Abstract
Human immunodeficiency virus type 1 (HIV-1) particle production, a process driven by the Gag polyprotein precursor, occurs on the plasma membrane in most cell types. The plasma membrane contains cholesterol-enriched microdomains termed lipid rafts, which can be isolated as detergent-resistant membrane (DRM). Previously, we and others demonstrated that HIV-1 Gag is associated with DRM and that disruption of Gag-raft interactions impairs HIV-1 particle production. However, the determinants of Gag-raft association remain undefined. In this study, we developed a novel epitope-based Gag multimerization assay to examine whether Gag assembly is essential for its association with lipid rafts. We observed that membrane-associated, full-length Gag is poorly detected by immunoprecipitation relative to non-membrane-bound Gag. This poor detection is due to assembly-driven masking of Gag epitopes, as denaturation greatly improves immunoprecipitation. Gag mutants lacking the Gag-Gag interaction domain located in the N terminus of the nucleocapsid (NC) were efficiently immunoprecipitated without denaturation, indicating that the epitope masking is caused by higher-order Gag multimerization. We used this assay to examine the relationship between Gag assembly and Gag binding to total cellular membrane and DRM. Importantly, a multimerization-defective NC mutant displayed wild-type levels of membrane binding and DRM association, indicating that NC-mediated Gag multimerization is dispensable for association of Gag with membrane or DRM. We also demonstrate that different properties of sucrose and iodixanol membrane flotation gradients may explain some discrepancies regarding Gag-raft interactions. This report offers new insights into the association of HIV-1 Gag with membrane and with lipid rafts.
Retrovirus particle production is driven by the viral Gag polyprotein (26, 83), which, for human immunodeficiency virus type 1 (HIV-1) has been termed Pr55Gag. Upon particle release from the cell, the viral protease (PR) cleaves Pr55Gag into the mature Gag proteins matrix (MA or p17), capsid (CA or p24), nucleocapsid (NC or p7), and p6 and two small spacer peptides, SP1 and SP2 (26, 83). Pr55Gag contains three functional domains responsible for mediating the major steps in virus particle assembly: the membrane-binding (M) domain, which targets Gag to the cellular membrane at which virus particles form, the interaction (I) domain, which promotes Gag multimerization, and the late (L) domain, which facilitates the release of nascent virions from the host cell. HIV-1 M domain function is exerted by the N-terminal portion of MA, to which a myristic acid moiety is covalently attached (26, 83). A Pro-Thr-Ala-Pro sequence in p6 acts as the predominant HIV-1 L domain (18, 62). A large region spanning CA, SP1, and NC functions as the HIV-1 I domain (for a review, see reference 26). Structural studies and protein-protein interaction analyses have demonstrated that CA and SP1 likely contribute to Gag multimerization by facilitating direct interactions between Gag molecules (24, 29, 31, 33, 59-61, 67, 96). Mutations and deletions in the C-terminal domain of CA (12, 21, 42, 50, 56, 58, 76, 82, 87, 88, 90) and SP1 (1, 45, 49, 93) cause defects in virus particle production, indicating that the Gag-Gag interaction mediated by this region is physiologically important for virus assembly. Unlike CA and SP1, NC is thought to promote Gag multimerization primarily through interaction with RNA. Basic amino acid residues located in the N-terminal region of NC mediate binding to RNA, an interaction that may allow RNA molecules to provide a scaffold for Gag-Gag interactions (7, 8, 14, 17, 39, 43, 77). A large number of biochemical and genetic studies support a role for NC, specifically its basic residues, in Gag multimerization and hence virus assembly (3, 7, 8, 14, 17, 22, 25, 30, 35, 37, 41, 43, 47, 53, 67, 77, 89, 97, 98). Studies of other retroviral Gag proteins also suggest that NC basic residues play an important role in Gag multimerization (2, 4, 9, 10, 40, 54, 55, 64, 95). However, whether NC-dependent multimerization is essential for virus particle production remains to be resolved (72, 91, 92).
Recent studies suggest that several retroviruses, including HIV-1, utilize specific plasma membrane microdomains known as lipid rafts for their assembly (for a review, see reference 71). Lipid rafts contain high concentrations of cholesterol and saturated lipids and can be isolated based on their resistance to low-temperature detergent treatment (5, 79). Such detergent-resistant membrane (DRM), which is typically recovered as low-density material in equilibrium flotation gradients, is unlikely to represent intact lipid rafts that exist in living cells (20, 23, 36, 57, 63, 71). Nevertheless, measuring DRM association provides valuable information regarding the preference for rafts by proteins or lipids of interest when used in combination with other methods. HIV-1 Gag associates with DRM (20, 34, 36, 51, 66, 70, 99), and microscopic analyses demonstrate colocalization of Gag with raft markers (36, 66, 71). Cholesterol depletion, or acylation of Gag with an unsaturated fatty acid that targets Gag to nonraft membrane, severely inhibits extracellular HIV-1 particle production (52, 70). Altogether, these studies suggest that HIV-1 Gag interacts with lipid rafts and that its association with these membrane microdomains promotes virus particle production. However, some reports questioned the validity of Gag-DRM interaction assays. It was observed that Gag could be recovered in low-density fractions of flotation gradients even after harsh detergent treatments that removed most of the Gag-associated lipid (36) and that Gag did not cofractionate with classical raft markers (20). It is noteworthy that these studies used iodixanol flotation gradients whereas we used sucrose gradients in our previous analyses of Gag-DRM interaction.
