Abstract
Most human herpesviruses, including Epstein-Barr virus (EBV), express a protein which functions primarily as an mRNA export factor. Previously, we deleted the gene for the Epstein-Barr virus mRNA export factor EB2 from the EBV genome and then introduced the mutated genome into 293 cells. Using a transcomplementation assay in which ectopic expression of the transcription factor EB1/ZEBRA was sufficient to induce the EBV productive cycle, we showed that Ori-Lyt-dependent replication of the EBV DNA occurs in the absence of EB2, indicating that EB2 is not essential for the expression and export of early mRNAs. However, in the absence of EB2, no infectious viral particles are produced (H. Gruffat, J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant, J. Virol. 76:9635-9644, 2002). In this report, we now show that EB2 is essential for the nuclear export of most, but not all, late mRNAs produced from intronless genes that translate into proteins involved in intranuclear capsid assembly and maturation. As a consequence, we show that EB2 is essential for the proper assembly of intranuclear capsids. Interestingly, the late BLLF1 gene contains an intron, and both unspliced and spliced mRNAs must be exported to the cytoplasm to be translated into gp350 and gp220, respectively (M. Hummel, D. A. Thorley-Lawson, and E. Kieff, J. Virol. 49:413-417, 1984). Our results also demonstrate that although BLLF1 spliced mRNAs are exported to the cytoplasm independently of EB2, EB2 is essential for the nuclear export of unspliced BLLF1 mRNA. In the same assay, herpes simplex virus 1 ICP27 completely inhibited the nuclear export of BLLF1 spliced mRNAs whereas unspliced BLLF1 mRNAs were exported, confirming that in a physiological assay, ICP27 inhibits splicing.
The Epstein-Barr virus (EBV) is a human gammaherpesvirus that infects over 90% of the adult human population. After the primary infection, EBV persists lifelong in resting recirculating memory B lymphocytes, which express either one viral gene or none (2). Persistence is strongly linked to certain EBV-associated malignancies, i.e., Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, gastric cancer, posttransplant lymphoproliferative disorders, AIDS-associated lymphomas, and leiomyosarcomas (reviewed in reference 42). The association of EBV with cancers is strengthened by the fact that in vitro, EBV drives resting B lymphocytes to proliferate as activated lymphoblasts. In such proliferating blasts, the EBV transcriptome is restricted to few genes defining a type III latency, with no expression of the genes required for a complete productive cycle (reviewed in reference 34). However, in vivo, the EBV productive cycle occurs, since infectious viruses are found in the saliva of healthy carriers (13). Recently, in vivo reactivation has been linked to the terminal differentiation of infected B cells into plasma cells, but the signals and the molecular mechanisms that trigger the switch from latency to the productive cycle are as yet unknown (29). The productive cycle can be induced, however, in blasts latently infected in vitro, by a variety of treatments such as TPA (46), transforming growth factor β (6), and anti-surface immunoglobulin (41), without inducing terminal differentiation of EBV-infected B cells into plasma cells. These in vitro treatments induce the transcription of the two immediate-early genes, BZLF1 and BRLF1 (reviewed in reference 1), whose products are transcription factors called, respectively, EB1 (7) or ZEBRA (14) and R (17). The EB1/ZEBRA viral factor activates the transcription of all the early genes, including BRLF1 (7), some of them being essential to viral DNA replication, which is a prerequisite for the expression of most late genes, DNA encapsidation, and infectious virion production (reviewed in reference 26).
Not all the early gene products are involved in DNA replication. The BSLF2/BMLF1 early gene encodes a posttranscriptional regulatory protein originally called EB2 (7) but later renamed Mta (12) or SM (8). The BSLF2/BMLF1 open reading frame (ORF) is conserved in other human herpesviruses (HHVs), suggesting a conserved function: the herpes simplex virus 1 (HSV-1) ICP27 protein (37), the human cytomegalovirus (CMV) UL69 protein (30), and the Kaposi's sarcoma-associated virus (KSHV [HHV-8]) ORF57 protein (31).
HSV-1 ICP27, EBV EB2, and HHV-8 ORF57 are nuclear proteins that have properties of mRNA nuclear export factors. They shuttle between the nucleus and the cytoplasm in a CRM1-independent manner (11, 28, 30). They bind to RNA in vitro and in vivo (18, 32), interact with essential components of the nuclear export pathway RNA export factor (REF)/Aly (scYra1) and transport activating protein (TAP)/p15 (scMex67p/Mtr2p) (19, 28, 31), and induce cytoplasmic accumulation of some viral and cellular mRNAs (4, 27, 36). In line with the above-described results, deletion of the major part of the BSLF2/BMLF1 gene in the EBV genome abolished the production of infectious viral particles (15), demonstrating that EB2 is an essential viral factor whose function cannot be transcomplemented by cellular factors. Moreover, EB2 did not efficiently complement ICP27 when inserted into an ICP27-null mutant (3), and neither ICP27 nor UL69 transcomplemented EB2 in inducing the production of infectious virions from the EB2-null mutant (15), strongly suggesting a specific viral function for these factors.
Using our EB2 deletion virus, we show here that EB2 is required for the nuclear export of most late mRNAs translated into proteins required for the maturation and assembly of viral capsids. Accordingly, we show that, in the absence of EB2, the newly replicated viral DNA is not protected against DNase I digestion in the nuclei, strongly suggesting that it is not properly encapsidated. However, not all late mRNAs are exported by EB2. Exceptions include the BALF4 and the BBRF1 mRNAs. In addition, we show that cytoplasmic accumulation of the unspliced form of BLLF1 mRNA, which encodes the gp350 protein, is strongly dependent on EB2. However, the spliced version of this mRNA, which encodes for gp220 protein, accumulates in the cytoplasm, even in the absence of EB2. In our transcomplementation assay, ICP27 also exported some unspliced BLLF1 mRNA, but as expected, it abolished the appearance of spliced BLLF1 mRNA in the cytoplasm.
MATERIALS AND METHODS
Viral DNA analysis.
