Abstract
Herpes simplex virus VP22 is a major tegument protein of unknown function. Very recently, we reported that the predominant effect of deleting the VP22 gene was on the expression, localization, and virion incorporation of ICP0. In addition, the Δ22 virus replicated poorly in epithelial MDBK cells. We have also previously shown that VP22 interacts with the tegument protein VP16 and the cellular microtubule network. While the majority of VP22 in infected cells is highly phosphorylated, the nonphosphorylated form of VP22 is the predominant species in the virion, suggesting a differential requirement for phosphorylation through virus replication. Hence, to study the significance of VP22 phosphorylation, we have now constructed two recombinant viruses expressing green fluorescent protein-VP22 (G22) in which the previously identified serine phosphorylation sites have been mutated either to alanine to abolish the phosphorylation status of VP22 (G22P−) or to glutamic acid to mimic permanent phosphorylation (G22P+). Localization studies indicated that the G22P− protein associated tightly with microtubules in some infected cells, suggesting that VP22 phosphorylation may control its interaction with the microtubule network. By contrast, VP22 phosphorylation had no effect on its ability to interact with VP16 and, importantly, had no effect on the relative packaging of VP22. Intriguingly, virion packaging of ICP0 was reduced in the G22P+ virus while ICP0 expression was reduced in the G22P− virus, suggesting that these two ICP0 defects, previously observed in the Δ22 virus, were attributable to different forms of VP22. Furthermore, the Δ22 virus replication defect in MDBK cells correlated with the expression of constitutively charged VP22 in the G22P+ virus. Taken together, these results suggest an important role for VP22 phosphorylation in its relationship with ICP0.
Herpes simplex virus type 1 (HSV-1) is a large DNA virus composed of a capsid containing the double-stranded DNA genome that is surrounded by the tegument and an envelope that contains viral glycoproteins (6, 35). The tegument compartment of the virion is poorly characterized in relation to the function of its individual components, its role in virus entry, and its mechanism of assembly into the maturing virion. It contains a number of major structural proteins that are phosphorylated to different levels during virus infection (28), but the significance of these modifications is not yet clear for any of its components. Nonetheless, it has been suggested that the phosphorylation of tegument proteins shortly after virus entry causes their dissociation from the capsid (32).
The major tegument protein VP22 is highly phosphorylated during HSV-1 infection (15, 25). Studies on the HSV-2 homologue of VP22 have indicated that the protein undergoes highly regulated, virus-dependent phosphorylation and dephosphorylation events in infected cells (18). Moreover, we and others have shown that the nonphosphorylated form of the protein is the predominant form present in the virus particle, suggesting that the level of phosphorylation might regulate the assembly of VP22 into the virion (15, 18). In support of this, it has been shown for the bovine herpesvirus type 1 (BHV-1) homologue of VP22 that the presence of tyrosine phosphorylation sites on the protein are crucial for the efficient assembly of VP22 into the BHV-1 virion (34).
While the function of VP22 in virus infection has not yet been definitively established, the protein exhibits a number of characteristics that may be controlled by differential phosphorylation. For example, we have previously shown that VP22 is capable of interacting with another tegument protein, VP16 (9), an interaction that may provide insights into the process of tegument assembly. In addition, we have also shown that VP22 stabilizes the cellular microtubule (MT) network (11). It has been suggested that phosphorylation of VP22 is involved in its targeting to the nucleus late in infection (33), although in our own studies, we have not seen such dramatic nuclear localization of VP22 at late times in infection. In addition, it has recently been reported that VP22 interacts with the cytoplasmic tail of the envelope glycoprotein D (gD) (3). Finally, we have very recently reported on the characterization of a VP22 knockout virus in HSV-1 and have shown that the major effect of deleting VP22 is on the expression, localization, and virion incorporation of ICP0 (8). This virus also exhibits a cell type-specific replication defect in epithelial MDBK cells, suggesting that there may be a differential requirement for VP22 in virus replication that potentially involves its relationship with ICP0. While it is not yet clear if this relationship is the result of a direct interaction between these two proteins, it is conceivable that phosphorylation may play a role in its regulation.
In a previous report, we demonstrated that when expressed in virus-infected cells or in isolation, HSV-1 VP22 is phosphorylated only on serine residues (15). We have identified the specific sites that are phosphorylated by mutating individual serine residues in the VP22 open reading frame and expressing these proteins by transient transfection (14). These studies have indicated that the major serine phosphorylation site on VP22 is a substrate for the cellular kinase casein kinase II. Furthermore, a minor site at the extreme C terminus of the protein may be specific for an as yet unidentified cellular and/or virus-encoded kinase. Previous in vitro studies have implicated the virus kinase encoded by the UL13 gene in the phosphorylation of VP22 (4), and a recent report has identified a specific site in the HSV-2 homologue of VP22 that is phosphorylated by HSV-2 UL13 (17).
The construction of a virus lacking the VP22 gene has now enabled us to introduce point mutations of the serine phosphorylation sites into the VP22 open reading frame and insert these variant forms back into the virus genome to investigate the requirement for VP22 to be phosphorylated during the HSV infectious cycle. Our results demonstrate that, unlike BHV-1, VP22 phosphorylation is not required for incorporation of this protein into the virion. However, alterations in VP22 phosphorylation resulted in a number of subtle changes in the biology of HSV-1. In particular, our studies have revealed that the level of phosphate present on VP22 alters the expression and localization of ICP0 in the infected cell and affects the replication of HSV-1 in epithelial MDBK cells. Thus, we believe that the role of VP22 phosphorylation during HSV-1 infection is not to dictate the assembly of VP22 into the virus particle but to regulate its relationship with ICP0 at different stages of infection.
MATERIALS AND METHODS
Cells and viruses.
Vero, BHK, MDBK, and Cos-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum. Viruses were routinely grown in BHK or Vero cells and titrated on Vero cells. Extracellular virions were purified on Ficoll gradients from the infected cell medium of 5 × 108 BHK cells as described previously (13). Infectious virus DNA was also produced from extracellular virus as described previously (13).
The parental virus strain used in this study was strain 17 (s17) of HSV-1. The recombinant virus expressing green fluorescent protein (GFP)-tagged VP22 (G22v) has been described previously (13). The VP22 deletion mutant (169v) that expresses GFP in place of VP22 has also been described recently (8). In the course of studying the VP22 deletion mutant, we isolated a virus that expressed a nonfluorescing version of GFP due to a single point mutation in the GFP open reading frame. This new virus, called 169vc, was utilized in the isolation of the recombinant viruses described below.
Plasmids.
