Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is the causative agent of three malignancies associated with AIDS and immunosuppression. Tumor cells harbor latent virus and express kaposin (open reading frame [ORF] K12), v-FLIP (ORF 71), v-Cyclin (ORF 72), and latency-associated nuclear antigen (LANA; ORF 73). ORFs 71 to 73 are transcribed as multicistronic RNAs initiating from adjacent constitutive and inducible promoters upstream of ORF 73. Here we characterize a third promoter embedded within the ORF 71-to-73 transcription unit specifying transcripts that encode ORF 71/72 or K12. These transcripts may also be the source of 11 microRNAs arranged as a cluster between K12 and ORF 71. Our studies reveal a complex arrangement of interlaced transcription units, incorporating four important protein-encoding genes required for latency and pathogenesis and the entire KSHV microRNA repertoire.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV, or human herpesvirus 8) is a gamma-2-herpesvirus associated with three human malignancies: Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (7, 15, 24). Typically, KSHV is present in the majority of tumor cells and maintains a state of latency in which the viral genome persists as a circular episome in the host cell nucleus, but only a tiny fraction of the 90 or so viral open reading frames (ORFs) are expressed. Four KSHV proteins are consistently detected in all latently infected cells: kaposin (several isoforms encoded by ORF K12), v-FLIP (ORF 71), v-Cyclin (ORF 72), and the latency-associated nuclear antigen (LANA; ORF 73). Numerous lines of evidence suggest these gene products drive proliferation of the host cell, prevent apoptosis, facilitate immune evasion, and maintain the extrachromosomal viral genome during repeated cell divisions (3, 4, 17, 18, 20, 26, 28, 32, 36, 37, 39, 42, 49, 51). Each of these functions is likely to be critical for KSHV pathogenesis, prompting diverse efforts to understand how the gene cluster is regulated and to elucidate the molecular function of each protein (8, 15).
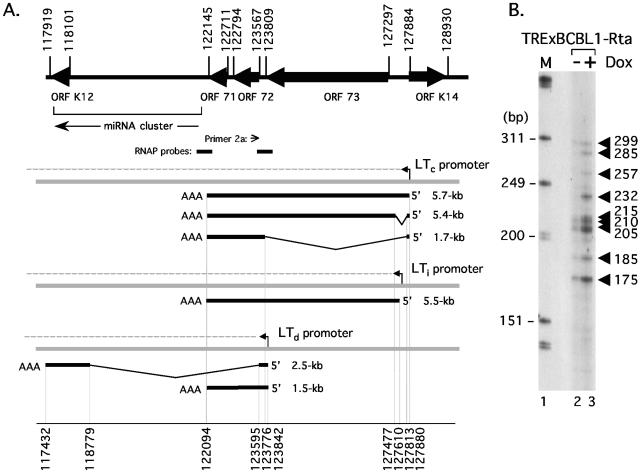
The genomic organization of the major latency cluster is shown in Fig. 1A. ORFs 71, 72, and 73 belong to a multicistronic transcriptional unit, known as the latency transcript (LT) cluster (14, 50). The LT cluster is transcribed from a constitutively active promoter (LTc) and through alternative splicing gives rise to a 5.4-kb mRNA containing ORFs 71, 72, and 73 and a 1.7-kb transcript containing ORFs 71 and 72. It is likely that LANA (ORF 73) is the principal translation product of the longer mRNA, whereas both v-Cyclin and v-FLIP are synthesized from the shorter transcript, the latter by way of an internal ribosome entry site upstream of ORF 71 (5, 6, 22, 34). Although not shown in the figure, a 1.1-kb monocistronic (ORF 71) transcript can also be detected in PELs at low abundance compared to the 1.7-kb mRNA and is derived from LTc by additional splicing (22). Expression of the K12 locus is unusual because translation initiates at non-methionine codons that are located within a repetitive and polymorphic sequence, giving rise to three different polypeptides (45). K12 is separated from the LT cluster by an ∼4-kb intergenic region that includes one of two origins of lytic DNA replication (2, 31). Recent studies have also revealed a cluster of microRNAs (miRNAs; see Fig. 1A) within this so-called intergenic region (9, 40, 46). Although their function is not yet known, all 11 miRNAs are encoded on the same strand and are expressed constitutively in latently infected PEL cells.