293BMLF1-ko cells were transfected with an EB1 expression vector to activate the EBV productive cycle and transcomplemented or not transcomplemented with an EB2 expression vector. At 48 h after transfection, the medium was removed, and the cells were resuspended in Tris-buffered saline (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 5.5 mM glucose, 25 mM Tris-HCl, pH 7.4) and divided into two equal samples, which were then used to prepare total cellular and DNase-resistant (encapsidated) DNAs. The cells from both samples were pelleted and resuspended in 184 μl of reticulocyte standard buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2) containing 0.5% NP-40. An equal volume of 2× CLB (20 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1.2% sodium dodecyl sulfate, 1 mg/ml of proteinase K) was added either immediately (total cellular DNA) or after incubation in the presence of 200 μg of DNase I/ml, with occasional mixing, for 20 min at 37°C (encapsidated DNA). After the addition of proteinase K, all samples were incubated for 1 h at 37°C, extracted sequentially with phenol and chloroform, and precipitated with ethanol, and then the nucleic acids were redissolved in 10 mM Tris-HCl (pH 7.5)-1 mM EDTA containing 5 μg of RNase A/ml and 50 U of RNase T1/ml. Gel analysis of the DNAs was performed as follows. Samples of DNA corresponding to the yield from 4.105 cells were cleaved with BamHI enzyme, and the resulting fragments were separated by agarose gel electrophoresis, transferred to a Hybond-N membrane (Amersham), and detected by hybridization to 32P-labeled BRRF1 probes. As a control for the amount of DNA loaded on the gel, a 32P-labeled β-globin probe was also used. Southern blots were analyzed using a PhosphorImager.
Cell lines and transfections.
The 293BMLF1-ko cell line has been described extensively previously (15). Cells were grown in RPMI 1640 medium supplemented with penicillin, streptomycin, and 10% fetal calf serum (Invitrogen). A total of 5.106 cells were transfected by electroporation (Bio-Rad GenePulser) (210 V, 950 μF) with 10 μg of each vector. HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with penicillin, streptomycin, and 5% fetal calf serum (Invitrogen). Transfections were performed using calcium phosphate with 15 μg of total DNA (1 μg of each vector and pUC18 to 15 μg), as described previously.
Expression vectors.
pCMV-BZLF1 contains the BZLF1 cDNA coding for the EB1 protein cloned under the control of the CMV promoter in the pRc-CMVΔneo vector (39). pCi-FlagEB2 contains the BSLF2-BMLF1 cDNA coding for the EB2 protein cloned under the control of the CMV promoter in the pCi vector (Promega). The Flag EB2 protein expressed from this vector has been tagged at its N terminus with the Flag epitope which can be detected by the M2 monoclonal antibody (Sigma reference F3165) (19). The ICP27- and UL69-expressing vectors were kindly provided by, respectively, A. Epstein and T. Stamminger (30). pCi-Flag-BdRF1+i and pCi-Flag-BFRF3+i contain, respectively, the BdRF1 and the BFRF3 ORFs obtained by reverse transcription-PCR (RT-PCR) with primers that are described in Table 1. In pCi-Flag-BdRF1-i or pCi-Flag-BFRF3-i, the original pCi intron has been deleted by digestion with Afl II. Plasmids used for transfection were prepared by the alkaline lysis method and purified through two CsCl-ethidium bromide gradients.
TABLE 1.
List of primers used
| Gene and primer | Sequence (5′ → 3′) | B95-8 relative position |
|---|---|---|
| U6 snRNA | ||
| Forward | CGCTTCGGCAGCACATATAC | |
| Reverse | AAAATATGGAACGCTTCACGA | |
| β-Actin | ||
| Forward | GCTGCGTGTGGCTCCCGAGGAG | |
| Reverse | ATCTTCATTGTGCTGGGTGCCAG | |
| BALF4 | ||
| Forward | GGAGTCGTAGGCAAATTGGA | 157922-157941 |
| Reverse | TCAAGAACCTGACGGAGCTT | 158125-158145 |
| BBRF1 | ||
| Forward | AAGATCTGCCAGCTCCTGAA | 115017-115036 |
| Reverse | ATGAACTCCTCCACCACGTC | 115197-115216 |
| BcLF1 | ||
| Forward | GACAATTATCAAAAAACCACC | 133313-133332 |
| Reverse | GGTGCAGTTTGTAGTG | 133528-133543 |
| BDLF1 | ||
| Forward | CAGATTTGAAAGTGGTAGTGTC | 133283-133301 |
| Reverse | TTATCTTAACCAGCAAGTGGCCG | 132400-132422 |
| BdRF1/BVRF2 | ||
| Forward | CACTATCAGGTAACGCAGGAG | 148710-148728 |
| Reverse | TCAAGCCAGCGTTTATTCAGC | 149723-149744 |
| BFRF3 | ||
| Forward | GGGAGGCTCAAAGAAGTTACCTG | 61628-61650 |
| Reverse | ATGAAGAAACAGAGGGGGTCGC | 61882-61903 |
| BFRF3 (for cloning) | ||
| Forward | GCCCAGATCTCGAGCAGCACGCCGGCTGCCC | 61510-61524 |
| Reverse | GCCCTCTAGACTACTGTTTCTTACGTGCCCCGCG | 62014-62037 |
| BLLF1 | ||
| Forwarda | TGTGCTGATAGAGGCTGGTG | 90018-90037 |
| Reverseb | TGACACCAAGTCCATCTCCA | 90672-90691 |
| BMRF1 | ||
| Forward | CCAGACATACGGTCAGTCCATCTC | 80881-80904 |
| Reverse | TGCTTCACTTTCTTGGGGTGC | 81071-81091 |
| BORF1 | ||
| Forward | GCCCCTCGAGCAAGGTCCAGGGGTCCGTCG | 75242-75259 |
| Reverse | GCCCTCTAGAGAATCACCTCCCAGTCAGAG | 76308-76333 |
| BVRF1 | ||
| Forward | GGGCAACCTGAACTTTACCA | 145991-146010 |
| Reverse | CGGCGATGATTTTCTCTAGG | 146219-146238 |
Primer 299.
Primer 300.
Western blotting analysis.