Plasmid pGE120 containing 800-bp flanking sequences of HSV-1 UL49, plasmid pGE155 expressing GFP-VP22 under the control of the human cytomegalovirus immediate-early (IE) promoter, and plasmid pGE168 expressing a mutant VP22 in which the serines at residues 35, 71, 72, 73, 292, and 294 in the VP22 open reading frame had been mutated to alanine residues have been described previously (13, 14). The plasmid pCP1 encoding GFP-VP22 containing the mutated serine residues under the control of the human cytomegalovirus immediate-early promoter was constructed by amplifying the VP22 open reading frame from pGE168 and inserting it as a BamHI-BglII fragment into plasmid pEGFP-C1 (BD Biosciences Clontech). To construct a transfer vector for recombination into the virus, the AseI-BsrGI fragment from pEGFP-N1 (BD Biosciences Clontech) was then inserted into AseI-BsrGI-digested pCP1, resulting in plasmid pCP2. The BamHI cassette of GFP-VP22P− from pCP2 was inserted into the BamHI site of pGE120 to produce pCP3, which consists of GFP-VP22P− carrying the mutations S35A, S71A, S72A, S73A, S292A, and S294A, surrounded by the UL49 flanking sequences, and hence driven by the UL49 promoter. The plasmid pCP4 carrying the GFP-VP22P+ gene in VP22 flanking sequences was obtained by mutagenesis overlap extension from plasmid pCP1. The resulting PCR product, consisting of GFP-VP22P+ carrying the mutations S35A, S71E, S72E, S73E, S292E, and S294E, was inserted into plasmid pGE166 as a BsrGI-KpnI fragment to produce plasmid pCP4 encoding GFP-VP22P+ surrounded by the UL49 flanking sequences and hence driven by the UL49 promoter.
Construction of recombinant viruses.
To construct the G22P−v and G22P+v recombinant viruses, subconfluent monolayers of Vero cells were cotransfected with infectious genomic DNA from 169vc and pCP3 or pCP4, respectively, using the calcium phosphate precipitation technique modified with BES [N,N-bis (2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline. The day after, medium was changed and 1% human serum added. Infected cells were screened 4 days after cotransfection for the presence of GFP plaques. Isolated G22P−v and G22P+v recombinants were plaque purified three times on Vero cells and virus stocks prepared. Each virus was sequenced across the UL49 gene to ensure that the correct mutations had been introduced.
Antibodies.
The rabbit polyclonal anti-VP22 antibody AGV031 has been described previously (12). The antibodies against VP16 (LP1) and gD (LP14) were kindly provided by Tony Minson (University of Cambridge). Mouse monoclonal anti-GFP antibody was purchased from BD Biosciences. Monoclonal antibodies against ICP4 and ICP0 were obtained from Virusys. An additional anti-ICP0 monoclonal antibody (11060) was kindly provided by Roger Everett, MRC Virology Unit, Glasgow, Scotland. The anti-VP5 monoclonal antibody was obtained from Autogen Bioclear. Monoclonal anti-α-tubulin and monoclonal anti-β-actin were obtained from Sigma and AbCam, respectively.
Virus growth curves.
Confluent Vero or MDBK cells grown in a six-well plate were infected at low multiplicity of infection (0.1 or 0.02 PFU/cell). One hour postinfection, the inoculum was removed, the cells were washed three times with phosphate-buffered saline (PBS), and 1 ml of fresh medium was added to each well. At various times postinfection, either extracellular virus from the cell medium or intracellular virus from the cells was harvested from one well of infected cells and scraped into 1 ml of PBS or total virus was harvested bycombining the cells and medium. Each virus sample was then titrated on Verocells.
SDS-PAGE and Western blot analysis.
Solubilized proteins were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the gels were either stained with Coomassie blue or transferred to nitrocellulose filters. The filters were incubated with the appropriate primary antibody, processed using horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Bio-Rad), and developed using an enhanced chemiluminescence kit (Pierce).
In vivo radiolabeling assay.
Confluent monolayers of Vero cells in 6-cm- diameter dishes were infected at a multiplicity of infection of 10 PFU per cell and incubated at 37°C. One hour postinfection, the inoculum was removed and cells were washed twice with phosphate-free DMEM (ICN) and starved for phosphates by incubating them in phosphate-free DMEM supplemented with 10% dialyzed calf serum for 1 h at 37°C. Radioactive labeling was conducted by the addition of 100 μCi of [32P]orthophosphate (Amersham) per dish, and the cells were incubated at 37°C for a further 13 h. After washing off the unincorporated label with PBS, proteins were extracted with 1 ml of radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Na deoxycholate, 0.5% Triton X-100) containing protease inhibitors (Miniprotease inhibitors; Roche Diagnostics Ltd.). Proteins from the soluble fraction were immunoprecipitated with the anti-VP22 polyclonal antibody AGV031, and after extensive washing of the protein A Sepharose beads (Amersham), proteins were analyzed by SDS-PAGE and autoradiography.
Live-cell microscopy analysis.
Vero cells for live analysis of GFP expression were plated into two-well Lab-Tek coverglass chambers (Quadrachem Laboratories), infected with HSV-1 at a low multiplicity of infection (0.1 PFU/cell), and examined using a Zeiss LSM 410 inverted confocal microscope. Images were collected every 2 h for 24 h after infection and processed using Adobe Photoshop software.
Immunofluorescence.
Vero cells were grown on 16-mm coverslips in the individual wells of a six-well plate. Cells were fixed for 10 min in 100% methanol, or 20 min in 4% paraformaldehyde followed by permeablization for 5 min with 0.1% Triton X-100. The fixed cells were blocked by incubation for 30 min in PBS containing 10% newborn calf serum. Primary antibody was added in block solution for 1 h. Following extensive washing in PBS, Texas Red-conjugated anti-mouse immunoglobulin G (Vector Labs) was added in block solution and the mixture was incubated for a further 45 min. The coverslips were washed in PBS and mounted in Vectashield (Vector Labs). Images were acquired using a Zeiss LSM 410 inverted confocal microscope and processed using Adobe Photoshop.
RESULTS
Construction of recombinant viruses expressing phosphorylation variants of VP22.
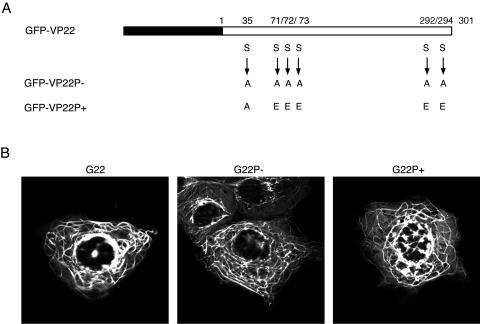
We have previously shown that VP22 expressed by transient transfection colocalizes with cellular MTs, resulting in their reorganization and stabilization in bundles (11). Furthermore, this phenotype is maintained in a GFP-VP22 fusion protein (10). To determine if the phosphorylation status of VP22 had any effect on MT bundling, we constructed two variants of GFP-VP22 for analysis by transient transfection: one in which the previously identified serine phosphorylation sites had been replaced with alanine residues in order to abolish the phosphorylation of VP22 (G22P−) and one in which the serines had been replaced with glutamic acid residues in an attempt to mimic a permanent phosphorylation status of VP22 (G22P+) (Fig. 1A). These plasmids were transfected into Cos-1 cells, and the localization of the VP22 variants was compared to that of wild-type (WT) GFP-VP22 (G22) (Fig. 1B). Live-cell analysis of cells expressing each variant of VP22 indicated that they both localized to the characteristic cytoplasmic bundles observed for GFP-VP22, suggesting that the mutation of the phosphorylation sites had little effect on the subcellular localization of VP22 when expressed in isolation (Fig. 1B).