FIG. 1.
The latency gene cluster is expressed via a network of overlapping transcripts. (A) Organization of known ORFs, miRNAs, and mRNA transcripts within the major latency cluster of KSHV. Coordinates are based on the prototype BC-1 sequence (44) and correspond to initiation/termination codons (upper set) and exon sequences (lower set). A cluster of 11 miRNA genes, orientated in the direction indicated by an arrow, are located between the body of the K12 ORF (nucleotide 117,970) and the 3′ end of ORF 71 (nucleotide 121,911) (9, 40, 46). RNAP, RNase protection assay probe. A constitutively active promoter (LTc) gives rise to a precursor RNA (dotted line) that undergoes polyadenylation/cleavage and alternative splicing to produce ∼5.7-kb and ∼5.4-kb tricistronic (ORFs 71, 72, and 73) and ∼1.7-kb dicistronic (ORFs 71 and 72) mRNAs (14, 47, 50). Expression of the lytic activator RTA induces a second promoter (LTi) located between ORF 73 and LTc, giving rise to a 5.5-kb mRNA spanning all three ORFs (35). A 2.3- to 2.5-kb spliced mRNA corresponding to ORF K12, encoding multiple isoforms of kaposin, is transcribed during latency from a promoter at the 3′ end of ORF 73 and is induced further during lytic replication (27, 45). Shorter K12 transcripts initiating at 118,758 have also been reported (45). ORF K14 is essentially silent during latency but strongly induced by RTA utilizing promoter elements shared by LTi (11, 25, 30, 35). (B) Primer extension analysis to detect transcripts initiating upstream of ORF 72. Poly(A)+ RNA was isolated from mock (−Dox; lane 2) or Dox-treated (+Dox; lane 3) TRExBCBL1-Rta cells and reverse transcribed in the presence of radiolabeled oligonucleotide primer 2a. Extension products were resolved on an 8% acrylamide-7 M urea denaturing gel and visualized by autoradiography. Lane 1, size marker (in base pairs) prepared from end-labeled HinfI-digested ΦX174 DNA (Promega).
Recent work from our laboratory has characterized a second promoter (LTi) that is located between LTc and the initiation codon of ORF 73 (35). In latent cells, LTi is inactive but can be induced by the lytic switch protein RTA encoded by ORF 50. Using a PEL-derived cell line (TRExBCBL1-Rta) that has been engineered to allow regulated expression of the lytic switch protein RTA in the presence of doxycycline (38), we observed a five- to sevenfold induction of both the 5.4- and 1.7-kb LT mRNAs, accompanied by a small but appreciable decrease in average transcript length to approximately 1.5 kb (35).
