a twofold decrease in promoter efficiency. Notably, combined mutation of sites 2 and 3 was almost as dramatic as the triple mutation and reduced promoter activity in P19 cells to the level of the wild-type LTR promoter activity observed in NIH 3T3 cells (Fig. 6A and B). The promoter activity of the triple mutant was only 9% of the wild-type level in P19 cells, while the same combination of mutations reduced promoter activity in NIH 3T3 cells to a lesser degree. NIH 3T3 cells are clearly less responsive to the abolition of Sp1/Sp3 recognition sequences than P19 cells, confirming the crucial role of these regions in the transcriptional activation of ETnII LTRs in the P19 cell line and, presumably, in ES cells and embryonic tissues. The results of these deletion and mutation analyses are suggestive of activation of LTR promoter ability in P19 cells versus its suppression in NIH 3T3 cells, since we registered no increase in the promoter efficiency in NIH 3T3 cells upon the mutation of putative transcriptional repressor binding sites.
FIG. 6.
Promoter activities of Sp-binding site mutant LTRs in P19 and NIH 3T3 cells. (A) Graphical representation of the promoter efficiencies of LTR#6 constructs mutated at Sp1/Sp3 recognition sites. The putative TATA box and the transcription start site identified previously (20) are shown. Ovals represent possible Sp1 binding sites. Crossed ovals represent mutated Sp1 binding sites. The cores of the wild-type (w/t) Sp1 recognition sequences are boxed, and the mutated nucleotides are lowercased. (B) Promoter activities of the mutated LTR constructs in P19 and NIH 3T3 cells as percentages of the promoter activities of wild-type LTR#6.
Despite the high responsiveness of the LTR reporter constructs to the mutation of Sp1/Sp3 binding sites, cotransfection of P19 and NIH 3T3 cell lines with expression plasmids encoding Sp1 and Sp3 proteins (kind gifts from Paul Gardner, Tom Shenk, and Guntram Suske) together and separately in varying proportions did not significantly alter the promoter efficiencies of ETnII or MusD LTRs (data not shown). This result suggests that ubiquitously expressed Sp1 and Sp3 are not limiting factors for LTR transcription efficiency and indicates possible involvement of other tissue-specific coregulating factors. This is confirmed by our unpublished data showing that Sp1 and Sp3 expression determined by reverse transcription-PCR is similar in both cell lines. It is also possible that other transcription factors which employ the same or overlapping binding sites as the Sp family proteins are specifically expressed in undifferentiated cells and are those responsible for transcriptional activation of ETn and MusD elements.
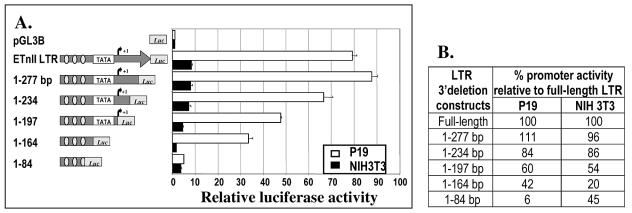
High promoter activity of the 3′-deleted ETnII LTR lacking the putative TATA box.
To determine whether LTR sequences 3′ to the previously reported transcription start site (56) play a role in transcriptional activation, functional analysis of the 3′-deleted constructs of the ETnII LTR#6 was carried out. The promoter activities of the LTR 3′ deletion constructs were analyzed in P19 and NIH 3T3 cells. Deletions of the LTR from the 3′ end caused a mild drop in the promoter efficiency of the LTR in both the P19 and NIH 3T3 cell lines, suggesting a lack of cell-specific regulatory sequences in the region (Fig. 7). Notably, the construct spanning bp 1 to 164 of the LTR had surprisingly high promoter activity in the P19 cell line, despite the deletion of its 3′ sequence including a previously predicted noncanonical TATA box (20). This may be due to an alternative transcription start site 5′ to the weak TATA box, since GC-rich sequences present in the 5′ end of the LTR may serve as a TATA-less promoter. Previous studies identified a single site of transcription initiation at the border of the U3 and R regions in the ETn LTR in primary acute myeloid leukemia (56) and carcinoma (20) cells. However, the elements investigated in those studies belong to the ETnI family and thus differ significantly from the ETnII elements studied here in the 3′ ends of their LTRs and part of the downstream sequence (4, 49). Although the presumed U3 region is nearly identical in both families, highly divergent U5 and downstream sequences may play a role in variable transcription initiation between ETnI and ETnII elements. Our data are suggestive of an alternative transcription start site in ETnII LTRs that functions in the P19 cell line.