Several studies comparing full-length and C-terminally truncated Gag constructs (e.g., MA-CA) have suggested that Gag multimerization can enhance both membrane binding and DRM association (51, 74, 75, 78, 100). A myristyl switch model for Gag membrane binding has been proposed in which the N-terminally attached myristate moiety can alternate between a conformational state in which it is sequestered within the globular fold of MA and one in which it is highly exposed and available for interaction with membrane (68, 73, 80, 100). It has been suggested that Gag multimerization enhances myristate exposure thereby increasing Gag-membrane binding (74, 78, 84).
We previously observed that a Gag derivative that contains only MA and the N-terminal region of CA displays levels of steady-state membrane binding and DRM association similar to those of full-length Gag (67, 70). These results indicate that Gag multimerization is not essential for Gag-membrane and Gag-DRM interactions. Nevertheless, kinetic analyses suggest that the presence of SP1 and NC sequences accelerates or stabilizes the binding of Gag to total cellular membrane and DRM (70). To determine whether this enhancement of Gag-membrane association is caused by increased Gag-Gag interaction, multimerization levels of membrane- and DRM-bound Gag need to be assessed in kinetic analyses. However, most mammalian-cell-based Gag multimerization assays developed thus far are not well suited for this purpose. One such assay measures the rescue of nonmyristylated Gag derivatives into extracellular virus particles upon their coexpression with wild-type (WT) Gag (13, 67). Equilibrium density gradient centrifugation of Gag-containing cell lysates is also widely used to measure Gag multimerization (48, 61, 85). However, in general, these methods cannot distinguish between Gag multimerization in the cytosol and on the membrane. Use of fluorescence resonance energy transfer between chimeras of Gag and green fluorescent protein derivatives (19, 46) is a powerful method to measure Gag-Gag interaction in a location-specific manner, but slow maturation of green fluorescent protein variants (15, 65, 86) may hamper kinetic analyses. Because of the inherent limitations of the above assays, we sought to develop a method with which Gag multimerization and membrane binding can be examined simultaneously.
In this report, we establish that Gag multimerization mediated by NC basic amino acids causes masking of Gag epitopes recognized by anti-Gag antisera. By using this epitope masking to measure higher-order Gag multimerization, we investigated the kinetic aspects of NC-dependent Gag assembly and the relationship between Gag assembly and Gag membrane binding and DRM association. We observed that replacement of all the basic residues within the N terminus of NC blocks epitope masking without affecting Gag binding to total membrane or to DRM. These results indicate that the Gag-Gag interaction mediated by the highly basic domain of NC is not needed for association of Gag with total cellular membrane or with lipid rafts. We also demonstrate that, in contrast to results obtained with sucrose, Gag complexes remaining after solubilization of lipid rafts with octylglucoside still float in iodixanol gradients, suggesting that iodixanol may be poorly suited for analyzing the interaction of large protein complexes with membrane.
MATERIALS AND METHODS
Plasmids.
Molecular clones pNL4-3/Myr− (1GA) (28), pNL4-3/CA146 (67), pNL4-3/p41 (68), and pNL4-3/NC35 (67), encoding Gag mutants, and pNL4-3/PR− (38), encoding an inactive PR, were described previously. A molecular clone encoding the CA amino acid substitution mutant, pNL4-3/WM184,185AA, was constructed by QuikChange (Stratagene) mutagenesis using the following mutagenic oligonucleotides: CA184WA,185MA-F (5′-CACAAGAGGTAAAAAATGCGGCGACAGAAACCTTGTTGGTCC-3′) and CA184WA,185MA-R (5′-GGACCAACAAGGTTTCTGTCGCCGCATTTTTTACCTCTTGTG-3′). The PR− version of pNL4-3/WM184,185AA (pNL4-3/WM184,185AA/PR−) was constructed by introducing the ApaI (pNL4-3 nt 2006)-to-EcoRI (pNL4-3 nt. 5743) fragment from pNL4-3/PR− into pNL4-3/WM184,185AA. A molecular clone encoding the NC amino acid substitution mutant, pNLHX15A, was a kind gift from J. Luban, Columbia University (13). The isogenic molecular clone encoding WT Gag (pNLSE/HXB2) was constructed by introducing the SpeI (pNL4-3 nt. 1507)-to-EcoRV (pNL4-3 nt. 2977) fragment from pHXB2R3 into pNL4-3. PR− versions of pNLSE/HXB2 (pNLHX/PR−) and pNLHX15A (pNLHX15A/PR−) were constructed by introducing the BglII (pNL4-3 nt 2096)-to-EcoRI (pNL4-3 nt. 5743) fragment from pNL4-3/PR− into pNLSE/HXB2 and pNLHX15A. Construction of pCMVNLGagPolRRE using pCMVGagPolRRE (a kind gift from D. Rekosh, University of Virginia [81]) was described previously (70). The vesicular stomatitis virus G glycoprotein (VSV-G) expression vector pHCMV-G (94) was generously provided by J. Burns (University of California, San Diego).
Cells, transfections, and infections.