Cells were collected by centrifugation, lysed on ice for 30 min in 100 μl of HNTG buffer (50 mM HEPES [pH 7.5]; 150 mM NaCl; 1% Triton; 10% glycerol; 1 mM EDTA; 1 mM phenylmethylsulfonyl fluoride). Proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide electrophoresis gels and then transferred to a nitrocellulose membrane by electroblotting (Hybond-ECL; Amersham Biosciences). The membranes were incubated with either the anti-protease monoclonal antibody (8E9E11) (a gift from P. Morand) or the rat monoclonal antibody anti-BFRF3 (OT15E) (44). Membranes were also incubated with an anti-EB2-specific polyclonal antibody (dilution, 1:500) (15). Goat anti-mouse or goat anti-rabbit horseradish peroxidase conjugate (Amersham) was used at a dilution of 1:5,000 as a secondary antibody. For immunoblot detection, the ECL system (Amersham) was used.
Immunofluorescence assay.
Cells were seeded on polylysine pretreated glass coverslips and then transfected with the appropriate plasmid. At 48 h after transfection, cells were washed twice with phosphate-buffered saline and fixed with 4% paraformaldehyde, and then indirect immunofluorescence was performed as described before (11). The glass coverslips were incubated with either the anti-BdRF1 monoclonal antibody (OT41A) (45) or the anti-BcLF1 monoclonal antibody (MAB8185 Chemicon) or the anti-gB monoclonal antibody (MAB8184 Chemicon).
RNA extraction and RT-PCR analysis.
Transfected cells were washed twice with phosphate-buffered saline, and cytoplasmic RNAs were extracted as previously described (16). Briefly, cells were resuspended in 475 μl of lysis buffer (10 mM Tris-HCl [pH 7.8], 10 mM NaCl, 2 mM MgCl2, 5 mM dithiothreitol), and after 5 min on ice, 25 μl of 10% NP40 was added. After centrifugation to collect the nuclei (2 min, 1,200 rpm, 4°C), cytoplasmic RNAs contained in the supernatant were purified by phenol chloroform extraction and ethanol precipitation. Nuclear RNAs were purified from the isolated nuclei with TRIzol reagent (Invitrogen). Then, a DNase treatment master mix containing AMV/RT reaction buffer (Biolabs), MgSO4 (2.5 mM final concentration), and 1 U DNase/RNase free (Roche Diagnostics) was prepared to digest any DNA contamination from RNA preparations for 1 h at 37°C. This was followed by 15 min of DNase inactivation at 65°C. Reverse transcription were performed with 2 μg of purified nuclear or cytoplasmic RNA by use of Stratascript reverse transcriptase (Stratagene) and 0.5 μg of oligo(dT18) (Roche Diagnostics) for 1 h at 42°C. PCRs were performed using a Taq core kit (Q-Biogen) in the presence of [α32-P]dCTP (0.1 μCi), with a set of specific primer pairs (see Table 1), on various amounts of the RT reaction mixtures (0.2, 0.4, 1 or 2 μl) to have a linearly increasing signal after 25 PCR cycles. The PCR-amplified fragments were then analyzed on 2% agarose gels and autoradiographed. Each of the PCR-amplified bands was subsequently quantified with a Storm PhosphorImager. To control our RT-PCR experiments, we evaluated the endogenous expression of β-actin mRNA by RT-PCR (see Table 1). Amplification of a 690-bp DNA fragment corresponding to β-actin mRNA showed that no DNA contamination was present in our RNA preparations. We have also used RT-PCR to look for the presence of the U6 snRNA in the nuclear and cytoplasmic extracts to control for the purity of our cytoplasmic RNAs.
RESULTS
EB2 is encoded by an extension of the BSLF2 ORF and not by the BMLF1 ORF.
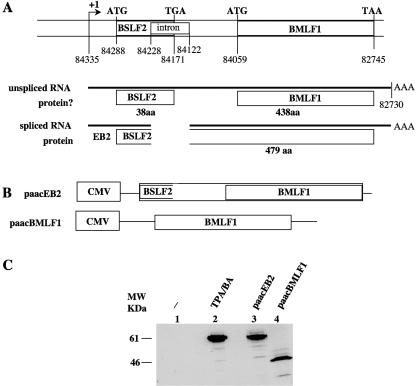
It has been reported that the EBV BSLF2 and BMLF1 ORFs are joined to encode a protein originally called EB2 (7) and later renamed Mta (12) or SM (8) (Fig. 1A). cDNA cloning allowed us to characterize two cytoplasmic mRNAs covering the BSLF2/BMLF1 region (5), one unspliced and one spliced. Sequencing of the cDNAs demonstrated that in the spliced mRNA, the BSLF2 ORF termination codon is deleted, and that the BSLF2 ORF starting at position 84228 is prolonged until the next termination codon, at position 82745 (B95-8 sequence), encoding a putative polypeptide of 479 amino acids (aa) (Fig. 1A). Since no internal ribosomal entry site has been described upstream of the BMLF1 ORF, only the BSLF2 ORF should be used in unspliced mRNAs, thus encoding a putative polypeptide of 38 aa. In addition, in the unspliced mRNA, the BSLF2 ORF and the BMLF1 ORF are not in frame. The polypeptide encoded by the BMLF1 ORF should be 438 aa long. To confirm that the extended BSLF2 ORF from the spliced mRNAs encodes EB2, EB2 expression was induced in Raji cells by treatment with TPA/butyric acid (BA) or by transfecting either a cDNA corresponding to the spliced RNA (paacEB2; Fig. 1B) or a truncated cDNA corresponding to a putative mRNA containing only the BMLF1 ORF (paacBMLF1; Fig. 1B). As shown in Fig. 1C, a polyclonal antibody raised against the EB2 protein detected a polypeptide of about 60 kDa in TPA/BA-induced Raji cells (Fig. 1C, lane 2). A polypeptide with the same apparent molecular weight was detected in Raji cells transfected with paacEB2 (Fig. 1C, lane 3), while a polypeptide of about 46 kDa was detected in Raji cells transfected with paacBMLF1 (Fig. 1C, lane 4). This clearly demonstrated that EB2 is the product of the extended BSLF2 ORF and that the BMLF1 ORF exists only theoretically.
FIG. 1.
EB2 is the product of the BSLF2/BMLF1 gene. (A) Schematic structure of the BSLF2 and BMLF1 open reading frames on the EBV genome. The relative positions of the initiation codons (ATG), stop codons, the borders of the intron, and the putative ORFs are given according to the sequence of the B95-8 viral strain. (B) Schematic representation of the expression vectors used. The cDNA sequence from the BSLF2/BMLF1 gene was inserted in the paac vector to generate the paacEB2 vector. paacBMLF1 contains the BMLF1 ORF. CMV, cytomegalovirus promoter. (C) Western blot analysis of Raji cells transfected with the expression vector paacEB2 (lane 3) or paacBMLF1 (lane 4) or induced into the productive cycle by treatment with TPA/BA (lane 2). A rabbit polyclonal serum directed against the EB2 protein was used.