FIG. 1.
Phosphorylation variants of VP22 as GFP-VP22 fusion proteins. (A) Line drawing of the GFP-VP22 (G22) open reading frame showing GFP (black box) and the 301 amino acids of the VP22 WT (white box). The serine (S)-to-alanine (A) or S-to-glutamic acid (E) substitutions present in the plasmids expressing the GFP-VP22P− (G22P−) or GFP-VP22P+ (G22P+) mutant are shown. These substitutions were introduced by PCR mutagenesis. (B) Live transfected Cos-1 cells expressing either the G22, G22P−, or G22P+ proteins, examined by confocal microscopy 24 h posttransfection.
We next constructed recombinant viruses expressing each of these VP22 variants as GFP fusion proteins. Plasmids encoding the GFP-VP22 variants in the background of the UL49 flanking sequences were cotransfected into Vero cells with genomic viral DNA purified from cells infected with our VP22 deletion mutant. The cells were incubated for 4 days until plaques had formed and were screened for GFP fluorescence to identify recombinant viruses. One fluorescent plaque for each cotransfection was chosen, and the recombinant viruses were each plaque purified three times to obtain the recombinant virus G22P−v expressing a phosphorylation-deficient variant of GFP-VP22 and the recombinant virus G22P+v expressing a constitutively charged GFP-VP22.
Phosphorylation status of the GFP-VP22 variant proteins.
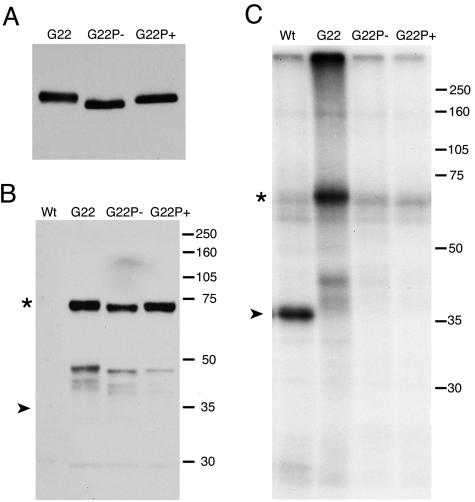
To ensure that G22P−v and G22P+v were correctly expressing the phosphorylation variants of GFP-VP22, we first analyzed the expression of GFP-VP22 in cells infected with the three viruses. Vero cells infected at a high multiplicity of infection were harvested 10 h after infection, and total cell extracts were analyzed by Western blotting with an anti-VP22 antibody. The results show that both phosphorylation variants express VP22 at levels similar to those of the GFP-VP22 virus (Fig. 2A). Interestingly, the P− version of VP22 ran with a faster mobility than WT VP22, confirming that this protein was less negatively charged than the protein expressed in a WT infection (Fig. 2A). In addition, the P+ version of VP22 ran with the same mobility as parental VP22, suggesting that the correct charge in this protein had been maintained by introduction of the glutamic acid residues in place of serines (Fig. 2A).
FIG. 2.
Characterization of VP22 expressed in G22, G22P− or G22P+ virus infections. (A) Monolayers of Vero cells were infected with G22v, G22P−v, or G22P+v at a multiplicity of infection of 10, and total cell lysates were harvested after 10 h. Equal amounts of lysate were analyzed by SDS-PAGE followed by Western blotting with an antibody against VP22. (B) Vero cells were infected at a multiplicity of infection of 10 with the WT viral s17, GFP-VP22 expressing G22v, or the recombinant viruses G22P−v and G22P+v and incubated in a labeling medium containing [32P]orthophosphate. Protein cell lysates were immunoprecipitated using an antibody directed against VP22 and then analyzed by Western blotting using an antibody directed against GFP. (C) SDS-PAGE analysis of the immunoprecipitated proteins and autoradiography. The positions of VP22 (➤) and GFP-VP22 (*) are shown.
To ensure that we had targeted all the potential phosphorylation sites in VP22, we next analyzed the in vivo phosphorylation levels of VP22 expressed from our parental virus (s17), the GFP-VP22-expressing virus (G22v), and the two phosphorylation variants (G22P−v and G22P+v). Vero cells infected with these viruses were labeled in vivo with [32P]orthophosphate, cell extracts were prepared, and immunoprecipitations were carried out with the anti-VP22 antibody (Fig. 2B and C). Western blotting of the immunoprecipitated extracts with an anti-GFP antibody demonstrated that both phosphorylation variants of the GFP-VP22 protein were precipitated in approximately similar amounts to the G22 protein (Fig. 2B). Nevertheless, while [32P]phosphate was efficiently incorporated into WT untagged and GFP-tagged VP22, there was no detectable phosphate present in the phosphorylation variant proteins G22P− and G22P+ (Fig. 2C). While we cannot rule out the possibility that a poorly utilized, and therefore undetectable, phosphorylation site was still present in these variants of VP22, we nonetheless interpret these results to mean that neither of the GFP-VP22 variants is phosphorylated during infection and that we have successfully mutated all the major utilized phosphorylation sites.
Live-cell analysis of infected cells.