HeLa cells were cultured as previously described (27). Gag was expressed either by transfecting cells with molecular clones or by infecting with high-titer vector virus stocks. Transfection of HeLa cells by the calcium phosphate method was performed as previously described (27). Infection of HeLa cells with virus stocks pseudotyped with VSV-G was performed by culturing cells for 9 h with high-titer virus stocks at 30 counts/min of reverse transcriptase activity/cell. VSV-G-pseudotyped virus stocks were prepared by transfecting HeLa cells with pCMVNLGagPolRRE, pHCMV-G, and the molecular clone pNLHX/PR− or pNLHX15A/PR−.
Cell fractionation and denaturation of Gag.
Separation of denucleated cell homogenates into pellet and supernatant fractions was performed as described previously (68). For this experiment, HeLa cells were labeled overnight with [35S]Met/Cys in the presence of 5% fetal bovine serum as previously described (27). For detection of metabolically labeled Gag, fractions obtained either by cell fractionation or by equilibrium flotation centrifugation (see below) were mixed with 2× radioimmunoprecipitation assay (RIPA) buffer (280 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4, 2% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 20 mM iodoacetoamide, and protease inhibitors). Prior to immunoprecipitation, RIPA-treated fractions were aliquoted to two portions and either not denatured or denatured by mixing with 0.057 volume of 2× sample buffer (125 mM Tris-HCl [pH 6.8] containing 6% SDS, 10% 2-mercaptoethanol, and 20% glycerol) and boiled for 5 min. Both portions were then diluted with 6 volumes of 50 mM Tris-HCl (pH 7.5) containing 300 mM NaCl and 0.5% Triton X-100, incubated with protein A-agarose, and clarified by centrifugation at 13,200 rpm (16,100 × g) for 1 min in a microcentrifuge or at 1,500 × g for 5 min in a benchtop low-speed centrifuge. Immunoprecipitation with HIV immunoglobulin (HIV Ig; obtained from the NIH AIDS Research and Reference Reagent Program) was performed as described previously (27). Detection of Gag by Western blotting was performed as previously described (44).
Assays for membrane binding and DRM association.
For analyses of membrane binding and DRM association of Gag, HeLa cells expressing Gag proteins were either pulse-labeled for 5 min and chased for 0, 15, or 30 min or labeled for 90 min with [35S]Met/Cys. Postnuclear supernatants (PNSs) of cell homogenates were subjected to equilibrium flotation centrifugation using sucrose gradients as detailed previously (67, 68, 70). For DRM association, PNSs were treated with Triton X-100 (final concentration, 0.25%) for 20 min on ice prior to flotation centrifugation. Fractions were either not denatured or denatured and analyzed by immunoprecipitation as described above.
For equilibrium flotation centrifugation using iodixanol gradients, PNSs not treated or treated with the indicated concentrations (Fig. 6 legend) of detergents were mixed with TNE (25 mM Tris-HCl [pH 7.5] containing 150 mM NaCl and 4 mM EDTA) containing 60% (wt/vol) iodixanol (Axis-Shield PoC, Oslo, Norway) (final concentration, 50%). On top of this mixture was layered 40% (wt/vol) and 10% (wt/vol) iodixanol in TNE. Centrifugation of gradients and analyses of fractions were performed in the same manner as sucrose gradient flotation experiments except that iodixanol gradients were centrifuged for 2 h.
FIG. 6.
Octylglucoside-treated Gag complexes are recovered in low-density fractions (Fr.) in iodixanol flotation gradients. PNSs derived from HeLa cells expressing WT Pr55Gag were incubated in the absence or presence of 0.25% Triton X-100 or 30 mM octylglucoside on ice and fractionated on equilibrium flotation gradients using sucrose (A) or iodixanol (B). Distribution of WT Pr55Gag in these gradients was analyzed by Western blotting. (C) Octylglucoside-treated PNS was loaded on the top of a 20 to 73% sucrose step gradient. Note that octylglucoside treatment solubilizes membrane but does not disrupt Gag multimers.
Equilibrium sucrose density gradient centrifugation was performed as described previously (68).
Immunofluorescence microscopy.
Immunostaining of HeLa cells was performed as previously described (69). For detection of Gag, cells were labeled either with anti-p17 or -p24 monoclonal antibody (Advanced Biotechnologies, Columbia, MD) prelabeled with Zenon One Alexa Fluor reagent (Molecular Probes, Eugene, OR) and observed with a Zeiss 510 laser scanning confocal microscope.
RESULTS
Membrane-bound Pr55Gag requires denaturation for efficient recovery by immunoprecipitation.
In the course of studies designed to evaluate the binding of Gag to membrane, we separated denucleated homogenates of Pr55Gag-expressing cells into pellet and supernatant fractions. In these assays, we observed that immunoprecipitation versus immunoblotting with HIV Ig produced apparently conflicting results regarding the amount of Gag detected in these two fractions. In contrast to immunoblotting, which showed a large amount of WT Pr55Gag in the pellet relative to the supernatant fraction, immunoprecipitation only poorly detected WT Pr55Gag in the pellet fraction (Fig. 1A). This discrepancy between immunoblotting and immunoprecipitation was not observed with a nonmyristylated Gag derivative (Myr−). When samples were denatured prior to immunoprecipitation, we observed a marked increase in the amount of WT Gag in the pellet relative to the supernatant fraction (Fig. 1B). These results suggest that epitopes in WT Gag molecules recovered in the pellet fraction are masked and are thus not available for immunoprecipitation. Similar results were obtained with other pools of HIV-infected patient sera and with several anti-Gag antibodies (data not shown).
FIG. 1.