EB2 is essential for the proper encapsidation of the EBV genome.
We have previously reported the generation of an EBV mutant in which the BSLF2/BMLF1 gene encoding the EB2 protein has been partly deleted. The mutated viral genome was introduced into 293 cells, and then a clonal population called 293BMLF1-ko was selected (15). We reported that such cells produced infectious EBV particles only when transfected with both an EB1 and an EB2 expression vector (15). In this transcomplementation assay, EBV DNA replication occurs in the absence of EB2, but there is no production of infectious viral particles, suggesting that the newly replicated intranuclear EBV DNA is either poorly encapsidated or improperly encapsidated in the absence of EB2. Herpesvirus encapsidation proceeds by a complex mechanism in which cleavage of viral DNA concatemers into monomeric units is tightly coupled with their insertion into the procapsid (for a review, see reference 20). The intranuclear encapsidated viral DNA is protected against the action of exogenously added DNase I. This property was used to test whether the replicated intranuclear viral DNA was encapsidated in the absence of EB2. 293BMLF1-ko cells were mock transfected, transfected with an EB1 expression vector, or transfected with both an EB1 and an EB2 expression vector. Three days after transfection, nuclei were prepared, and the DNA was extracted, digested with BamHI, and analyzed by Southern blotting using an EBV BRRF1 probe. As shown in Fig. 2, the intranuclear resident viral DNA (lane 1) was amplified when the cells were transfected with an EB1 expression plasmid (lane 2); as already shown, EB2 enhanced the replication efficiency (lane 3). As a control for the amount of DNA loaded on the gel, the Southern blot was incubated with a β-globin probe. The result showed that, indeed, in each lane a comparable amount of total DNA was used. We next asked whether the replicated DNA was encapsidated. When intact nuclei were treated with DNase I, the resident intranuclear viral DNA was completely digested (Fig. 2, lane 4), as well as the viral DNA whose replication was induced by EB1 (Fig. 2, lane 5). However, in the presence of EB1 and EB2, conditions required for the production of infectious EBV particles by 293BMLF1-ko cells (15), part of the intranuclear replicated viral DNA was protected against digestion by DNase I (Fig. 2, lane 6). These results strongly suggest that EB2 is essential for the proper encapsidation of replicated intranuclear viral DNA, probably through its action on the export of some late mRNAs.
FIG. 2.
Packaging of the EBV DNA in 293BMLF1-ko cells. 293BMLF1-ko cells were induced into the productive cycle by transfection with an EB1 expression vector and transcomplemented or not transcomplemented with an EB2 expression vector as indicated above the lanes. Total (/) and DNase-resistant (DNase I) DNAs were prepared 48 h after transfection. Samples were cleaved with BamHI, and the fragments were resolved by electrophoresis through a 0.8% agarose gel, transferred to a nylon membrane, and hybridized to 32P-labeled BRRF1 DNA. The numbers below the lanes containing total DNA indicate the ratios of newly replicated viral DNA quantified after PhosphorImager exposure of the membrane. As a control for the amount of DNA loaded on the gel, the Southern blot was also incubated with a β-globin probe.
EB2 is essential for the production of viral capsid proteins.
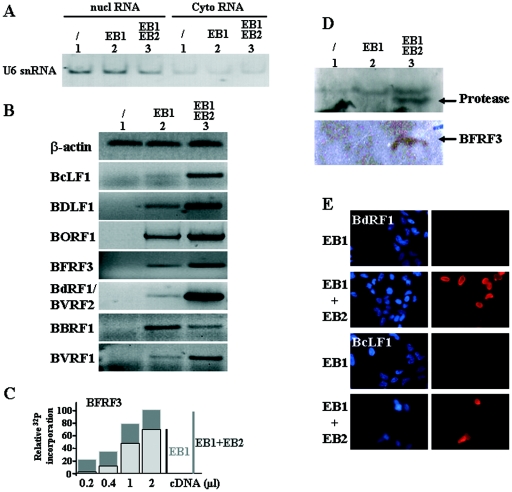
We reasoned that EB2 could be essential for the nuclear export of viral mRNAs translated into proteins required for the assembly and maturation of virions. Therefore, we looked at the EBV genes encoding capsid components or capsid assembly factors. EBV protein composition has not been systematically studied, as in the case of HSV-1, except for the glycoproteins. EBV protein annotations have usually been based on DNA sequence homology to a characterized herpesvirus ORF, with verification for EBV in specific instances. Accordingly, EBV BcLF1, BDLF1, BORF1, BFRF3, BVRF2, BdRF1, BBRF1, and BVRF1 ORFs are likely to encode, respectively, the major capsid protein (MCP), minor capsid protein (mCP), the mCP-binding protein (mCP-BP), the smallest capsid protein (sCP), the protease (Pr), the protease-assemblin (Pr/AP), the portal protein, and the capsid-associated cork (Table 2) (10, 25). On the basis of this nomenclature we have used RT-PCR to look for the expression of the corresponding mRNAs in 293BMLF1-ko cells that were mock transfected, transfected with an EB1 expression vector, or transfected with both an EB1 and an EB2 expression vector. Cytoplasmic mRNAs were extracted from the transfected cells, reverse transcribed, and quantified by PCR using different primers specific for the genes studied (see Table 1). We also analyzed by RT-PCR the presence of U6 snRNA, which cannot be exported, to control the purity of our cytoplasmic RNAs. As shown in Fig. 3A, although U6 snRNA was mainly detected in the nuclear fraction, our cytoplasmic RNAs contained some U6 snRNA, likely due to nuclear leakage during nuclear-cytoplasmic fractionation. β-Actin RNA was used to confirm that the same amount of RNA was used in each RT-PCR experiment and that there was no DNA contaminating the RNA preparation (Fig. 3B). In mock-transfected 293BMLF1-ko cells, the proteins necessary for viral capsid formation are not expressed; accordingly, their corresponding mRNAs were not detectable by RT-PCR (Fig. 3B, lanes 1). In 293BMLF1-ko cells transfected with an EB1 expression vector, the RNAs synthesized from the capsid genes were detected by RT-PCR to various degrees ranging from not detected (BcLF1) to weakly detected (BDLF1, BFRF3, BdRF1/BVRF2, and BVRF1) and clearly detected (BORF1 and BBRF1) (Fig. 3B, lanes 2). BcLF1 expression was not detected even when more RT product was used in the PCR (data not shown). As expected, in 293BMLF1-ko cells expressing both EB1 and EB2, there was an increase in the cytoplasmic accumulation of all the mRNAs encoding the capsid proteins except for the BBRF1 mRNA (Fig. 3B, lanes 3). Each PCR was done with various amounts of RT products, and linearly increasing signals were observed. Such semiquantitative analysis is presented for the BFRF3 mRNA (Fig. 3C) and for the BALF4 mRNA (Fig. 4). The PCR results presented in Fig. 3B are in the linear range, since they correspond to 1 μl of cDNA and 25 cycles of PCR. The above-described results were also confirmed by Western blotting or indirect immunofluorescence at the protein level (Fig. 3D and 3E). Hence, the viral protease (product of the BVRF2 gene) and the smallest capsid protein (BFRF3 protein) were detected by Western blotting with specific antibody only when EB2 was expressed in 293BMLF1-ko cells (Fig. 3D). Similarly, the BdRF1 protein (assemblin) and the BcLF1 protein (MCP) were detected by immunofluorescence in 293BMLF1-ko cells only when the EB2 protein was expressed (Fig. 3E). Taken together, the above-described results demonstrate that EB2 is indirectly—probably through its mRNA export function—necessary for EBV capsid formation.