FIG. 7.
Promoter activities of 3′-deleted ETnII LTRs in P19 and NIH 3T3 cells. (A) Graphical representation of the promoter efficiencies of 3′-deleted LTR#6 constructs. The putative TATA box and the transcription start site identified previously (20) are shown. Ovals represent possible Sp1 binding sites. (B) Promoter activities of the LTR deletion constructs in P19 and NIH 3T3 cells as percentages of the promoter activities of full-length LTR#6.
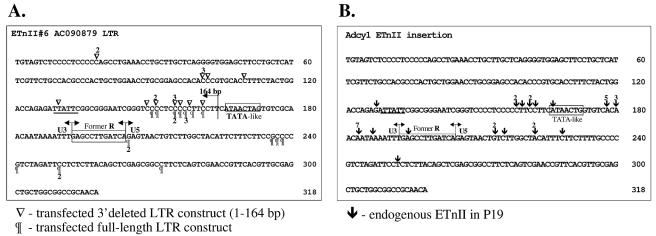
Determination of ETnII transcription initiation sites.
To determine transcription start sites, we performed 5′-RACE of full-length and 3′-deleted ETnII LTR constructs transfected into P19 cells (Fig. 8A) and endogenous ETnII elements in P19 cells (Fig. 8B). The 3′ deletion construct spanning bp 1 to 164 of the LTR appeared to possess multiple transcription start sites upstream of the previously predicted nonconsensus TATA-like sequence, explaining its high promoter activity in the P19 cells. The transcription initiation sites of the bp-1-to-164 construct largely overlap with those of the transfected full-length LTR#6 and fall primarily into the region spanning bp 149 to 161 of the LTR (Fig. 8B). It is therefore possible that a band detected in an EMSA experiment by Tanaka and Ishihara (56) with an oligonucleotide spanning bp 147 to 169 of the LTR contains polymerase complexes involved in initiation of transcription.
FIG. 8.
Mapping of transcription initiation sites in ETnII LTRs by 5′-RACE. (A) Transcription initiation sites of transfected full-length and 3′-deleted ETnII LTR#6 in P19 cells. Symbols represent 5′ termini of individual RACE clones, and numbers indicate that certain clones were found multiple times. The 3′-deleted construct (bp 1 to 164) is indicated by an arrow. The previously noted TATA-like element (20) is boxed, and a second such motif is underlined. (B) Transcription initiation sites of endogenous ETnII elements in P19 cells. Arrows represent 5′ termini of RACE clones, and numbers indicate that certain clones were found multiple times. The LTR sequence is that of the new ETnII insertion into the Adcy1 locus (25). The previously noted TATA-like element (20) is boxed, and a second such motif is underlined.
We have also observed multiple transcription initiation sites within the LTRs of endogenously expressed ETnII elements (Fig. 8B). This is a very common trait of promoters containing a weak TATA box and a GC-rich upstream sequence; in fact, as many as 85% of all human genes have been reported to possess highly variable transcription start sites (54). The transcription start site most frequently used by endogenous ETnII elements in P19 cells does not exactly correspond to the sites employed by the transfected full-length LTR#6 reporter construct, though the overlap does exist. This may be due to the fact that ETnII elements with LTRs other than LTR#6 account for the majority of transcripts in P19 cells, as seen from the sequences of 5′-RACE clones. The sequence differences between the LTRs may result in variable efficacies of transcription initiation sites. It is also possible that the ETnII sequence downstream of the LTR, present in the endogenous ETnII elements but not in the LTR reporter constructs, may affect transcription initiation.