To explore the origins of these shorter transcripts, we performed primer extension analysis using a synthetic oligonucleotide (primer 2a) complementary to the 5′ end of ORF 72 (indicated in Fig. 1A). TRExBCBL1-Rta cells were treated with water (−Dox, Fig. 1B, lane 2) or 1 μg/ml doxycycline (+Dox, lane 3) for 18 h, and then poly(A)+ RNA was isolated using oligonucleotide-coated beads (Oligotex Direct; QIAGEN). Cells were cultured in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 20% Neugem serum (Gemini Bio-Products), 100 μg/ml hygromycin B (Invitrogen), 2 mM l-glutamine, and antibiotics. After elution from the resin, equal amounts of RNA were mixed with the 32P-labeled oligonucleotide primer 2a (5′-GGGCGGGTTATTGGCAGTTGCCAT-3′, corresponding to KSHV nucleotides 123,544 to 123,567) (44), annealed by incubation of the mixture at 58°C for 20 min, and then slow cooled to room temperature. The annealed primers were extended using avian myeloblastosis virus reverse transcriptase (RT; Primer Extension System; Promega) at 41°C for 1 h before the reaction was stopped by ethanol precipitation, and the extension products were resolved on an 8% acrylamide-7 M urea denaturing gel followed by autoradiography. At least nine bands were detected, ranging in size from 175 to 299 bp. Six of the products (175, 185, 205, 210, 215, and 299 bp) were clearly represented in the −Dox and +Dox RNA samples, whereas three others (232, 257, and 285 bp) were specific to the Dox-treated cells. Only the longest 299-bp product was consistent with the 1.7-kb spliced transcript initiating at LTc. The remaining extension products appeared to be too short to have initiated at LTc or LTi using the known splice donor or acceptor sites. From this result, we hypothesized that the smaller products were generated by an uncharacterized RNA splice or, alternatively, represented transcripts that had initiated within the intergenic region between ORF 72 and ORF 73 (see Fig. 2A).
FIG. 2.
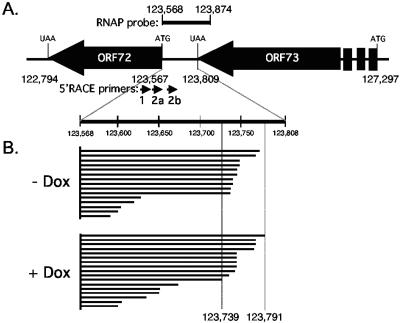
Characterization of transcripts initiating in the LTd region. (A) Schematic showing the intergenic region between ORFs 72 and 73. The probe used for RNase protection is shown above the ORF map and the primers used for 5′-RLM-RACE are shown below the map. For simplicity, all 5′-RLM-RACE products in panel B are depicted as originating from the common point of origin at nucleotide 123,568. (B) Summary of 5′-RLM-RACE products obtained from uninduced (−Dox) and induced (+Dox) TRExBCBL1-Rta cells using the KSHV-specific primers described for panel A. Twenty-one out of 38 5′-RLM-RACE products sequenced terminated in the highlighted region between nucleotides 123,739 and 123,791. An additional six clones (not shown) contained products corresponding to the splice from 123,776 to 127,813 and terminated at 127,880.
To distinguish between these two possibilities, we next performed 5′-RNA ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACE; Ambion) using primers from the same region of ORF 72 (Fig. 2A and B). In addition to primer 2a described above, we used two KSHV-specific primers: primer 1 (5′-ACATAGCGTGGGATCCAGAAGTCC-3′; nucleotides 123,517 to 123,540) and primer 2b (5′-TGGAGTTTCAGGGTGCTCT-3′; nucleotides 123,605 to 123,623). Primer 1 was used for the first round of nested PCR and then followed by either primer 2a or 2b in the second round. These were combined with primers complementary to an oligonucleotide RNA adaptor that was ligated to the 5′ end of the RNA template prior to reverse transcription. An important feature of the 5′-RLM-RACE protocol is that the adaptor is specifically ligated to the decapped 5′ end of the RNA, thereby excluding truncated products generated by RNA breakage or premature termination of the reverse transcriptase (33). 5′-RLM-RACE products were subcloned into pCR2.1Topo (Invitrogen) without selection for insert size, and 38 clones were chosen at random for further analysis. Upon DNA sequencing, the −Dox and +Dox samples yielded three clones each that contained 5′-RLM-RACE products corresponding to the previously described 1.7-kb spliced mRNA initiating at nucleotide 127,880 in LTc (Fig. 1A). The remaining 32 clones had captured extension products that terminated downstream of ORF 73, and these are shown schematically in Fig. 2B. Twenty-one clones (55% of the total) terminated within a 53-bp region (nucleotides 123,739 to 123,791) and were similarly represented in the −Dox and +Dox samples. The sheer variety of extension products obtained by 5′-RLM-RACE was surprising given that this methodology is specifically designed to amplify products that correspond to the 5′ ends of mRNAs only. However, the result was strongly reminiscent of the ladder of primer extension products, and nearly half of the primer extension products are the correct length to have initiated in the region between nucleotides 123,739 and 123,791. Both RNA preparations yielded products (two from −Dox samples, four from +Dox samples) that had apparently initiated at residue 123,757, suggesting this is a more frequently utilized start site. Five additional 5′-RLM-RACE products initiated within 6 bp on either side of this residue, consistent with a loose clustering of initiation sites. These results imply the presence of a constitutive promoter, which we will refer to as “LT downstream” (LTd), located within the intergenic region immediately downstream of the primary ORF 73 termination codon.