TABLE 2.
EBV genes and their encoded proteins involved in the capsid structure
| EBV gene | HSV-1 homologue | Protein description | MWa |
|---|---|---|---|
| BcLF1 | UL19 (VP5) | Major capsid protein (MCP) | 155 |
| BDLF1 | UL18 (VP23) | Minor capsid protein (mCP) | 30 |
| BORF1 | UL38 (VP19C) | Minor capsid protein-BP (mCP-BP) | 40 |
| BFRF3 | UL35 (VP26) | Smallest capsid protein (sCP) | 18 |
| BVRF2 | UL26 (VP24) | Protease (Pr) | 26 |
| BdRF1 | UL26.5 (VP22a) | Protease-assemblin (Pr/AP) | 80 |
| BBRF1 | UL6 | Portal | 68 |
| BVRF1 | UL25 | Capsid-associated “Cork” | 58 |
MW, molecular weight.
FIG. 3.
The EB2 protein induces the cytoplasmic accumulation of specific mRNA encoding EBV capsid protein. (A) Control of the purity of our cytoplasmic (Cyto) RNA fraction: RNA from 293BMLF1-ko cells transiently transfected with a control vector or expression plasmids encoding proteins as indicated at the top of the figure were submitted to RT-PCR analysis using specific primers to detect the U6 snRNA. nucl, nuclear. (B) RNA from 293BMLF1-ko cells transiently transfected as indicated at the top of the figure were submitted to RT-PCR analysis using specific primers to detect the β-actin cellular mRNA or EBV late mRNA encoding BcLF1, BDLF1, BORF1, BFRF3, BdRF1, BBRF1, and BVRF1. The PCR products were loaded on a 2% agarose gel and visualized by ethidium bromide staining. (C) Example from the results of our semiquantitative experiments done using the BFRF3 mRNA. Each PCR (25 cycles) was done with various amounts of RT product and in the presence of [α-32P]dCTP. The gel was quantified after PhosphorImager exposure. (D) Western blot analysis of the 293BMLF1-ko cells transiently transfected with a control vector or expression plasmids encoding proteins as indicated at the top of the figure. The membranes were incubated with either an anti-protease or an anti-BFRF3 monoclonal antibody. (E) Expression of the BdRF1 and BcLF1 proteins was monitored by immunofluorescence. The 293BMLF1-ko cells were transiently transfected with expression plasmids encoding proteins as indicated on the left of the figure. After the cells were labeled with the appropriate antibody, they were stained with bisBENZIMIDE to label their nuclei and observed on a UV fluorescence microscope. The nuclei stained blue; the BdRF1 and BcLF1 proteins stained red.
FIG. 4.
Cytoplasmic accumulation of late BALF4 mRNA is EB2 independent. (A) RNA from 293BMLF1-ko cells transiently transfected with a control vector or expression plasmids encoding proteins as indicated at the top of the figure were submitted to RT-PCR analysis using specific primers to detect the BALF4 mRNA. The PCR (25 cycles) was done with various amounts of RT product and in the presence of [α-32P]dCTP. The PCR products were loaded on a 6% polyacrylamide gel and visualized by autoradiography. (B) Expression of the BALF4 protein (gp110) was monitored by immunofluorescence. The 293BMLF1-ko cells were transiently transfected with expression plasmids encoding proteins as indicated on the left of the figure. After the cells were labeled with the anti-BALF4 antibody, they were observed on a UV fluorescence microscope. The BALF4 protein stained red.
Not all late mRNAs are EB2 targets.
As shown above, in 293BMLF1-ko cells, EB2 increases the cytoplasmic accumulation of all EBV late mRNAs encoding the proteins required for assembly and processing of the viral capsid except BBRF1 mRNA. We therefore asked whether or not EB2 increased the cytoplasmic accumulation of another late mRNA essential for the formation of infectious virions. To answer this question, we analyzed the cytoplasmic accumulation of the mRNA encoding the viral glycoprotein gB (or gp110), the product of the BALF4 gene. 293BMLF1-ko cells were transfected as described above. Three days after transfection, cytoplasmic mRNAs were isolated and reverse transcribed and increasing amounts of the RT products were analyzed by PCR. As shown in Fig. 4A, the PCR signals increased linearly with the amount of RT product used. As expected, the BALF4 gene was not expressed in mock-transfected cells (lanes 1, 4, 7, and 10), and EB1 induced the expression of the BALF4 mRNA (lanes 2, 5, 8, and 11). However, EB2 had no effect on the amount of BALF4 mRNA exported to the cytoplasm (lanes 3, 6, 9, and 12). This was also seen at the level of gp110 expression in cells (Fig. 4B). The above-described results confirm that EB2 is not required for the export of all late mRNAs.