The majority of the transcription initiation sites of endogenous ETnII elements in P19 cells are clustered in the region TCACAACAAT (bp 176 to 185), with all three C−1A+1 dinucleotides in this sequence employed for initiation of transcription (Fig. 8B). This sequence resembles a loose initiator element consensus, YYA+1N(T/A)YY, where Y is a pyrimidine (for reviews, see references 26 and 50). In the absence of a strong TATA box, initiator sequences are known to effectively drive transcription in the presence of upstream activator Sp1 binding sites (for a review, see reference 50). Thus, ETnII elements appear to rely on Sp1/Sp3 recognition sequences in the absence of a strict TATA box for transcription from multiple initiation sites. However, we have also noted another potential TATA box at nucleotide positions 124 to 133 of the LTR (underlined in Fig. 8) that may play a role in initiation of some transcripts. Although the 5′-RACE procedure used is designed to detect only transcripts with a 5′ cap (32), it is possible that some of the transcription start sites downstream of the site previously identified (20) may be due to failure of PCR or reverse transcription during the RACE procedure. However, we are confident that the majority of the transcription start sites identified, especially those found more than once and those mapping 5′ to the previously defined beginning of the R region, are legitimate.
Most well-studied exogenous retroviruses have strong TATA boxes and a single major transcriptional initiation site which defines the beginning of the R region, though multiple transcription start sites have been found to be functional in exogenous and endogenous retroviruses (13, 45, 52). In addition, multiple transcription initiation sites in the ETnII LTR were found in the ES cell line R1 (data not shown), suggesting that this may be a common feature of ETnII elements in different cell lines. It is possible that families of retroviral elements that reside in the genome gradually assume more flexibility in transcriptional control than their exogenous counterparts.
It is worth noting that, despite the multitude of copies of ETnII elements in the genome, the transcribed elements are highly similar to one another, based on the sequences derived from the 5′-RACE clones. Most of the transcripts in P19 cells were identical to the newly retrotransposed ETnII-β element which caused a mutation in the Adcy1 gene in an ICR mouse (25) (GenBank accession no. AF108230), suggesting that they belong to a small subfamily of the most active and evolutionarily young retroviral elements. Since the P19 cell line is derived from the C3H/HeJ mouse background, and ETn elements are highly polymorphic among mouse strains (3), it is difficult to positively identify the ETn element responsible for the majority of the transcripts based on the published C57BL/6 genome. The 5′-RACE sequences are most similar to an ETn element on chromosome 13 at position Ch13:110665922 to -110671471 in the May 2005 draft of the mouse C57BL/6 genome. This is the same element we identified previously as being identical to ETnII sequences highly transcribed in the early embryo from different mouse strains, LM and SELH (GenBank accession number AC079540) (4). However, using genomic PCR analysis, we have found that this particular element is polymorphic in different mouse strains (3) and is not present in P19 cells or in the parental strain C3H/HeJ (data not shown).
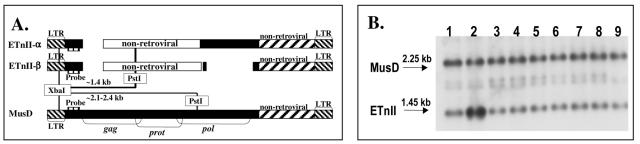
Apparent amplification of ETnII elements in the permissive P19 cell line.
To determine whether the high transcription rate of ETnII and MusD elements resulted in copy number elevation in permissive cell lines, we performed genomic Southern blotting to compare ETnII and MusD copy numbers in the DNAs of P19, R1, and NIH 3T3 cells and their respective parental mouse strains. DNA from the NIH/Swiss parental mouse strain of the NIH 3T3 cell line was not analyzed due to unavailability. The approximate expected sizes of the fragments, given the heterogeneous nature of ETns, were 1,400 bp for the ETnII elements and 2,100 to 2,350 bp for the MusD elements (Fig. 9A). Southern blotting results (Fig. 9B) indicate a significant increase in ETnII copy number in the P19 cell line, likely due to high levels of transcription (see Fig. 1) and retrotransposition. Though the majority of transcripts in the P19 cell line are very similar (see the preceding section), it is not clear whether the new ETnII copies are the products of a single, highly transcribed source element or are derived from one or multiple proviruses that originated from this element. No increase in ETnII proviruses is evident in any of the ES cell lines at this resolution. These data suggest, however, that commonly used ES cell lines may accumulate new ETn copies if grown for many generations. Methods capable of detecting new single ETn integrations are necessary to assess the frequency of such events in ES cells.