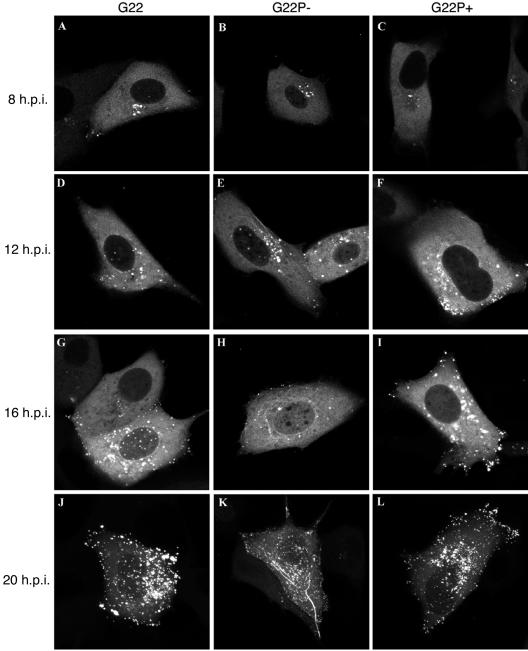
We have previously shown that incorporation of the GFP-VP22 protein into the virus particle does not alter the growth properties of the virus, allowing us to use this virus to monitor the trafficking of VP22 in living cells (13). To investigate if there is any requirement for VP22 to be phosphorylated during virus infection for its correct subcellular targeting, we compared the localization of the phosphorylation variant G22P− and G22P+ proteins with the parental GFP-VP22 protein. Vero cells were infected with G22v, G22P−v, or G22P+v at a low multiplicity of infection (0.1 PFU/cell), and representative images were collected at 8, 12, 16, and 20 h postinfection. As previously described, the GFP-VP22 protein exhibits a complex and sequential localization within the infected cell (13, 21). It appeared initially in a diffuse cytoplasmic pattern as early as 6 h after infection at a low multiplicity of infection (data not shown). Over the next hours, the cytoplasmic fluorescence increased in intensity while the GFP-VP22 accumulated in a Golgi-like pattern (Fig. 3A). Slightly later, a minor population of GFP-VP22 appeared in discrete dots within the nucleus, as described previously (21), while the cytoplasmic population took on a strong vesicle-like localization (Fig. 3D and G). At later stages of infection, GFP-VP22 accumulated at the cell periphery (Fig. 3J). In a similar manner to GFP-VP22, the two phosphorylation variants were first observed in a diffuse cytoplasmic pattern with an accumulation in a Golgi-like position (Fig. 3B and C). The localization of the G22P+ protein throughout the infectious cycle proceeded in a manner similar to that of the WT protein with the exception that VP22-containing nuclear dots could not be detected in cells infected with this virus (Fig. 3, compare panel F with panels D and E). However, a striking difference was observed in cells infected with the G22P− virus whereby VP22 was easily detectable in cytoplasmic filamentous bundles that we presume to be MTs from our previous results (Fig. 3E, H, and K). Nonetheless, vesicle-like structures containing VP22 were also often observed within the cytoplasm of these cells and at the cell periphery at later times, suggesting that the G22P− protein has the capacity to localize in a pattern similar to that of the WT protein.
FIG. 3.
Live-cell analysis of the VP22 phosphorylation variants in infected cells. Monolayers of Vero cells grown in coverslip chambers were infected with G22v (A, D, G, and J), G22P−v (B, E, H, and K), or G22P+v (C, F, I, and L) at a multiplicity of infection of 0.1 and were examined by confocal microscopy for GFP fluorescence every 2 h up to 20 h postinfection (h.p.i.). Representative images taken at 8 (A to C), 12 (Dto F), 16 (G-I), or 20 (J to L) h.p.i. are shown.
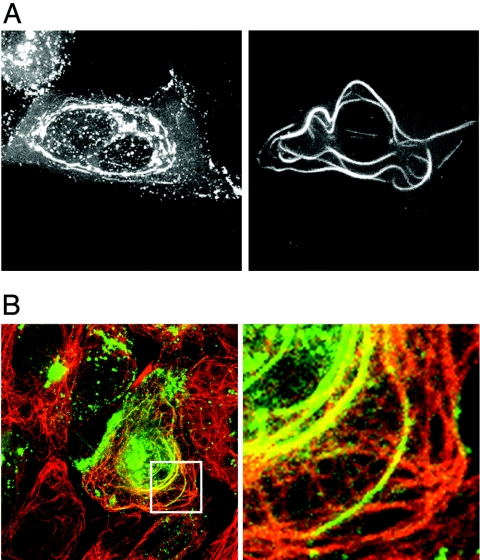
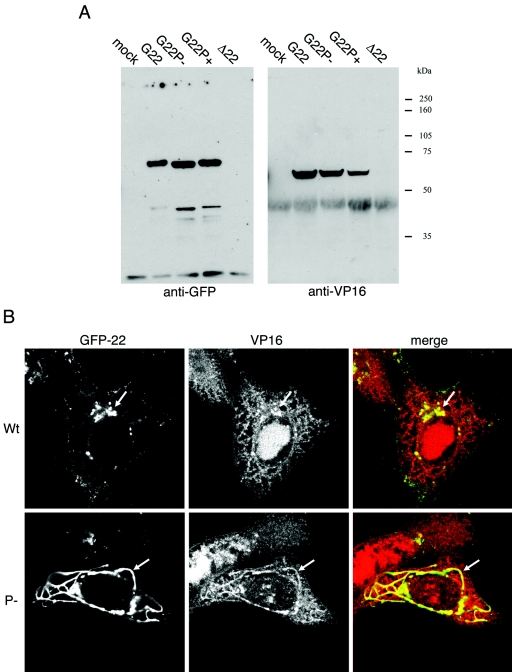
On further examination of a large number of cells infected with the G22P− mutant virus, it became clear that in a small population of cells, the filamentous localization of VP22 was particularly obvious (Fig. 4A) and resembled the localization of VP22 observed in transfected cells. To confirm that the G22P-containing bundles were indeed associated with MTs, we next carried out indirect immunofluorescence of infected cells with an anti-α-tubulin antibody. The resulting images show that the majority of the GFP-VP22-containing bundles colocalized with cellular MT bundles in G22P−v-infected cells (Fig. 4B), and while some of the bundles appear green rather than yellow in this image, this simply reflects the presence of a high concentration of VP22 compared to tubulin.
FIG. 4.
The G22P− protein forms MT bundles in infected cells. Vero cells infected with G22P-v at a multiplicity of infection of 0.1 were examined live at 20 h postinfection (A) or were fixed with methanol and processed for immunofluorescence with an anti-α-tubulin antibody. (B) Cells were examined by confocal microscopy for GFP-VP22 (green) and α-tubulin fluorescence (red). Right-hand panels show magnified images of the region in the white box.
The nonphosphorylated form of VP22 interacts efficiently with VP16 and relocalizes it to MT bundles.
We have previously shown that VP22 interacts directly with a second tegument protein, VP16 (9), an interaction that we and others have suggested may be involved in the process of tegument assembly (30). Hence, to determine whether our VP22 phosphorylation variants were capable of interacting with VP16 as efficiently as parental VP22, we carried out coimmunoprecipitation assays from infected cell extracts. VP22 was first immunoprecipitated with a polyclonal antibody against VP22 and the efficiency of precipitation determined by Western blotting with a monoclonal antibody against GFP (Fig. 5A, anti-GFP). These results indicated that all three VP22 proteins had been precipitated to similar levels. Next, the same samples were blotted with a monoclonal anti-VP16 antibody to determine the ability of each variant of VP22 to interact with VP16 (Fig. 5A, anti-VP16). Once again, the results showed that VP16 interacted efficiently with all three variants of VP22, suggesting that the charge on VP22 does not significantly affect its ability to interact with VP16. Furthermore, immunoprecipitation with an extract from cells infected with our Δ22 virus demonstrated that there was no nonspecific association of VP16 with the anti-VP22 antibody (Fig. 5A, Δ22 lanes).