WT Pr55Gag in P100 fraction requires denaturation for efficient recovery by immunoprecipitation. HeLa cells expressing WT or Myr− Pr55Gag were metabolically labeled overnight, and PNSs of cell homogenates were fractionated by ultracentrifugation at 100,000 × g for 1 h into pellets (P) and supernatants (S). Fractions were treated with RIPA buffer and analyzed by Western blotting (WB) or by immunoprecipitation (IP). Comparisons of Western blotting and immunoprecipitation were performed using the same samples either without (A) or with (B) prior denaturation.
Since pellets in cell fractionation assays contain not only membrane-bound material but also non-membrane-bound macromolecular complexes, which population of Gag contains masked epitopes was unclear. To address this question, we subjected PNSs from metabolically labeled, Pr55Gag-expressing HeLa cells to equilibrium sucrose flotation centrifugation. Membrane-bound and non-membrane-bound Pr55Gag molecules were recovered in fractions 1 to 5 and 6 to 10, respectively (Fig. 2). These fractions were treated with RIPA buffer, pooled, and divided into two portions, one of which was denatured prior to immunoprecipitation with HIV Ig. When not denatured, the majority of Pr55Gag in membrane fractions failed to be immunoprecipitated, whereas recovery of Pr55Gag in nonmembrane fractions was equally efficient regardless of the denaturation status. These results suggest that membrane-bound but not cytosolic Pr55Gag molecules harbor masked epitopes.
FIG. 2.
Membrane-bound Pr55Gag requires denaturation for efficient recovery by immunoprecipitation. HeLa cells expressing WT Pr55Gag were metabolically labeled for 90 min, and PNSs of cell homogenates were fractionated by membrane flotation. Fractions were treated with RIPA buffer and membrane (M) and nonmembrane (NM) fractions were pooled. Labeled Pr55Gag in each pooled fraction was recovered by immunoprecipitation either without (−) or with (+) prior denaturation. Sixteen percent of membrane-bound Gag was recovered from nondenatured fractions relative to denatured fractions, whereas 72% of non-membrane-bound Gag was detected without versus with denaturation.
A functional I domain is necessary for epitope masking of membrane-bound Gag.
To define the region of Gag responsible for epitope masking, we examined whether denaturation is needed for efficient immunoprecipitation of a series of C-terminally truncated Gag mutants (Fig. 3A). HeLa cells expressing full-length Pr55Gag or the NC35 or p41 truncation mutants were metabolically labeled for 90 min and pooled before membrane flotation. Like full-length Pr55Gag, a Gag mutant (NC35) containing MA, CA, SP1, and the first 35 amino acids of NC required denaturation for efficient recovery from RIPA buffer-treated membrane fractions (Fig. 3B). In contrast, a Gag mutant (p41) that consists of MA and CA was fully immunoprecipitated by HIV Ig with or without denaturation. Since the N-terminal portion of NC promotes Gag-Gag interaction, these results suggest that Gag multimerization may cause the observed epitope masking.
FIG. 3.
A functional I domain is necessary for epitope masking of membrane-bound Gag. (A) Schematic representation of Gag mutants. Positions of N-terminal myristate (m) and the two zinc finger motifs in NC (Zn) are shown. (B) HeLa cells expressing full-length Pr55Gag or the NC35 or p41 truncation mutants were metabolically labeled for 90 min and pooled before membrane flotation. Fractions were analyzed as for Fig. 2. Note that, in contrast to full-length Pr55Gag and NC35, membrane-bound p41 was efficiently immunoprecipitated even in the absence of prior denaturation. In a typical experiment, the efficiencies of Gag recovery from nondenatured membrane (M) fractions relative to denatured membrane fractions were 29% for full-length Pr55Gag, 51% for NC35, and 108% for p41. NM, nonmembrane. (C) HeLa cells expressing either WT or 15A mutant Pr55Gag were pulse-labeled for 5 min and chased for 30 min before membrane flotation. Fractions were analyzed as for Fig. 2. Note that 15A mutant Gag in membrane fractions was efficiently immunoprecipitated in the absence of prior denaturation. (D) HeLa cells expressing either WT or 15A-mutant Pr55Gag in the presence of active PR were immunostained either with monoclonal anti-p17 or -p24 antibody prelabeled with Zenon One Alexa Fluor reagent.
To assess the influence of Gag multimerization on epitope exposure in the context of full-length Gag, we examined a Gag derivative (15A) in which all NC basic amino acids critical for Gag-Gag interaction were replaced with Ala (13). Unlike WT Pr55Gag, both non-membrane-bound and membrane-bound 15A Pr55Gag molecules were efficiently immunoprecipitated by HIV Ig without denaturation (Fig. 3C). Consistent with defective Gag multimerization, 15A Gag did not display the punctate pattern observed with WT Gag even though, like WT, 15A Gag localized to the plasma membrane (Fig. 3D). These results strongly suggest that Gag multimerization facilitated by NC basic amino acids causes epitope masking of membrane-bound Gag.
Pr55Gag in immature but not mature virions requires denaturation for efficient recovery by immunoprecipitation.