EBV EB2 and CMV UL69 export unspliced BLLF1 mRNA.
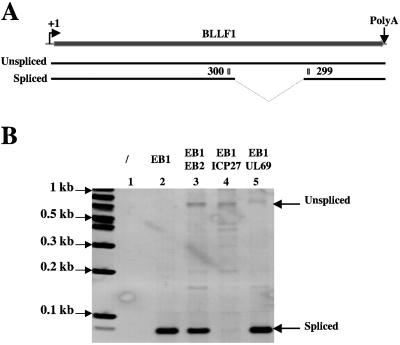
Most of the EBV and HSV1 late genes are intronless. It has been suggested that, among other functions, EBV EB2 and HSV1 ICP27 have a unique function in the viral productive cycle that cannot be transcomplemented by cellular factors, i.e., to export mRNAs generated from intronless genes (11, 28, 36). In addition, EB2 exports unspliced mRNAs of different origins (4, 11), and ICP27 inhibits splicing (reference 38 and references therein). In this context, it was interesting to analyze the cytoplasmic accumulation of the BLLF1 mRNA because, as shown in Fig. 5A, the EBV BLLF1 late gene contains an intron. Both spliced and unspliced mRNAs produced by facultative splicing are exported to the cytoplasm and are translated in 100-kDa and 130-kDa proteins, precursors of the EBV glycoproteins gp220 and gp350, respectively (24). We therefore looked at the cytoplasmic accumulation of the spliced and unspliced BLLF1 late mRNAs to analyze the functions of EB2 and ICP27. As shown in Fig. 5B, BLLF1 mRNAs were not expressed in mock-transfected 293BMLF1-ko cells (Fig. 5B, lane 1). Interestingly, only the spliced BLLF1 mRNAs accumulated efficiently in the cytoplasm of 293BMLF1-ko cells transfected with an expression vector for EB1 (Fig. 5B, lane 2), whereas in 293BMLF1-ko cells transfected with vectors expressing EB1 and EB2, both spliced and unspliced BLLF1 mRNAs were exported to the cytoplasm (Fig. 5B, lane 3). It is noteworthy that EB2 had no effect on the cytoplasmic accumulation of spliced BLLF1 mRNAs (Fig. 5B, lane 3), whereas, and as expected, coexpression of EB1 and ICP27 was followed by a near-complete absence of cytoplasmic spliced BLLF1 mRNAs (Fig. 5B, lane 4). However, some unspliced BLLF1 mRNAs accumulated in the cytoplasm of 293BMLF1-ko cells transfected with expression vectors for EB1 and ICP27 (Fig. 5B, lane 4). Since the human cytomegalovirus UL69 appears to be an mRNA export factor (15, 30), we also looked at its effect on the export of BLLF1 mRNAs. As shown in Fig. 5B, lane 5, UL69 had no effect on the cytoplasmic accumulation of spliced BLLF1 mRNAs but induced the nuclear export of unspliced BLLF1 mRNAs. Several low-abundance forms of unspliced BLLF1 migrating with a lower molecular weight were observed on the gel; their presence was probably due to the fact that the amplified fragment is constituted by several repeats. Taken together, these results suggest that UL69 has functions similar to that of EB2 and ICP27, i.e., facilitating the export of unspliced mRNAs, but these three proteins probably act by a different mechanism. For example, ICP27, but not EB2 and UL69, inhibits splicing.
FIG. 5.
Expression of the late BLLF1 gene. (A) Schematic representation of the BLLF1 gene. +1, mRNA initiation site; PolyA, polyadenylation signal. From the BLLF1 gene, by alternative splicing, two mRNAs are synthesized encoding, respectively, the gp350 and gp220 proteins. 299 and 300 are the primers used for the PCR. (B) RNA from 293BMLF1-ko cells transiently transfected with a control vector or expression plasmids encoding proteins as indicated at the top of the panel were submitted to RT-PCR analysis using specific primers (299 and 300) to detect the two BLLF1 mRNAs. The PCR products were loaded on a 2% agarose gel and visualized by ethidium bromide staining.
The EB2-dependent nuclear export of intronless EBV mRNAs can be reproduced in a transient expression assay, with nonreplicating plasmids.
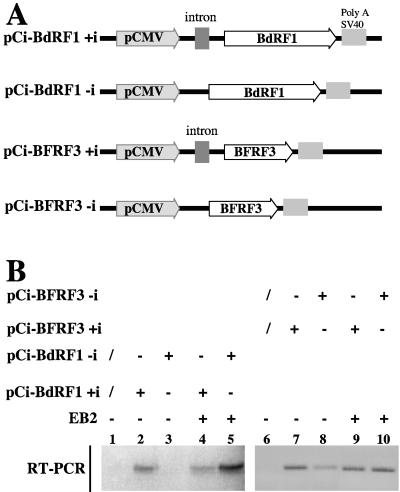
As shown above, EB2 increases the export of some EBV mRNAs generated from late intronless genes but has no effect on the export of mRNAs generated from genes containing a functional intron. To rule out any putative effect of EBV DNA replication and as a first attempt to determine what makes an mRNA an EB2 target, we cloned the BFRF3 and the BdRF1 ORFs under the control of the CMV promoter in nonreplicative plasmids. Two types of constructions were made. In the first type, BFRF3 and BdRF1 were expressed from precursor mRNAs containing an artificial intron localized in the 5′ untranslated region (BFRF3+i and BdRF1+i, respectively). In the second type, BFRF3 or BdRF1 was expressed from intronless precursor mRNAs (Fig. 6A). In all these constructions, the transcripts are initiated at the CMV promoter and contained only the viral ORF; viral RNA sequences corresponding to 5′ methylated cap the 5′ untranslated region, the 3′ untranslated region, and the cleavage-polyadenylation signal were excluded. As shown in Fig. 6B, only small amounts of BFRF3-i mRNA (lane 8) and no BdRF1-i mRNA (lane 3) accumulated in the cytoplasm of transiently transfected HeLa cells. However, in the presence of EB2, the amount of cytoplasmic BdRF1-i (lane 5) and BFRF3-i (lane 10) mRNAs strongly increased. As expected, BdRF1+i (lane 2) and BFRF3+i (lane 7) mRNAs were efficiently exported to the cytoplasm and EB2 did not significantly affect the nuclear export of these mRNAs (compare lanes 2 to 4 and lanes 7 to 9). These results confirm that EB2 has no effect on the nuclear export of mRNAs generated from intron-containing genes. In contrast, EB2 increases the export of mRNAs generated from late viral intronless genes, an effect independent of 5′ and 3′ noncoding viral RNA sequences. In addition, these results show that the effect of EB2 observed on the late mRNAs is direct and could not be attributable to the effect of EB2 on DNA replication, since this experiment was done using HeLa cells that were not EBV infected and by transfection of nonreplicative plasmids.