Both RACE and primer extension rely on the processivity of the RT enzyme, and we were concerned that the new initiation sites might be caused by premature termination during the RT step. To address this, we performed RNase protection analysis, an unrelated method that is not dependent on the processivity of a polymerase (Fig. 3A). A 373-nucleotide 32P-labeled antisense ribonucleotide complementary to KSHV nucleotides 123,568 to 123,874 (indicated Fig. 1A and 2A), together with some additional vector sequence, was transcribed in vitro using bacteriophage T7 RNA polymerase, mixed with 10 μg of total RNA prepared from induced and uninduced TRExBCBL1-Rta cells (Fig. 3A, lanes 3 and 4, respectively), and hybridized at 55°C overnight. As a negative control, we also hybridized the probe to 10 μg of total RNA isolated from KSHV-negative human HeLa cells (lane 2). HeLa cells were maintained in Dulbecco's modified Eagle medium (Gibco) supplemented with 2 mM l-glutamine and antibiotics. RNA duplexes were digested with a mixture of RNase A and RNase T1 for 30 min at 37°C to remove single-stranded probe, and the protected fragments were analyzed by 8% denaturing polyacrylamide gel electrophoresis. A series of protected probe fragments were observed with both of the TRExBCBL1-Rta RNA samples but not with the HeLa RNA. Approximate sizes of the protected products were estimated by comparison to a sequencing ladder (not shown) run alongside. A prominent protected fragment (estimated as 320 nucleotides in length) was detected in the TRExBCBL1-Rta samples but not in the HeLa sample. This is likely to correspond to the entire KSHV portion of the probe (307 nucleotides), which would be protected by the 5.4- to 5.7-kb tricistronic mRNAs. Compression of the sequencing ladder made it difficult to accurately calculate the sizes of the smaller products, and the numbers given in the figure represent relatively crude estimates. Regardless of the precise sizes, the majority of the smaller protected fragments were of an appropriate size for the 5′ ends of transcripts that had initiated within a region between KSHV nucleotides 123,737 and 123,793. Thus the series of bands detected by RNase protection appears to be in general agreement with the end points of LTd-derived transcripts mapped previously by primer extension (Fig. 1B) and 5′RLM-RACE (Fig. 2B). It is worth noting that the spliced 1.7-kb mRNA derived from LTc should generate a 209-nucleotide protected fragment (nucleotides 123,568 to 123,776) and would therefore also fall within the cluster of protected products.
FIG. 3.