FIG. 5.
The phosphorylation variants of VP22 interact with VP16. (A) Monolayers of Vero cells were either mock infected or infected at a multiplicity of infection of 10 with G22v, G22P−v, G22P+v, or the Δ22 virus (169v) and lysed in radioimmunoprecipitation assay buffer 15 h postinfection. Cell lysates were immunoprecipitated using an antibody directed against VP22 and then analyzed by SDS-PAGE and Western blotting with antibodies against GFP and VP16. (B) Monolayers of Vero cells were infected with either G22v or G22P−v at a multiplicity of infection of 0.1. At 20 h postinfection, the cells were fixed with methanol and processed for immunofluorescence with an anti-VP16 antibody. Cells were examined by confocal microscopy for GFP-VP22 (green) and VP16 (red) fluorescence.
Because the phosphorylation-deficient version of VP22 was able to interact with VP16, we next investigated the localization of VP16 in cells infected with this virus. Indirect immunofluorescence of Vero cells infected with either the G22-expressing virus or the G22P-expressing virus was carried out with an anti-VP16 antibody. In cells expressing G22, cytoplasmic VP16 was clearly concentrated in the VP22-containing vesicles that in the example cell are localized close to the nucleus (Fig. 5B, G22v). Furthermore, in cells expressing the P− version of VP22, in which MT bundles had been formed, VP16 was also concentrated at the major site of VP22 localization within the cytoplasmic bundles (Fig. 5B, G22P−v). Thus, these results show that MT-bound VP22 can recruit VP16 into the MT bundles, suggesting that the VP22-MT interaction may be dominant over the VP22-VP16 interaction in infected cells.
VP22 phosphorylation levels affect ICP0 expression and localization.
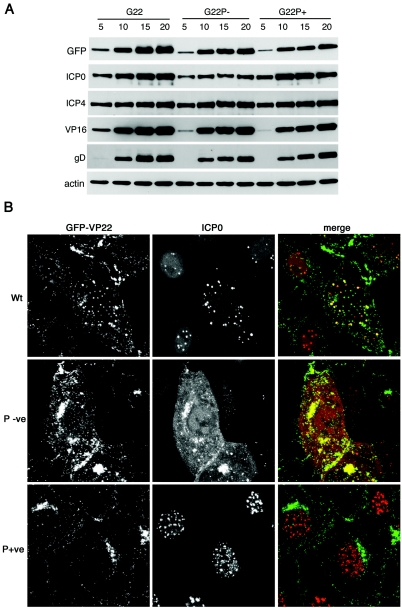
In light of our recent observation that virus grown in the absence of VP22 displays a delay in the initiation of viral protein synthesis, especially for the immediate-early protein ICP0 (8), we next wished to investigate the role that VP22 phosphorylation might play in such a phenotype. Thus, we examined the kinetics of expression of a range of virus-encoded proteins in cells infected with the three GFP-VP22-expressing viruses. Vero cells infected at a high multiplicity of infection were harvested at various times after infection, total cell extracts were analyzed by Western blotting with a range of antibodies against virus-encoded proteins, and an anti-β-actin antibody was used as a loading control (Fig. 6A). Interestingly, the results show that while there is a very slight reduction in the kinetics of expression of the late proteins VP16, gD, and VP22 itself in both variant viruses (Fig. 6, VP16, gD, and GFP and compare G22P− and G22P+ with G22), there was a clear difference in the expression of ICP0 that was specific to the G22P−v infection (Fig. 6A, ICP0 and compare G22P− with G22 and G22P+). Thus, while ICP0 was expressed in the G22P+v-infected cells in the same manner as G22v-infected cells, the level of ICP0 present in G22P−v-infected cells was clearly reduced. Taken together with our previous observation that ICP0 levels are also reduced in cells infected with a VP22 deletion virus (8), this suggests that it is specifically the phosphorylated version of VP22 that is required to allow optimal levels of ICP0 to be present during HSV-1 infection. In addition, there was no apparent difference in the expression levels of ICP4 between the three viruses, whereas cells infected with the VP22 deletion virus also exhibit a reduction in the levels of this IE protein.
FIG. 6.
ICP0 expression and localization in cells infected with the VP22 phosphorylation mutants. (A) Monolayers of Vero cells were infected with G22v, G22P−v, or G22P+v at a multiplicity of infection of 10, and total cell lysates were harvested every 5 h after infection up to 20 h. Equal amounts of total cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies against GFP, ICP4, ICP0, VP16, gD, and cellular β-actin. (B) Monolayers of Vero cells were infected with G22v, G22P−v, or G22P+v at a multiplicity of infection of 10 and fixed 7 h later in 4% paraformaldehyde. The cells were then processed for immunofluorescence with a monoclonal anti-ICP0 antibody and examined by confocal microscopy for GFP-VP22 (green) and ICP0 fluorescence (red). Note that the ICP0 image for G22P−v was imaged using 10-fold more laser power because of its weak expression in infected cells.
In our previous studies on the VP22 deletion mutant, we have shown that the absence of VP22 affects the cytoplasmic localization of ICP0 and subsequently its incorporation into the virion. Hence, we next addressed the localization of ICP0 in cells infected with our VP22 phosphorylation mutants. Vero cells infected with the three GFP-VP22-expressing viruses at a multiplicity of infection of 10 were fixed 6 h after infection, and immunofluorescence was carried out with an ICP0 antibody (Fig. 6B). As we have shown before, at this time in a WT infection, ICP0 shifts from the nucleus into the cytoplasm, where it colocalizes with VP22 (8) (Fig. 6B, Wt). Interestingly, ICP0 expressed in the G22P− virus-infected cells had also shifted to the cytoplasm by this time, where it was also colocalized with the G22P− protein in spite of the much lower level of ICP0 in infected cells (Fig. 6B, P-ve). By contrast, there was a dramatic alteration in the localization of ICP0 expressed in the G22P+-virus-infected cells, with the protein being located entirely in the nucleus, where it was present in multiple intense speckles (Fig. 6B, P+ve). This apparent retention of ICP0 in the nucleus was also maintained at later times in infection (data not shown), demonstrating that the effect was not simply a delay in ICP0 translocation from the nucleus to the cytoplasm.
Incorporation of the VP22 phosphorylation variants into virus particles.