FIG. 6.
The EB2-dependent cytoplasmic accumulation of intronless EBV mRNAs can be reproduced in a transient expression assay with nonreplicating plasmids. (A) Schematic representation of the reporter plasmids used. The BFRF3 or BdRF1 ORF was cloned under the control of the CMV promoter into the pCI vector containing or not containing an artificial intron. (B) RNA from HeLa cells transiently transfected with a control vector or expression plasmids encoding proteins as indicated at the top of the panel were submitted to RT-PCR analysis using specific primers to detect the BFRF3 or the BdRF1 mRNA. The PCR products were loaded on a 2% agarose gel and visualized by ethidium bromide staining.
DISCUSSION
Using a clonal population of 293 cells carrying an EBV genome from which the EB2 gene has been deleted, we show that upon induction of the productive cycle by transfection of an EB1 expression vector, transcomplementation with the EB2 protein is essential for efficient nuclear export of a subpopulation of EBV late mRNAs generated from intronless genes that are translated into proteins involved in intranuclear capsid assembly and maturation (Fig. 3). The EB2-induced increase in the cytoplasmic accumulation of some late mRNAs is unlikely to be due to the EB2-induced increase in the number of intranuclear viral DNA copies (Fig. 2). Indeed, some late mRNAs are efficiently exported in the absence of EB2, and EB2 did not increase their cytoplasmic accumulation. Moreover, the spliced mRNAs made from the intron-containing BLLF1 late gene were also efficiently exported in the absence of EB2, and again EB2 had no effect on its export. Finally, the EB2-dependent export of some late mRNAs could be reproduced in a transient expression assay, using nonreplicating plasmids (Fig. 6), in line with the work of Serio et al. (40) showing that late gene expression from EBV BcLF1 and BFRF3 promoters does not require DNA replication in cis. Taken together, the above-described results demonstrate that EB2 is required for efficient nuclear export of a subpopulation of EBV late mRNAs and that this effect is not linked to the EB2-enhanced replication of the viral genome.
Importantly however, Ori-Lyt-replicated nuclear viral DNA was completely digested by DNase I in the absence of EB2 (Fig. 2), demonstrating that a correct balance of expression of proteins involved in the intranuclear capsid assembly and maturation is obtained only in the presence of EB2. The correct balance of these proteins cannot be obtained when HSV-1 ICP27 is used rather than EB2 in our transcomplementation assay (15), demonstrating that EB2 and ICP27 are not functionally equivalent in the production of EBV infectious virions. This is in clear contrast with experimental data showing that EB2 can complement the growth defect of an HSV-1 ICP27-null virus, which suggested that EB2 is functionally similar to ICP27 (3). This result is probably due to the fact that ICP27, in contrast to EB2, inhibits splicing.
The effect of EB2 on the nuclear export of the late mRNAs examined ranges from essential to facilitating (Fig. 3). Our results identify some viral transcripts that are dependent on EB2 for efficient cytoplasmic accumulation during the productive cycle. We have also looked at the nuclear accumulation of these transcripts in the absence and presence of EB2, and we observed that without EB2 there is no nuclear accumulation of the viral mRNAs (data not shown). This observation is in agreement with results obtained with HSV-1 (33), suggesting that unexported mRNAs are degraded.
Most of the EBV late genes are intronless, but EB2 does not export all the mRNAs generated from these intronless genes. If some viral late mRNAs depend on EB2 to be exported, it is evident that others such as BBRF1 (Fig. 3) and BALF4 (Fig. 4) are efficiently exported by cellular factors, likely SR proteins (for references, see reference 22) and/or REF (35). This is also true for the nuclear export of HSV-1 transcripts (33). We do not as yet know what makes an RNA a target for EB2-dependent nuclear export. EB2 does not seem to bind specifically to RNA in vitro (18) but specifically exports some but not all EBV mRNAs. Interestingly, EB2 coimmunoprecipitates, in vivo, with REF/Aly and TAP/NXF1 (19). As the coimmunoprecipitation is abolished by RNase treatment (19) this could suggest that EB2 is recruited to certain viral late mRNA nucleoproteins (mRNPs) by contacting both REF and the RNA and that the EB2-loaded viral mRNP is exported by RNA-bound TAP/NXF1. However, it could also be that EB2 is recruited to mRNAs by as-yet-unidentified nuclear factors but interacts with REF and/or TAP.
Indeed, EB2 also assists in the export of mRNAs carrying introns flanked by “ill-defined” splice sites. These include unspliced β-thalassemia (4), in which a mutation in the first exon 5′ splice site has caused the activation of three cryptic 5′ splice sites (43), pUC18 “cryptic” mRNAs (4), and the RNA export reporter gene pDM128, which is a human immunodeficiency virus-derived reporter gene carrying “suboptimal splice sites,” allowing export of unspliced RNAs (9). However, these studies have been made with artificial reporter genes, in transient expression assays. In this study, we confirm the above-described findings by demonstrating that when the productive cycle is induced in cells carrying the EBV genome with the EB2 gene deleted, the unspliced EBV late BLLF1 mRNA, which is translated into a 130-kDa protein, the precursor of the major gp350 glycoprotein, is exported to the cytoplasm only when EB2 is expressed by transcomplementation (Fig. 5). It is noteworthy that in the absence of EB2, the correctly spliced BLLF1 mRNAs are exported to the cytoplasm, and EB2 has no effect on the cytoplasmic accumulation of these spliced BLLF1 mRNAs, whereas ICP27 completely abolished the appearance of spliced BLLF1 mRNAs in the cytoplasm, probably by inhibiting their splicing (38 and references therein). However, EBV EB2, HSV-1 ICP27, and CMV UL69, even if they differ by their effect on BLLF1-spliced mRNAs, all export unspliced BLLF1 mRNAs (Fig. 5).