FIG. 9.
Amplification of ETnII elements in the P19 cell line. (A) Digestion scheme. One microgram of genomic DNA was digested with restriction endonucleases PstI and XbaI; XbaI cuts inside the LTR, and PstI cuts in the body of the element, resulting in different fragment lengths for ETnII (approximately 1.4 kb) and MusD (approximately 2.1 to 2.35 kb) elements. The probe used is specific for both the ETnII and MusD elements and is located in the common region just 3′ of the 5′ LTR. (B) Genomic Southern blotting and hybridization. Genomic DNAs from the parental mouse strain of the EC cell line P19, C3H/HeJ (lane1), the EC cell line P19 (lane 2), parental mouse strains of the ES cell lines, 129X1/SvJ (lane 3) and 129S1/Sv-+p +Tyr-c KitlSl-J/+ (lane 4), the ES cell lines R1 (lane 5), EK.ECC (lane 6), AB-1 (lane 7), and 671 (lane 8), and the fibroblast cell line NIH 3T3 (lane 9) were digested with PstI and XbaI. The blot was hybridized to the probe specific for both the ETnII and MusD elements.
One possible reason for amplification of ETnII elements in P19 versus ES cells is an increased probability of retrotransposition due to a longer culturing period, given that P19 was established in 1982 (33) whereas ES cell lines derived from the 129 mouse strain were established in the early 1990s (11, 34, 36) and are generally maintained in culture for very short periods. Accumulation of ETnII copies in P19 cells could also be explained by expression of one or more specific coding-competent MusD elements in this cell line. Only a small number of MusD elements are fully intact (2) and able to facilitate ETn retrotransposition (44). The finding that ETnII elements have amplified in preference to their MusD relatives is likely due to the fact that ETnII transcripts are generally much more abundant than MusD transcripts (4), making the former more likely to retrotranspose.
Finally, it is possible that the observed ETnII amplification is due to inherent genomic instability of the teratocarcinoma cell line P19, which may have led to DNA amplifications via segmental duplications or other rearrangements. In this case, it is expected that MusD elements would also be amplified, since both ETnII and MusD elements are distributed randomly in the genome (unpublished observations). Given that the MusD copy number has not noticeably increased in these cells, thus serving as an internal control, it is unlikely that DNA amplifications unrelated to retrotransposition explain the increase in the number of ETnII elements. However, the remote possibility that only a small DNA region containing an ETnII element(s) and no MusD elements has selectively amplified in the P19 genome cannot be ruled out.
Role of Sp1 and Sp3 transcription factors in ETn expression.
This study has identified three regions containing Sp1/Sp3 binding sites as indispensable for high promoter activity of the ETnII LTR in the ETn-expressing cell line P19. Mutation of all three of these sites reduces LTR promoter activity 10-fold, to the level observed in the nonexpressing cell line NIH 3T3.
While Sp1 is a ubiquitously expressed transcription factor, its expression may differ more than 100-fold between different tissues and developmental stages (47). It binds GC- and GT-rich boxes and is involved in the transcriptional regulation of many housekeeping, tissue-specific, and developmental genes (for a review, see reference 53) as well as some viral genes (14, 42, 46). Sp1 binding sites have been implicated not only in protection of CpG islands from de novo methylation but also in providing signals for direct DNA demodification (7, 12, 30). Another member of the Sp family of transcription factors, Sp3, which has been shown to bind the same recognition sequences, either compensates for Sp1 or competes with it. Since some of the Sp3 isoforms may function as transcriptional repressors, the ratio of Sp1 to Sp3 is frequently responsible for the degree of gene activation (for a review, see reference 53).