To determine whether Gag multimers in which epitopes are masked are physiologically relevant to the virus assembly process, we examined the epitope availability of virion-associated Gag. Virus particles derived from HeLa cells expressing Pr55Gag in the presence or absence of active PR were solubilized in RIPA buffer. Viral lysates were aliquoted into two portions, one of which was denatured prior to immunoprecipitation with HIV Ig (Fig. 4). We found that denaturation of virus lysates did not affect immunoprecipitation of p24(CA) associated with mature virus particles. In contrast, recovery of Pr55Gag from immature particles released in the absence of active PR was greatly increased by denaturation. These results indicate that Gag epitopes in immature virus particles are masked like those of cell-associated, membrane-bound Gag, suggesting that epitope masking is physiologically relevant to virus particle production. Masked epitopes in the CA domain of Pr55Gag become exposed upon Gag cleavage presumably because the tightly packed Gag multimer reorganizes during maturation.
FIG. 4.

WT Gag proteins in lysates from immature but not mature virions require denaturation for efficient recovery by immunoprecipitation. HeLa cells transfected with molecular clones encoding WT Pr55Gag with active (PR+) or inactive (PR−) viral PR were metabolically labeled overnight. Virus particles were pelleted from culture supernatants, and virus lysates were analyzed by immunoprecipitation either without (−) or with (+) prior denaturation. Signal intensity was quantified by phosphorimager analysis, and epitope exposure of Gag in nondenatured samples relative to denatured samples was calculated.
Pr55Gag undergoes epitope masking gradually after it binds membrane.
To characterize further the epitope-masked Gag multimer, we performed a kinetic analysis of Gag epitope exposure (Fig. 5). HeLa cells expressing Pr55Gag were pulse-labeled for 5 min and chased for up to 30 min; PNSs were separated into membrane and nonmembrane fractions. Amounts of labeled Pr55Gag immunoprecipitated with or without denaturation were compared. We observed that epitopes on non-membrane-bound Gag remained exposed throughout the chase period. In contrast, epitopes on membrane-bound Gag were highly exposed immediately after pulse-labeling but became substantially masked within 30 min. These results suggest that higher-order Gag multimerization takes place after Gag binds membrane. The observed time-dependent increase in epitope masking indicates that the higher-order Gag complexes do not form during sample preparation but arise during the process of virus assembly.
FIG. 5.

Pr55Gag undergoes epitope masking gradually after it binds membrane. HeLa cells expressing WT Pr55Gag were pulse-labeled for 5 min and chased for 0, 15, and 30 min before membrane flotation. Fractions were analyzed as for Fig. 2, and epitope exposure was calculated as for Fig. 4. Note that the membrane-bound Gag epitopes is highly exposed even without denaturation immediately after the pulse but becomes masked within 15 min.
Octylglucoside-treated Gag complexes are recovered in low-density fractions upon equilibrium flotation centrifugation in iodixanol but not in sucrose.
Since the kinetics of Gag epitope masking are similar to those of Gag association with DRM in that both occur more slowly than the initial binding of Gag to membrane (70), we sought to examine the relationship between higher-order Gag multimerization and Gag-raft association. Equilibrium flotation centrifugation of detergent-treated PNSs in either sucrose or iodixanol gradients has been widely used for studying protein-DRM association (5, 79). As mentioned in the introduction, we previously demonstrated that non-membrane-bound complexes do not float in sucrose gradients (70), whereas others, using iodixanol gradients, have reported that Gag complexes from which most of the lipid has been removed can be recovered in low-density fractions (36). To directly compare which type of gradient is better suited for analyzing Gag-DRM association, we performed flotation centrifugation of PNSs derived from HeLa cells expressing WT Gag using either sucrose or iodixanol gradients. Prior to loading the gradients, PNSs were either treated with cold Triton X-100 or another nonionic detergent, octylglucoside, or not treated. In sucrose gradients, substantial amounts of Gag in untreated and Triton X-100-treated PNSs were recovered in fractions 2 to 4 while virtually no Gag was detected in these fractions after octylglucoside treatment (Fig. 6A). This observation is consistent with the finding that lipid rafts are solubilized by treatment with octylglucoside (6). As we have shown previously (70), when octylglucoside-treated PNSs are analyzed by equilibrium density gradient centrifugation using a 20 to 73% sucrose gradient, Gag-containing complexes are readily detected across a wide density range (Fig. 6C). Disruption of these Gag complexes by denaturation results in Gag remaining at the top of the 20 to 73% sucrose gradient (data not shown). These results indicate that octylglucoside-treated Gag complexes are not recovered in low-density fractions after flotation centrifugation using sucrose gradients.
When iodixanol gradients were used for flotation centrifugation, the majority of Gag in untreated and Triton X-100-treated PNSs was found in top fractions (Fig. 6B). Even after octylglucoside treatment, a large amount of Gag was recovered in the low-density fractions (Fig. 6B). Notably, when the bottom fractions of sucrose flotation gradients were examined by equilibrium density gradient centrifugation following octylglucoside treatment, Gag-containing complexes were readily detectable in the 20 to 73% sucrose gradient (data not shown). Therefore, non-membrane-bound Gag complexes were still present in the bottom fractions of sucrose flotation gradients but did not float to the low-density fractions. These results suggest that Gag complexes not associated with lipid rafts can float in iodixanol but not in sucrose gradients.
15A-mutant Gag can still bind membrane and DRM efficiently without forming higher-order multimers.