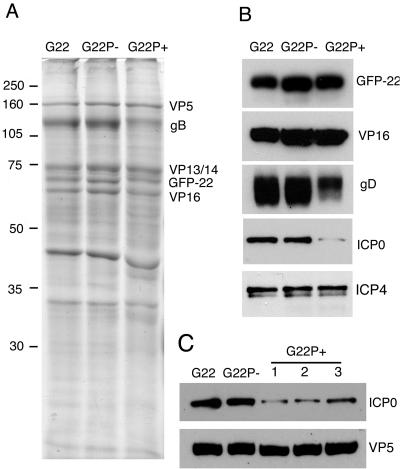
We have previously shown that during infection, the nonphosphorylated form of VP22 is the predominant species to be incorporated into the virus structure (15). To determine if the incorporation of GFP-VP22 is affected by the phosphorylation status of the protein, we analyzed extracellular virions from all three GFP-VP22-expressing viruses by SDS-PAGE, followed by Coomassie blue staining or Western blotting with equal amounts of the virion preparations loaded according to their VP5 content (Fig. 7A). The results clearly demonstrate that both phosphorylation variants of VP22 are packaged into virions in amounts similar to the WT VP22, as seen in the virion profiles (Fig. 7A). This result was confirmed by Western blotting of the same virion preparations with an antibody against VP22 (Fig. 7B). Comparison of the Coomassie blue-stained virion profiles revealed no major differences in the virion content of the major structural proteins for the two variant viruses, and in particular, there was no difference in the levels of VP16 (Fig. 7A and B). Interestingly, however, we detected a reduction in the levels of the envelope gD in the G22P+ virions as determined by Western blotting (Fig. 7B, gD) and a reduction in the levels of gB as determined by Coomassie blue staining (Fig. 7A, gB). We have recently shown that virus particles assembled in the absence of VP22 also exhibit a similar reduction in glycoprotein levels, in particular, of gD (8), and hence the results presented here suggest that efficient incorporation of the glycoproteins may require the nonphosphorylated form of VP22. Finally, our previous data on VP22-deficient virions have indicated that VP22 is absolutely required for the incorporation of the two immediate-early proteins, ICP0 and ICP4, into the virus. Because we have shown above that expression of the G22P+ version of VP22 has a dramatic effect on the localization of ICP0 while the G22P− version has an effect on the levels of ICP0 in the infected cell, we wished to determine the relative packaging of ICP0 into the corresponding virions. The relative charge of VP22 does not seem to play a role in the assembly of ICP4 into the virion, as both phosphorylation variant viruses were able to package ICP4 to the levels of WT virus (Fig. 7B, ICP4). Likewise, ICP0 was packaged efficiently into G22P− virions, suggesting that although the levels of ICP0 were reduced in cells infected with this virus, the fact that it colocalized with VP22 in the cytoplasm was indicative of its assembly into the virion. By contrast, we detected a clear reduction in the levels of ICP0 present in the G22P+ virions (Fig. 7B) that correlated with the lack of cytoplasmic ICP0 in cells infected with this virus. Moreover, this effect was reproducible, with three separate virion preparations of G22P+ virus particles exhibiting the same reduction in ICP0 content (Fig. 7C).
FIG. 7.
Incorporation of the phosphorylation variants of VP22 into virions. Approximately equivalent amounts of purified G22v, G22P−v, and G22P+v virions were solubilized and analyzed by SDS-PAGE followed by either Coomassie blue staining (A) or Western blotting with antibodies directed against VP22, VP16, gD, ICP0, and ICP4 (B). (C) Equivalent amounts of purifed G22v, G22P−v, and three different preparations of G22P+v virions were analyzed by Western blotting for ICP0 and VP5.
VP22 phosphorylation level affects HSV-1 replication.
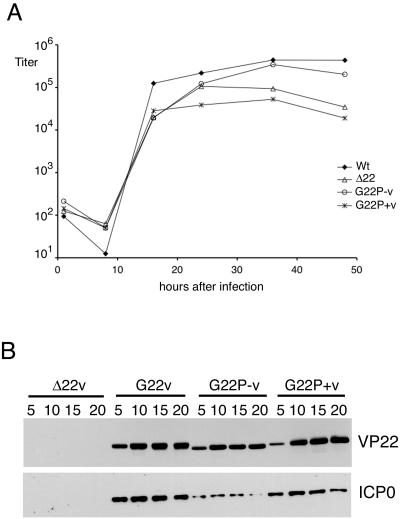
We have previously shown that the HSV-1 VP22 deletion mutant grows efficiently in Vero cells but displays reduced replication in MDBK cells. Thus, we next assessed the rate of virus replication of our VP22 phosphorylation variants by conducting low multiplicity time courses of infection in both Vero and MDBK cells. Our results indicate that, as for the VP22 deletion mutant, both phosphorylation mutant viruses grew as efficiently as the WT virus on Vero cells (data not shown). In addition, the G22P− virus grew as efficiently as WT virus in MDBK cells (Fig. 8A). By contrast, however, the G22P+ virus exhibited a clear reduction in replication efficiency in MDBK cells that was comparable to that observed for the VP22 deletion mutant (Δ22) (Fig. 8A). To confirm that the trends in ICP0 expression levels in these infections were similar in MDBK cells and in Vero cells, we examined the kinetics of expression of ICP0 in cells infected with the three GFP-VP22-expressing viruses compared to cells infected with the Δ22 virus. MDBK cells infected at a high multiplicity of infection were harvested at various times after infection, and total cell extracts were analyzed by Western blotting with antibodies against VP22 and ICP0. In both cases, comparable results were obtained in MDBK cells compared to Vero cells. Hence, there was little effect on VP22 expression in cells infected with the phosphorylation mutants compared to WT VP22, but ICP0 levels were greatly reduced in cells infected with the G22P− virus compared to cells infected with the G22P+ virus or WT virus (Fig. 8B). Furthermore, the reduced ICP0 levels in the G22P− virus-infected cells were still higher than those obtained in Δ22 virus-infected MDBK cells, suggesting that for this feature at least, the G22P− virus had an intermediate phenotype compared to the WT and the deletion virus. In summary, the reduced replication efficiency in MDBK cells correlates with the altered localization and assembly of ICP0 observed in G22P+ virus-infected cells and not the reduced levels of ICP0 observed in G22P mutant virus-infected cells.
FIG. 8.
HSV-1 expressing the P+ version of VP22 displays a growth defect in MDBK cells. (A) Monolayers of MDBK cells were infected with WT, Δ22 G22P−, or G22P+ viruses at a multiplicity of infection of 0.02. At various times after infection, total virus was harvested by combining the cells and medium and the samples were titrated on Vero cells. (B) Monolayers of MDBK cells were infected with Δ22, G22, G22P−, or G22P+ viruses at a multiplicity of infection of 10. At various times after infection, total cell extracts were made from infected cells and analyzed by Western blotting for VP22 and ICP0.