The Sp1 transcription factor is a candidate for being a major regulator of the onset of zygotic genome activation, since it is required for activation of the hsp70 promoter in two-cell mouse embryos (5). Despite being ubiquitously expressed, Sp1 seems to be involved in transcriptional activation of some embryo-specific genes, such as Oct3/4 (12, 38, 41), telomerase (39), and FGF-4 (23, 29), as well as ST8Sia-I (55), which is expressed in P19 cells but not in 3T3 cells. This versatility of Sp1 apparently stems from the multitude of its binding partners, since Sp1 can be transactivated by a number of other transcription factors (21, 40, 46). Given that Sp1 and Sp3 levels are comparable between the ETn-expressing cell line P19 and the non-ETn-expressing cell line NIH 3T3 (data not shown), and given that transient cotransfection experiments with Sp1 and Sp3 expression vectors did not significantly alter LTR promoter efficiency in either of the cell lines (data not shown), we suggest that the high transcription rate of ETn elements and the high LTR promoter activity in the P19 cell line are dependent on the availability of Sp1/Sp3-binding transcription factors. Presumably, high ETn transcriptional activity in other types of undifferentiated cells, including ES cells and embryonic tissues, relies on the same mechanism.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research.
We thank Lynn Mar and Laura Sly for providing RNA and DNA from ES cell lines and Guntram Suske, Tom Shenk, Ed Seto, and Paul Gardner for providing Sp1 and Sp3 expression constructs. We also thank Catherine Dunn and two anonymous reviewers for helpful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Asch, B. B., H. L. Asch, D. L. Stoler, and G. R. Anderson. 1993. De-regulation of endogenous retrotransposons in mouse mammary carcinomas of diverse etiologies. Int. J. Cancer 54:813-819. [DOI] [PubMed] [Google Scholar]
- 2.Baillie, G. J., L. N. van de Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 78:5784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baust, C., G. J. Baillie, and D. L. Mager. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13:423-428. [DOI] [PubMed] [Google Scholar]
- 4.Baust, C., L. Gagnier, G. J. Baillie, M. J. Harris, D. M. Juriloff, and D. L. Mager. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 77:11448-11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevilacqua, A., M. T. Fiorenza, and F. Mangia. 2000. A developmentally regulated GAGA box-binding factor and Sp1 are required for transcription of the hsp70.1 gene at the onset of mouse zygotic genome activation. Development 127:1541-1551. [DOI] [PubMed] [Google Scholar]
- 6.Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. [DOI] [PubMed] [Google Scholar]
- 7.Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. [DOI] [PubMed] [Google Scholar]
- 8.Brûlet, P., H. Condamine, and F. Jacob. 1985. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 82:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brulet, P., M. Kaghad, Y. S. Xu, O. Croissant, and F. Jacob. 1983. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc. Natl. Acad. Sci. USA 80:5641-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragani, T. A., G. Manenti, G. Della Porta, S. Gattoni-Celli, and I. B. Weinstein. 1986. Expression of retroviral sequences and oncogenes in murine hepatocellular tumors. Cancer Res. 46:1915-1919. [PubMed] [Google Scholar]
- 11.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 12.Gidekel, S., and Y. Bergman. 2002. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 277:34521-34530. [DOI] [PubMed] [Google Scholar]
- 13.Gunzburg, W. H., F. Heinemann, S. Wintersperger, T. Miethke, H. Wagner, V. Erfle, and B. Salmons. 1993. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature 364:154. [DOI] [PubMed] [Google Scholar]