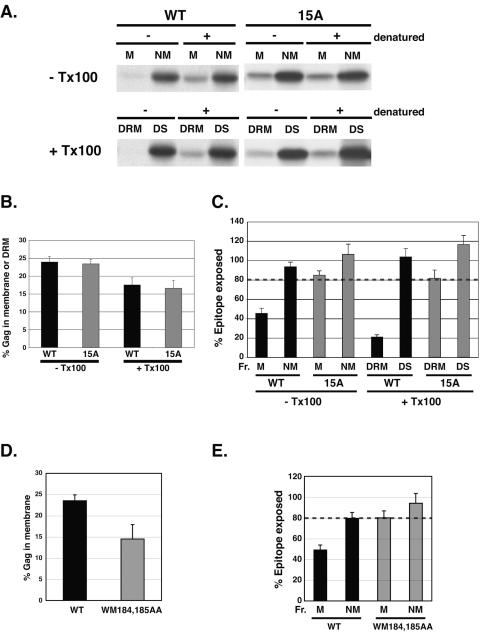
Based on the results presented above, we used sucrose gradients for analyses of Gag-DRM association. To examine whether higher-order Gag multimerization is necessary for efficient association of Gag with membrane and DRM, HeLa cells expressing WT or 15A Pr55Gag were pulse-labeled for 5 min and chased for 30 min. Levels of recovery of labeled Pr55Gag in total membrane and DRM fractions were compared by equilibrium flotation centrifugation (Fig. 7A and B). To assess the level of higher-order multimerization, Gag epitope exposure was calculated as a ratio of the amount of Gag immunoprecipitated with or without denaturation (Fig. 7A and C). We found that the efficiencies of membrane binding and DRM association were similar between WT and 15A Gag (Fig. 7B). Interestingly, WT Gag epitopes were more masked in DRM fractions than in total membrane fractions, suggesting that DRM association is accompanied by more-extensive assembly. In contrast to the results obtained with WT Gag, both membrane-bound and DRM-associated 15A Gag was readily immunoprecipitated (Fig. 7C). As a consequence, DRM-associated 15A Gag displayed a fourfold-greater degree of epitope exposure than did DRM-associated WT Gag. These results indicate that the higher-order Gag multimerization mediated by NC basic residues is not required for efficient binding of Gag to total membrane or to DRM.
FIG. 7.
Gag multimerization mediated by NC basic residues is not required for Gag membrane binding and DRM association. HeLa cells expressing either WT or 15A-mutant Pr55Gag were pulse-labeled for 5 min and chased for 30 min. PNSs were incubated in the absence (− Tx100) or presence (+ Tx100) of 0.25% Triton X-100 on ice for 30 min before membrane flotation. Fractions were analyzed as for Fig. 2. (A) Membrane (M), nonmembrane (NM), DRM, and detergent-soluble (DS) fractions are indicated. Similar results were obtained in four independent experiments. (B) Percentages of Gag in membrane and DRM fractions were calculated using the results obtained by immunoprecipitation with prior denaturation. Data are shown as means ± standard errors of the means. (C) Percentages of Gag recovered with or without denaturation were calculated as for Fig. 4. Data are shown as means ± standard errors of the means. Full epitope exposure was defined as >80% recovery of Gag without denaturation (dashed line). (D and E) HeLa cells expressing WT or WM184,185AA-mutant Pr55Gag were analyzed for total membrane binding (D) and epitope exposure (E) as for panels B and C, respectively. Data are shown as means ± standard errors of the means.
As not only NC but also CA is involved in Gag multimerization, we next examined the impact of a Gag derivative (WM184,185AA) containing a two-amino-acid substitution at the CA dimer interface (29, 88). Virus release efficiency of this mutant was approximately 10% that of WT (data not shown), and membrane binding was reduced approximately 40% relative to WT (Fig. 7D). Both membrane-bound and non-membrane-bound WM184,185AA Gag molecules were readily immunoprecipitated without denaturation. These results suggest that a defect in Gag dimerization impairs higher-order Gag multimerization and Gag membrane binding.
DISCUSSION
In this study, we demonstrate that epitope masking of membrane-bound Gag reflects the extent of Gag multimerization. Even though epitope masking is observed predominantly with membrane-bound Gag, Gag-membrane binding itself does not cause epitope masking. Rather, epitope sequestration is caused by higher-order Gag multimerization promoted by basic residues in NC (Fig. 3). Since Gag proteins in immature virus particles also display epitope masking (Fig. 4), this masking is likely caused by productive Gag multimerization during assembly rather than nonproductive Gag aggregation. Once Gag is cleaved by PR during virus particle maturation, Gag epitopes become available for immunoprecipitation (Fig. 4). Interestingly, epitope masking was observed not only with HIV Ig but also with several anti-Gag antibodies (A. Ono and E. O. Freed, unpublished data). The data presented here indicate that the level of epitope exposure can be used to monitor Gag multimerization under physiological conditions. In this study, we used this method to clarify the relationship between Gag multimerization mediated by NC basic residues and Gag association with total cellular membrane and DRM.
It is formally possible that nondenatured, higher-order Gag multimers may fail to be immunoprecipitated not because epitopes are masked by Gag multimerization but because the large size of Gag multimeric complexes blocks their retention on antibody-bound protein A beads. However, considering that immunoprecipitation can be performed to recover intact virus particles (11, 66), the size of Gag complexes is unlikely to prevent immunoprecipitation of higher-order Gag multimers. Indeed, it is possible that a portion of membrane-bound Gag recovered without denaturation may represent Gag complexes captured by antibodies whose epitopes remain exposed on the surface of Gag multimers.