RNase protection assays to characterize the 5′ and 3′ ends of ORF 71- and 72-associated transcripts. (A) A 373-nucleotide 32P-labeled antisense riboprobe (KSHV nucleotides 123,568 to 123,874 plus vector sequence) was transcribed in vitro using T7 RNA polymerase. Undigested probe is shown in lane 1. Probe was hybridized with 10 μg of total RNA from KSHV-negative human HeLa cells (lane 2), Dox-treated TRExBCBL1-Rta cells (lane 3), mock-treated TRExBCBL1-Rta cells (lane 4), BC-1 cells (lane 5), and BC3 cells (lane 6). After hybridization and RNase digestion, protected fragments were resolved on an 8 M urea-8% polyacrylamide gel and visualized by autoradiography. An open arrow indicates full-length probe. Fragment sizes were calculated from a DNA sequencing ladder run in adjacent lanes (not shown). (B) A 388-nucleotide antisense riboprobe (lane 1) corresponding to KSHV nucleotides 121,820 to 122,205 (plus 2 additional nucleotides), was hybridized to total RNA isolated from HeLa (lane 2) and Dox- or mock-treated TRExBCBL1-Rta cells (lanes 3 and 4).
RNase protection was also used to ask if similar LTd-derived transcripts could be detected in other KSHV-positive PEL cell lines. Total RNA was prepared from BC-1 (KSHV+ Epstein-Barr virus positive [EBV+]) (10) and BC3 (KSHV+ EBV−) (1) cells and hybridized to the same riboprobe. After digestion, both samples (lanes 5 and 6) yielded a similar ladder of protected fragments to the TRExBCBL1-Rta samples. The protected fragments were especially abundant in the BC3 sample (lane 6), consistent with previous reports of high levels of RNA and protein products from the LT gene cluster in this PEL line (12, 27, 41). This result shows that the transcripts at LTd are not peculiar to TRExBCBL1-Rta cells but can be readily detected in other PEL-derived cell lines.
Earlier studies of the LTs mapped the major site of polyadenylation to residues 122,070 and 122,066, less than 80 nucleotides downstream of ORF 71 (14, 43, 50). To map the 3′ end of the LTs, we constructed a 388-nucleotide 32P-labeled antisense riboprobe corresponding to genomic coordinates 121,820 to 122,205 (386 nucleotides, see Fig. 1A). This probe was transcribed directly from a PCR product that included the T7 promoter that added only 2 nucleotides of non-KSHV sequence. The labeled antisense riboprobe was hybridized to RNA from mock-treated and induced TRExBCBL1-Rta cells and processed as described above. Fewer protected species were observed compared to the 5′-end probe, and these consisted of three relatively abundant products in the 136- to 145-nucleotide range and a doublet at ∼300 nucleotides (Fig. 3B). The shorter products likely correspond to the previously mapped cleavage point that should give a 136-nucleotide product. The longer protected fragments might represent transcripts that have extended slightly further, ending near nucleotide 121,906 within the intergenic region. We also detected a weak signal corresponding to the full-length probe. This was absent in the HeLa sample, suggesting that it is a bona fide protected species signifying longer transcripts that extend the entire length of the probe. At least one miRNA is encoded with this protected region, and our mapping data do not preclude the possibility that primary transcripts extend across the entire intergenic region and beyond.