DISCUSSION
Protein phosphorylation is a fundamental regulatory mechanism within the cell, with the level of phosphate on a protein dictating the activity of that protein, its cellular location, and/or its interaction with other proteins. Many virus-encoded proteins are phosphorylated, and in some cases, these proteins are known to be substrates of cellular kinases (26, 27). Many viruses are also known to encode their own kinases so that they have the potential to regulate the phosphorylation status of their own proteins, and several have been shown to package kinases (either cellular or virus encoded) into the structure of the virus particle, thereby ensuring that the kinase is delivered to the cell at the site of virus entry (1, 20, 22, 29, 31). This may be important for virus-induced alteration of the local cellular environment (for example, rearrangement of the actin cytoskeleton) or for the regulation of proteins involved in early virus gene expression. In the case of alphaherpesviruses, the two virus kinases encoded by the UL13 and US3 genes are packaged into the virion (5, 19, 36). Although the exact role of these virion-associated kinases is not yet clear, it is noteworthy that the UL13 gene is conserved throughout the alpha-, beta-, and gammaherpesvirus families (24), suggesting that it plays an intrinsic role in virus replication.
One of the viral substrates of HSV-1 UL13 is the major tegument protein VP22 (4), which is also known to be an efficient substrate for the cellular kinase casein kinase II (14). VP22 is highly phosphorylated in cells but not in the virus particle (15), confirming the potential for phosphorylation of VP22 at virus entry. However, the exact role of VP22 in virus infection is not yet clear and hence the significance of VP22 phosphorylation is difficult to establish. Nonetheless, phosphorylation of the BHV-1 homologue of VP22 has been shown to be critical for assembly of this protein into the virus, implying a role for phosphorylation in the assembly process (34). In this report, we have investigated the role of VP22 phosphorylation in HSV-1 by generating viruses in which the phosphorylation sites within the open reading frame of VP22 have been mutated to either noncharged residues to generate a nonphosphorylated protein or negatively charged residues to mimic a permanent phosphorylation status. Although HSV-1 appears to preferentially package the nonphosphorylated form of VP22 when presented with both forms in a WT infection, our results show that both phosphorylation variants of VP22 examined in this study are able to be packaged in equivalent amounts, suggesting that there is no absolute requirement for either form of VP22 for its assembly into the tegument. We cannot rule out the possibility that the P+ variant of VP22 did not successfully mimic permanent phosphorylation, and hence, we cannot state definitively that phosphorylation of VP22 does not inhibit its assembly into the virion. Nonetheless, our results with the P− variant demonstrate unequivocally that unlike the situation in BHV-1 (34), HSV-1 VP22 does not require phosphorylation for efficient assembly into the tegument.
The UL13-specific phosphorylation site in HSV-2 VP22 has been mapped to serine 35, one of the serines targeted in the mutagenesis of HSV-1 VP22 performed here. Hence, we have probably mutated phosphorylation sites for both cellular and viral kinases in our P− variant of VP22. It has been proposed that UL13 is involved in the phosphorylation of the tegument at virus entry, thereby releasing tegument proteins into the cytosol and revealing the capsid structure for transport to the nucleus (32). However, although in vitro studies have shown that UL13 is required for the major tegument proteins to dissociate from the tegument in a biochemical assay (32), it has not yet been definitively shown to have the same effect in cells. As we have now constructed a virus expressing a variant of VP22 that is no longer able to be phosphorylated (by cellular or viral kinases), the premise that VP22 is phosphorylated by UL13 at virus entry in order to be released from the tegument should now be testable, in particular, because we have tagged VP22 with GFP.
Recruitment of tegument proteins into the maturing virion is likely to involve specific interactions with other virus components and/or cellular structures. A potential mechanism for recruitment of VP22 is through its interaction with another tegument protein, VP16 (9). Our results presented here show that both phosphorylation variants of VP22 bind efficiently to VP16, and hence, phosphorylation is not required for these two tegument proteins to form a complex. Interestingly, our virion profiles indicate that there is a clear reduction in the level of envelope gD and gB in the virus expressing the P+ version of VP22. This result extends our own recent observation that virions grown in the absence of VP22 also exhibit a reduction in glycoprotein content, in particular, gD (8), and the recent demonstration by others that VP22 can interact with the Cterminus of gD in vitro (3). Intriguingly, this may suggest that the gD-VP22 interaction requires that one or another of the VP22 phosphorylation sites is unmodified for efficient binding between the two proteins. In the pseudorabies virus system, it has been shown that the C terminus of VP22 interacts with the cytoplasmic tails of gE and gM (16), and therefore, by extrapolation, the gD-VP22 interaction in HSV-1 may also involve the C-terminal region of VP22, encompassing one of its previously identified phosphorylation sites (14).
The VP22 knockout virus that we described in a very recent publication (8) exhibits a clear delay in the onset of virus protein synthesis, and although we do not yet have a definitive explanation for this phenotype, it does suggest that VP22 plays a role at early times in infection, one that could be either at the level of capsid transport to the nucleus or at the level of IE gene expression. Interestingly, we have shown that the levels of the IE protein ICP0 were greatly reduced in cells infected with the Δ22 virus, and although not so dramatic, we now show a corresponding decrease in the intracellular levels of ICP0 present in cells infected with the virus expressing the P− variant of VP22. This implies that efficient involvement of VP22 with ICP0 at this stage of infection requires the phosphorylated form of VP22. Further studies must be done to determine if the reduction of ICP0 in cells infected with either of these VP22 mutant viruses is due to reduced expression rates or reduced stability of the ICP0 protein. An additional result of deleting the VP22 gene from HSV-1 was the abrogation of ICP0 incorporation into the virus particle, suggesting the involvement of VP22 in recruiting this IE protein to the tegument of the virion (8). In this current study, we have shown a clear but not complete reduction in the levels of ICP0 assembled into the virions purified from cells infected with the P+-variant-expressing virus, suggesting that, unlike the requirement for VP22 at early stages of infection, this process requires the nonphosphorylated form of VP22. Interestingly, this lack of ICP0 assembly into the virion correlates with an altered localization of ICP0 in the infected cell. In both WT- and Δ22-infected cells, ICP0 has been shown to translocate to the cytoplasm at 4 to 5 h after infection (8). In the case of WT virus, the ICP0 then localizes to VP22-containing complexes that we believe to be involved in assembly. However, in the case of the Δ22 virus, ICP0 remains diffuse in the cytoplasm and is not packaged into the virion. By contrast, ICP0 in G22P+-infected cells fails to translocate to the cytoplasm and is retained in the nucleus, where it localizes to subtly different nuclear structures than those present in WT-infected cell nuclei. While we believe the reason for the reduction in ICP0 content in the virion is due to this failure of ICP0 to move into the cytoplasm, the mechanism of such nuclear retention remains unclear, in particular, because VP22 is predominantly cytoplasmic. However, the fact that the behavior of ICP0 is quite different in the presence of a mutant version of VP22 than in the complete absence of VP22 implies that the P+ variant of VP22 has a dominant-negative activity on ICP0. Furthermore, the presence of the VP22P+ protein reduces HSV-1 replication efficiency in MDBK cells to the level of the Δ22 virus, suggesting that the defect in virus replication observed in these cells correlates with a failure in the localization and packaging of ICP0 rather than the reduction in ICP0 levels detected in VP22P−-expressing cells.