Kinetic analyses of Gag epitope masking indicate that Gag membrane binding precedes higher-order Gag multimerization (Fig. 5). Moreover, 15A Gag that is assembly defective still displays WT levels of membrane binding and DRM association (Fig. 7). These results indicate that higher-order multimerization promoted by basic residues in NC is not required for accelerating or stabilizing Gag membrane binding and raft association.
Several reports using different experimental systems show that membrane binding and DRM association of a truncated Gag construct consisting of MA and CA are increased upon addition of SP1 and NC in either steady-state (51, 70, 74, 78) or kinetic (70) analyses. However, it is important to point out that the levels of Gag multimerization may differ between MA-CA and 15A. The C-terminal domains of CA and SP1 have been shown to promote Gag-Gag interaction (see the introduction). This protein-protein interaction domain may be disrupted by truncation of Gag at the CA-SP1 boundary whereas the effect of 15A mutations on CA-SP1-mediated multimerization may be minimal. Consistent with this interpretation, the densities of virus-like particles formed by MA-CA and 15A Gag are different: MA-CA particles are less dense than WT virions while 15A particles possess WT density (13). Moreover, it was reported in some (50) but not other (58) studies that amino acid substitutions in CA close to the boundary with SP1 disrupt Gag membrane binding. Mutations in SP1 that inhibit Gag multimerization also block Gag membrane binding and DRM association (32, 33). In addition, it has been observed that low-order multimerization (i.e., trimerization) of Gag promoted by CA shifts the N-terminal myristate to a more exposed conformation in which it is better able to interact with membrane (84). Similarly, in vitro analyses of Rous sarcoma virus Gag membrane binding showed that dimerization enhanced the membrane affinity of the MA domain (16). Consistent with these reports, we observed that a Gag derivative with a double-amino-acid substitution at the CA dimer interface (CA residues 184 and 185) binds membrane less efficiently than WT Gag (Fig. 7D and E). Taken together, these results suggest that Gag-membrane and Gag-raft interactions may be enhanced or stabilized by lower-order Gag-Gag interactions mediated by CA and SP1. Our data further suggest that, after initial membrane binding and lower-order multimerization events take place, higher-order Gag multimerization proceeds, promoted by NC basic residues in conjunction with nucleic acid. Our results indicate that membrane binding and DRM association do not require this higher-order Gag multimerization.
The density of Gag-containing DRM has been observed to be higher than that of typical raft membranes in flotation gradients (20, 51), presumably due to the association of tightly packed Gag molecules with a limited amount of DRM. Because of the difference in density between Gag-containing, detergent-resistant, buoyant material and classical raft markers, the hypothesis that HIV-1 Gag associates with lipid rafts was questioned. It was suggested that Gag recovered in the floated Triton X-100-resistant fractions may be non-membrane-bound Gag multimers rather than DRM-associated Gag (20, 36). This concern was supported by the observation that Gag was recovered in low-density fractions in iodixanol gradients even after harsh treatments that largely depleted cellular membrane cholesterol (20) or removed the majority of Gag-associated lipid (36). In this study, however, we observed that a defect in higher-order Gag multimerization does not affect Gag-DRM association. In addition, in this and previous studies we have shown that, after treatment with octylglucoside, which disrupts rafts but preserves Gag complexes, no Gag is recovered in low-density fractions of sucrose gradients (Fig. 6) (70). Importantly, in contrast to results obtained with sucrose gradients, in iodixanol gradients Gag complexes can be recovered in the buoyant fractions even after octylglucoside treatment (Fig. 6). This particular property of iodixanol likely explains the recovery of non-lipid-associated Gag complexes in low-density fractions observed previously (20, 36). We therefore conclude that sucrose is preferable to iodixanol for separating DRM-associated Gag from non-membrane-bound, detergent-resistant Gag complexes in flotation gradients. More generally, we feel that caution should be used in interpreting data obtained with highly oligomerized proteins in iodixanol gradients.
Although Gag multimerization mediated by NC basic residues is not needed for Gag membrane binding and DRM association, Gag membrane binding appears to be important for higher-order Gag multimerization. This conclusion is supported by the findings that epitope masking is observed predominantly with membrane-bound Gag and that nonmyristylated Gag (which does not bind membrane) does not undergo extensive epitope masking. DRM association may facilitate Gag multimerization, as the level of epitope masking is increased in DRM-associated Gag (Fig. 7). By analogy with the role of lipid rafts in a variety of cellular functions, lipid rafts may serve as concentration platforms for Gag, thereby facilitating higher-order Gag multimerization. Whether lipid rafts play an active role in increasing Gag-Gag interaction is currently under investigation.