- 14.Henson, J., J. Saffer, and H. Furneaux. 1992. The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann. Neurol. 32:72-77. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, J. W., D. Steffen, J. Gusella, C. Tabin, S. Bird, D. Cowing, and R. A. Weinberg. 1982. DNA methylation affecting the expression of murine leukemia proviruses. J. Virol. 44:144-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Housey, G. M., P. Kirschmeier, S. J. Garte, F. Burns, W. Troll, and I. B. Weinstein. 1985. Expression of long terminal repeat (LTR) sequences in carcinogen-induced murine skin carcinomas. Biochem. Biophys. Res. Commun. 127:391-398. [DOI] [PubMed] [Google Scholar]
- 17.Ikuma, S., M. Kiyota, C. Setoyama, and K. Shimada. 1986. Isolation and characterization of the cDNAs corresponding to mRNAs abundant in undifferentiated mouse embryonal teratocarcinoma stem cells, but not in differentiated mouse parietal endoderm cells. J. Biochem. (Tokyo) 100: 1185-1192. [DOI] [PubMed] [Google Scholar]
- 18.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 19.International Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- 20.Kaghad, M., L. Maillet, and P. Brulet. 1985. Retroviral characteristics of the long terminal repeat of murine E.Tn sequences. EMBO J. 4:2911-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kardassis, D., P. Papakosta, K. Pardali, and A. Moustakas. 1999. c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 274:29572-29581. [DOI] [PubMed] [Google Scholar]
- 22.Kazazian, H. H. J. 1998. Mobile elements and disease. Curr. Opin. Genet. Dev. 8:343-350. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, K., E. Rosfjord, K. Brigman, and A. Rizzino. 1996. Binding of transcription factors to widely-separated cis-regulatory elements of the murine FGF-4 gene. Mol. Reprod. Dev. 44:460-471. [DOI] [PubMed] [Google Scholar]
- 24.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leong, W. L., M. J. Dobson, J. M. Logsdon, Jr., R. M. Abdel-Majid, L. C. Schalkwyk, D. L. Guernsey, and P. E. Neumann. 2000. ETn insertion in the mouse Adcy1 gene: transcriptional and phylogenetic analyses. Mamm. Genome 11:97-103. [DOI] [PubMed] [Google Scholar]
- 26.Lo, K., and S. T. Smale. 1996. Generality of a functional initiator consensus sequence. Gene 182:13-22. [DOI] [PubMed] [Google Scholar]
- 27.Loebel, D. A., B. Tsoi, N. Wong, M. P. O'Rourke, and P. P. Tam. 2004. Restricted expression of ETn-related sequences during post-implantation mouse development. Gene Expr. Patterns 4:467-471. [DOI] [PubMed] [Google Scholar]
- 28.Lueders, K. K., J. W. Fewell, V. E. Morozov, and E. L. Kuff. 1993. Selective expression of intracisternal A-particle genes in established mouse plasmacytomas. Mol. Cell. Biol. 13:7439-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luster, T. A., L. R. Johnson, T. K. Nowling, K. A. Lamb, S. Philipsen, and A. Rizzino. 2000. Effects of three Sp1 motifs on the transcription of the FGF-4 gene. Mol. Reprod. Dev. 57:4-15. [DOI] [PubMed] [Google Scholar]
- 30.Macleod, D., J. Charlton, J. Mullins, and A. P. Bird. 1994. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8:2282-2292. [DOI] [PubMed] [Google Scholar]
- 31.Mager, D. L., and J. D. Freeman. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruyama, K., and S. Sugano. 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138:171-174. [DOI] [PubMed] [Google Scholar]
- 33.McBurney, M. W., and B. J. Rogers. 1982. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev. Biol. 89:503-508. [DOI] [PubMed] [Google Scholar]
- 34.McMahon, A. P., and A. Bradley. 1990. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62: 1073-1085. [DOI] [PubMed] [Google Scholar]
- 35.Mietz, J. A., J. W. Fewell, and E. L. Kuff. 1992. Selective activation of a discrete family of endogenous proviral elements in normal BALB/c lymphocytes. Mol. Cell. Biol. 12:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa, O., and T. Sugahara. 1981. 5-Azacytidine induction of mouse endogenous type C virus and suppression of DNA methylation. Proc. Natl. Acad. Sci. USA 78:6290-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordhoff, V., K. Hubner, A. Bauer, I. Orlova, A. Malapetsa, and H. R. Scholer. 2001. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm. Genome 12:309-317. [DOI] [PubMed] [Google Scholar]