An additional interaction of VP22 that appears to be affected by its phosphorylation status is its association with cellular MTs, a relationship that we initially demonstrated in cells transiently expressing VP22 (11). Indeed, enhanced binding of the P− version of VP22 to MTs in the infected cells shown here is the first definitive demonstration that such a virus-cell interaction occurs during infection. It is possible that the increased association of this variant of VP22 with MTs is due simply to its increased basic charge, allowing it to bind more tightly to the negatively charged MTs. However, phosphorylation is a modification known to regulate the binding of many cellular MT-associated proteins (2, 7, 23), and hence, this may be an important mechanism that the virus uses to control the interaction of VP22 with MTs in the infected cell. Although all three forms of VP22 exhibit the same MT localization when expressed in transient transfection, further studies such as in vitro MT binding assays would be required to measure the relative affinity of each VP22 variant for polymerized MTs in the absence of other virus proteins. However, in the case of infected cells where VP22 is likely to be involved in multiple interactions, the existing balance between the VP22-MT interaction and, for instance, the VP22-VP16 interaction would prevent WT VP22 from assembling onto cellular MTs in an uncontrolled fashion. If this balance were altered such that the MT interaction was stronger than the VP16 interaction, it might be expected that VP22 would assemble onto MTs even during infection and bring VP16 and other potential interacting proteins with it. Indeed, this turned out to be true when we examined VP16 localization in cells infected with the G22P− virus. Hence, while the role of the VP22-MT interaction during infection has yet to be determined, it would seem that phosphorylation may play an important role in ensuring that this interaction is not dominant over the other activities of VP22 during infection.
In summary, our results would suggest that VP22 phosphorylation is a subtle mechanism utilized by the virus to maintain a balance between the various activities of VP22 during the virus replication cycle. Importantly, we have shown that the main consequence of abolishing phosphorylation occurs during early times of the replication cycle, when it is known that VP22 is phosphorylated, while the major consequence of mimicking phosphorylation occurs in the assembly of the virus particle that usually contains predominantly nonphosphorylated VP22. Nonetheless, as is the case for mutations in so many virus proteins, the true biological consequences of shifting the balance of VP22 activities may become apparent only when investigating the growth of these mutant viruses in an animal model.
Acknowledgments
We thank Tony Minson and Helena Browne for generously providing us with the anti-VP16 and anti-gD antibodies. We also thank Roger Everett for the ICP0 antibody.
C.P. was funded by a grant from GlaxoSmithKline and by a prize scholarship from the Foundation Bettencourt-Schueller. G.E. is funded by Marie Curie Cancer Care.
REFERENCES
- 1.Banham, A. H., and G. L. Smith. 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191:803-812. [DOI] [PubMed] [Google Scholar]
- 2.Cambiazo, V., E. Logarinho, H. Pottstock, and C. E. Sunkel. 2000. Microtubule binding of the drosophila DMAP-85 protein is regulated by phosphorylation in vitro. FEBS Lett. 483:37-42. [DOI] [PubMed] [Google Scholar]
- 3.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 4.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham, C., A. J. Davison, A. Dolan, M. C. Frame, D. J. McGeoch, D. M. Meredith, H. W. Moss, and A. C. Orr. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303-311. [DOI] [PubMed] [Google Scholar]
- 6.Dargin, D. 1986. The structure and assembly of herpes viruses, p. 359-437. In J. R. Harris and R. W. Horne (ed.), Electron microscopy of proteins, vol. 5. Viral structure. Academic Press, London, United Kingdom. [Google Scholar]
- 7.Ebneth, A., G. Drewes, E. M. Mandelkow, and E. Mandelkow. 1999. Phosphorylation of MAP2c and MAP4 by MARK kinases leads to the destabilization of microtubules in cells. Cell Motil. Cytoskeleton 44:209-224. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., D. O'Reilly, and P. O'Hare. 1999. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J. Virol. 73:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., D. O'Reilly, and P. O'Hare. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140-145. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss, B. J., G. L. Cano, J. E. Tavis, and L. A. Morrison. 2004. Herpes simplex virus 2 VP22 phosphorylation induced by cellular and viral kinases does not influence intracellular localization. Virology 330:74-81. [DOI] [PubMed] [Google Scholar]
- 18.Geiss, B. J., J. E. Tavis, L. M. Metzger, D. A. Leib, and L. A. Morrison. 2001. Temporal regulation of herpes simplex virus type 2 VP22 expression and phosphorylation. J. Virol. 75:10721-10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui, E. K. 2002. Virion-associated protein kinases. Cell Mol. Life Sci. 59:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacque, J. M., A. Mann, H. Enslen, N. Sharova, B. Brichacek, R. J. Davis, and M. Stevenson. 1998. Modulation of HIV-1 infectivity by MAPK, a virion-associated kinase. EMBO J. 17:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, G. V., and W. H. Stoothoff. 2004. Tau phosphorylation in neuronal cell function and dysfunction. J. Cell Sci. 117:5721-5729. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi, Y., and K. Kato. 2003. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev. Med. Virol. 13:331-340. [DOI] [PubMed] [Google Scholar]
- 25.Knopf, K. W., and H. C. Kaerner. 1980. Virus-specific basic phosphoproteins associated with herpes simplex virus type a (HSV-1) particles and the chromatin of HSV-1-infected cells. J. Gen. Virol. 46:405-414. [DOI] [PubMed] [Google Scholar]
- 26.Leader, D. P. 1993. Viral protein kinases and protein phosphatases. Pharmacol. Ther. 59:343-389. [DOI] [PubMed] [Google Scholar]
- 27.Leader, D. P., and M. Katan. 1988. Viral aspects of protein phosphorylation. J. Gen. Virol. 69:1441-1464. [DOI] [PubMed] [Google Scholar]
- 28.Lemaster, S., and B. Roizman. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J. Virol. 35:798-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenard, J. 1999. Host cell protein kinases in nonsegmented negative-strand virus (mononegavirales) infection. Pharmacol. Ther. 83:39-48. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76: 1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel, D., and T. Mertens. 2004. The UL97 protein kinase of human cytomegalovirus and homologues in other herpesviruses: impact on virus and host. Biochim. Biophys. Acta 1697:169-180. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, E. E., Y.-F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Tyrosine phosphorylation of bovine herpesvirus 1 tegument protein VP22 correlates with the incorporation of VP22 into virions. J. Virol. 75:9010-9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 36.Zhang, G., R. Stevens, and D. P. Leader. 1990. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J. Gen. Virol. 71:1757-1765. [DOI] [PubMed] [Google Scholar]