Acknowledgments
We thank Vineet KewalRamani and members of the Freed lab for critical reviews of the manuscript and S. Ablan for expert technical assistance. We thank J. Burns, J. Luban, and D. Rekosh for providing plasmids. HIV Ig was obtained through the NIH AIDS Research Reference and Reagent Program.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by the Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantin, R., G. Martin, and M. J. Tremblay. 2001. A novel virus capture assay reveals a differential acquisition of host HLA-DR by clinical isolates of human immunodeficiency virus type 1 expanded in primary human cells depending on the nature of producing cells and the donor source. J. Gen. Virol. 82:2979-2987. [DOI] [PubMed] [Google Scholar]
- 12.Chazal, N., C. Carriere, B. Gay, and P. Boulanger. 1994. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J. Virol. 68:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimarelli, A., and J. Luban. 2000. Human immunodeficiency virus type 1 virion density is not determined by nucleocapsid basic residues. J. Virol. 74:6734-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 16.Dalton, A. K., P. S. Murray, D. Murray, and V. M. Vogt. 2005. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J. Virol. 79:6227-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 18.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 19.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257-283. [DOI] [PubMed] [Google Scholar]
- 24.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng, Y. X., T. Li, S. Campbell, and A. Rein. 2002. Reversible binding of recombinant human immunodeficiency virus type 1 Gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 76:11757-11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 27.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 30.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 31.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Krausslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo, X., and C. Liang. 2005. Opposing effects of the M368A point mutation and deletion of the SP1 region on membrane binding of human immunodeficiency virus type 1 Gag. Virology 335:232-241. [DOI] [PubMed] [Google Scholar]
- 33.Guo, X., A. Roldan, J. Hu, M. A. Wainberg, and C. Liang. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75(Pt. 11):2985-2997. [DOI] [PubMed] [Google Scholar]
- 36.Holm, K., K. Weclewicz, R. Hewson, and M. Suomalainen. 2003. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 77:4805-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshikawa, N., A. Kojima, A. Yasuda, E. Takayashiki, S. Masuko, J. Chiba, T. Sata, and T. Kurata. 1991. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J. Gen. Virol. 72:2509-2517. [DOI] [PubMed] [Google Scholar]
- 38.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huseby, D., R. L. Barklis, A. Alfadhli, and E. Barklis. 2005. Assembly of human immunodeficiency virus precursor Gag proteins. J. Biol. Chem. 280:17664-17670. [DOI] [PubMed] [Google Scholar]
- 40.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. (Erratum, 74:943, 1993.) [DOI] [PubMed] [Google Scholar]
- 42.Kattenbeck, B., A. von Poblotzki, A. Rohrhofer, H. Wolf, and S. Modrow. 1997. Inhibition of human immunodeficiency virus type 1 particle formation by alterations of defined amino acids within the C terminus of the capsid protein. J Gen. Virol. 78:2489-2496. [DOI] [PubMed] [Google Scholar]
- 43.Khorchid, A., R. Halwani, M. A. Wainberg, and L. Kleiman. 2002. Role of RNA in facilitating Gag/Gag-Pol interaction. J. Virol. 76:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson, D. R., Y. M. Ma, V. M. Vogt, and W. W. Webb. 2003. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 162:1233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 49.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindwasser, O. W., and M. D. Resh. 2002. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. USA 99:13037-13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayor, S., and F. R. Maxfield. 1995. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol. Biol. Cell 6:929-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melamed, D., M. Mark-Danieli, M. Kenan-Eichler, O. Kraus, A. Castiel, N. Laham, T. Pupko, F. Glaser, N. Ben-Tal, and E. Bacharach. 2004. The conserved carboxy terminus of the capsid domain of human immunodeficiency virus type 1 Gag protein is important for virion assembly and release. J. Virol. 78:9675-9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 60.Morellet, N., S. Druillennec, C. Lenoir, S. Bouaziz, and B. P. Roques. 2005. Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 14:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 63.Munro, S. 2003. Lipid rafts: elusive or illusive? Cell 115:377-388. [DOI] [PubMed] [Google Scholar]
- 64.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 66.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 78:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ono, A., and E. O. Freed. 2005. The role of lipid rafts in virus replication, p. 311-358. In P. Roy (ed.) Advances in virus research, vol. 64. Elsevier Academic Press, San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 72.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, K. Nagashima, R. C. Sowder II, D. T. Poon, and R. J. Gorelick. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 77:5547-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paillart, J. C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 78:9560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Platt, E. J., and O. K. Haffar. 1994. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc. Natl. Acad. Sci. USA 91:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 80.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srinivasakumar, N., N. Chazal, C. Helga-Maria, S. Prasad, M. L. Hammarskjold, and D. Rekosh. 1997. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 71:5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasakumar, N., M. L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 84.Tang, C., E. Loeliger, P. Luncsford, I. Kinde, D. Beckett, and M. F. Summers. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 101:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 87.von Poblotzki, A., R. Wagner, M. Niedrig, G. Wanner, H. Wolf, and S. Modrow. 1993. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology 193:981-985. [DOI] [PubMed] [Google Scholar]
- 88.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, C. T., H. Y. Lai, and J. J. Li. 1998. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J. Virol. 72:7950-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, C. T., J. Stegeman-Olsen, Y. Zhang, and E. Barklis. 1994. Assembly of HIV GAG-B-galactosidase fusion proteins into virus particles. Virology 200:524-534. [DOI] [PubMed] [Google Scholar]
- 91.Wang, S. W., and A. Aldovini. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76:11853-11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, S. W., K. Noonan, and A. Aldovini. 2004. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 78:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Pt. A):99-112. [DOI] [PubMed] [Google Scholar]
- 95.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang, W. H., D. J. Hockley, M. V. Nermut, Y. Morikawa, and I. M. Jones. 1996. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J. Gen. Virol. 77:743-751. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng, Y. H., A. Plemenitas, T. Linnemann, O. T. Fackler, and B. M. Peterlin. 2001. Nef increases infectivity of HIV via lipid rafts. Curr. Biol. 11:875-879. [DOI] [PubMed] [Google Scholar]
- 100.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70:8540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]