Therefore, it can be concluded that EB2 inhibits cryptic splicing or facultative splicing and enhances export of unspliced mRNAs. This resembles the mechanism used by splicing factors 9G8 and SRp20, which enhance the export of β-globin cDNA transcripts (intronless) made from a plasmid (21). The splicing factors 9G8 and SRp20 bind to the 22-nucleotide mouse histone H2a “constitutive” export element. In the absence of this element, almost all of the β-globin sequences are deleted by cryptic splicing, rendering the β-globin RNA transcript undetectable, although the spliced transcripts are efficiently exported (23). Cryptic splicing is inhibited by the export element containing globin construct, but intron-containing β-globin mRNAs are exported (21). It could be that a determinant for the nuclear export of mRNAs derived from viral intronless genes is the inhibition of cryptic splicing and/or recruitment of export factors like EB2 by cellular factors bound to 5′ cryptic splice sites. Confocal dual-label microscopy and immunoprecipitation of EB2-loaded mRNPs from infected cells in which the productive cycle has been induced might allow the identification of proteins required for EB2-mediated mRNA export.
Although our results explain why EB2 is essential for the production of EBV infectious virions, the molecular pathways for EB2-dependent and EB2-independent export of viral mRNAs remain to be elucidated.
Acknowledgments
We thank R. Buckland for reading the manuscript. We thank P. Morand for the anti-protease monoclonal antibody.
This work was financially supported by the Association pour la Recherche sur le Cancer (ARC 3420), the Ligue contre le Cancer, and the Institut National de la Santé et de la Recherche Medicale (INSERM). J.B. is a recipient of an MRT fellowship. A.S. and E.M. are CNRS scientists.
REFERENCES
- 1.Amon, W., and P. J. Farrell. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149-156. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, J. L., S. Swaminathan, and S. J. Silverstein. 2002. The Epstein-Barr virus SM protein is functionally similar to ICP27 from herpes simplex virus in viral infections. J. Virol. 76:9420-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buisson, M., F. Hans, I. Kusters, N. Duran, and A. Sergeant. 1999. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J. Virol. 73:4090-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buisson, M., E. Manet, M. C. Biemont, H. Gruffat, B. Durand, and A. Sergeant. 1989. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J. Virol. 63:5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chasserot-Golaz, S., C. Schuster, J. B. Dietrich, G. Beck, and D. A. Lawrence. 1988. Antagonistic action of RU38486 on the activity of transforming growth factor β in fibroblasts and lymphoma cells. J. Steroid Biochem. 30:381-385. [DOI] [PubMed] [Google Scholar]
- 7.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, I. D., F. Shanahan, and P. J. Farrell. 1994. Epstein Barr virus SM protein. Virology 205:217-227. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 2004. Assaying nuclear messenger RNA export in human cells. Methods Mol. Biol. 257:85-92. [DOI] [PubMed] [Google Scholar]
- 10.de Jesus, O., P. R. Smith, L. C. Spender, C. Elgueta Karstegl, H. H. Niller, and P. J. Farrell. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 84:1443-1450. [DOI] [PubMed] [Google Scholar]
- 11.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1 independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. Trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66: 5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber, P., S. Lucas, M. Nonoyama, E. Perlin, and L. I. Goldstein. 1972. Oral excretion of Epstein-Barr viruses by healthy subjects and patients with infectious mononucleosis. Lancet 2:988-989. [DOI] [PubMed] [Google Scholar]
- 14.Grogan, E., H. Jenson, J. Countryman, L. Heston, L. Gradoville, and G. Miller. 1987. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc. Natl. Acad. Sci. USA 84:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruffat, H., E. Manet, A. Rigolet, and A. Sergeant. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwick, J. M., P. M. Lieberman, and D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278:37790-37798. [DOI] [PubMed] [Google Scholar]
- 19.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. P. Sergeant, and E. Manet. 2003. A novel NES and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 20.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 21.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 23.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18:1642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummel, M., D. A. Thorley-Lawson, and E. Kieff. 1984. An Epstein-Barr virus DNA fragment encodes messages for the two major envelope glycoproteins (gp350/300 and gp220/200). J. Virol. 49:413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen, E., M. Luftig, M. R. Chase, S. Weickseil, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2395. In B. N. Fields, D. Knipe, and P. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven publishers, Philadelphia, Pa.
- 27.Kirshner, J., D. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shutting of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik, P., D. Blackbourn, and B. Clements. 2004. The evolutionary conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 32.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson, A., D. M. Knipe, and D. M. Coen. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 78:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rickinson, A., B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. M. Knipe and P. Howley (ed.), Field's virology, vol. 2. Lippincott Williams and Wilkins, New York, N.Y. [Google Scholar]
- 35.Rodriguez, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98: 1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 22:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segouffin, C., H. Gruffat, and A. Sergeant. 1996. Repression by RAZ of Epstein-Barr virus bZIP transcription factor EB1 is dimerization independent. J. Gen. Virol. 77:1529-1536. [DOI] [PubMed] [Google Scholar]
- 40.Serio, T. R., J. L. Kolman, and G. Miller. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71:8726-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, M. P., and R. Kurzrock. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803-821. [DOI] [PubMed] [Google Scholar]
- 43.Treisman, R., S. H. Orkin, and T. Maniatis. 1983. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature 302:591-596. [DOI] [PubMed] [Google Scholar]
- 44.van Grunsven, W. M., A. Nabbe, and J. M. Middeldorp. 1993. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen complex. J. Med. Virol. 40:161-169. [DOI] [PubMed] [Google Scholar]
- 45.van Grunsven, W. M., E. C. van Heerde, H. J. de Haard, W. J. Spaan, J. M. Middeldorp, W. M. van Grunsven, A. Nabbe, and J. M. Middeldorp. 1993. Gene mapping and expression of two immunodominant Epstein-Barr virus capsid proteins. J. Virol. 67:3908-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zur Hausen, H., F. J. O'Neil, U. K. Freese, and E. Hecker. 1978. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature 27:373-375. [DOI] [PubMed] [Google Scholar]