- 39.Nozawa, K., K. Maehara, and K. Isobe. 2001. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J. Biol. Chem. 276:22016-22023. [DOI] [PubMed] [Google Scholar]
- 40.Okumura, K., Y. Hosoe, and N. Nakajima. 2004. c-Jun and Sp1 family are critical for retinoic acid induction of the lamin A/C retinoic acid-responsive element. Biochem. Biophys. Res. Commun. 320:487-492. [DOI] [PubMed] [Google Scholar]
- 41.Pikarsky, E., H. Sharir, E. Ben-Shushan, and Y. Bergman. 1994. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol. Cell. Biol. 14:1026-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince, V. E., and P. W. Rigby. 1991. Derivatives of Moloney murine sarcoma virus capable of being transcribed in embryonal carcinoma stem cells have gained a functional Sp1 binding site. J. Virol. 65:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed-Inderbitzin, E., and W. Maury. 2003. Cellular specificity of HIV-1 replication can be controlled by LTR sequences. Virology 314:680-695. [DOI] [PubMed] [Google Scholar]
- 44.Ribet, D., M. Dewannieux, and T. Heidmann. 2004. An active murine transposon family pair: retrotransposition of “master” MusD copies and ETn trans-mobilization. Genome Res. 14:2261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridgway, A. A., R. A. Swift, H. J. Kung, and D. J. Fujita. 1985. In vitro transcription analysis of the viral promoter involved in c-myc activation in chicken B lymphomas: detection and mapping of two RNA initiation sites within the reticuloendotheliosis virus long terminal repeat. J. Virol. 54: 161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohr, O., D. Aunis, and E. Schaeffer. 1997. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 272:31149-31155. [DOI] [PubMed] [Google Scholar]
- 47.Saffer, J. D., S. P. Jackson, and M. B. Annarella. 1991. Developmental expression of Sp1 in the mouse. Mol. Cell. Biol. 11:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shell, B., P. Szurek, and W. Dunnick. 1987. Interruption of two immunoglobulin heavy-chain switch regions in murine plasmacytoma P3.26Bu4 by insertion of retroviruslike element ETn. Mol. Cell. Biol. 7:1364-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shell, B. E., J. T. Collins, L. A. Elenich, P. F. Szurek, and W. A. Dunnick. 1990. Two subfamilies of murine retrotransposon ETn sequences. Gene 86: 269-274. [DOI] [PubMed] [Google Scholar]
- 50.Smale, S. T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta Gene Structure Expr. 1351:73-88. [DOI] [PubMed] [Google Scholar]
- 51.Sonigo, P., S. Wain-Hobson, L. Bougueleret, P. Tiollais, F. Jacob, and P. Brulet. 1987. Nucleotide sequence and evolution of ETn elements. Proc. Natl. Acad. Sci. USA 84:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strazzullo, M., B. Majello, L. Lania, and G. La Mantia. 1994. Mutational analysis of the human endogenous ERV9 proviruses promoter region. Virology 200:686-695. [DOI] [PubMed] [Google Scholar]
- 53.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, Y., H. Taira, T. Tsunoda, J. Mizushima-Sugano, J. Sese, H. Hata, T. Ota, T. Isogai, T. Tanaka, S. Morishita, K. Okubo, Y. Sakaki, Y. Nakamura, A. Suyama, and S. Sugano. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takashima, S., M. Kono, N. Kurosawa, Y. Yoshida, Y. Tachida, M. Inoue, T. Kanematsu, and S. Tsuji. 2000. Genomic organization and transcriptional regulation of the mouse GD3 synthase gene (ST8Sia I): comparison of genomic organization of the mouse sialyltransferase genes. J. Biochem. (Tokyo) 128:1033-1043. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, I., and H. Ishihara. 2001. Enhanced expression of the early retrotransposon in C3H mouse-derived myeloid leukemia cells. Virology 280: 107-114. [DOI] [PubMed] [Google Scholar]
- 57.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 58.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.