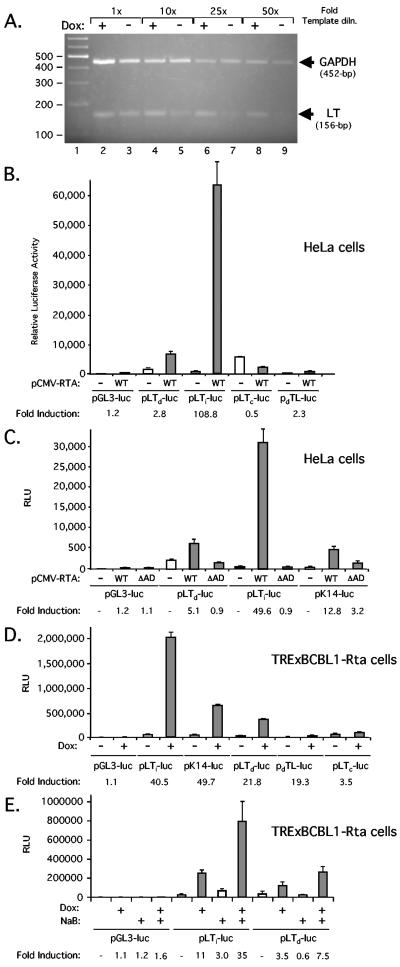
Our previous results indicated a five- to sevenfold increase in abundance of the ∼1.7-kb LT transcript following Dox treatment of TRExBCBL1-Rta cells (35). We have no evidence that the 5.5-kb transcript originating from LTi is spliced and suspect that the LTd promoter region might also be responsive to RTA. To verify this observation, we used semiquantitative RT-PCR to amplify the 5′ end of ORF 72 from poly(A)+ RNA isolated from TRExBCBL1-Rta cells that were either induced with Dox for 16 h or treated in parallel with water (Fig. 4A). RNA samples were treated with RNase-free DNase (Promega) to remove any contaminating viral DNA. Reverse transcription was carried out using AMV RT at 48°C for 45 min, and linear PCR amplification was achieved by using serial dilutions (1-, 10-, 25-, and 50-fold) of the resulting cDNA. The LT transcript primers were as follows: 123,434-GATTGTTGA AAGGTCCCAAA-123,453 and 123,589-CCACATACGCTCGCCACTCTA-123,569. The expected 156-bp product was obtained from both RNAs (lanes 2 and 3), and the dilutions confirmed that the template RNA was at least fivefold more abundant in the induced sample. To confirm that equal amounts of RNA were used in each pair of reactions, we included primers complementary to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-CCACATACGCTCGCCACTCTA-3′) (19) and yielded similar amounts of the expected 452-bp fragment.
FIG. 4.
LTd promoter can be induced by RTA. (A) RT-PCR analysis. Poly(A)+ mRNA was isolated from TRExBCBL1-Rta cells treated for 16 h with water (−Dox) or doxycycline (+Dox). PCR amplification was carried out using serial dilutions (1-, 10-, 25-, and 50-fold) of the cDNA with primers complementary to KSHV LT and human GAPDH, and the products were resolved on a 2% agarose gel stained with ethidium bromide. Size markers (lane 1) are shown in base pairs. (B) A total of 0.5 μg each of various reporter constructs was electroporated into 1 × 106 HeLa cells, together with 1 μg of an empty expression vector (−) or pCMV-RTA encoding full-length KSHV RTA (+). Luciferase activity was measured 24 h after transfection. Bars represent the mean and standard deviation of three independent transfections. (C) As in panel B, except that reporters were also cotransfected with pCMV-RTAΔAD, encoding a version of RTA truncated at residue 530 to remove the C-terminal activation domain. (D) A total of 1 × 107 TRExBCBL1-Rta cells were transiently transfected with 2 μg of the luciferase reporter construct indicated. After 24 h, the transfected cells were split into two flasks, and either mock or Dox treated. After a further 18 h, cell extracts were prepared and assayed for luciferase activity. (E) Addition of NaB amplifies the RTA response. Transfected TRExBCBL1-Rta cells were induced with Dox (1 μg/ml), NaB (3 mM), or a combination of both drugs for 16 h prior to harvest.
Next, we asked if a 700-bp genomic DNA fragment (nucleotides 123,568 to 124,267), encompassing the LTd initiation sites (centered on nucleotide 123,757) and sequences upstream, would be sufficient to drive expression of a luciferase reporter gene (Fig. 4B). The fragment was amplified from BC3 cell genomic DNA by high-fidelity PCR (Roche) and subcloned into a promoterless luciferase reporter plasmid (pGL3-basic; Promega) in either the correct (pLTd-luc) or incorrect (pdLT-luc) orientation relative to the luciferase cassette. Promoter activity of these two constructs was measured in transiently transfected HeLa cells and compared to that of the promoterless vector (pGL3-luc) or equivalent constructs containing the RTA-inducible LTi (pLTi-luc, nucleotides 127,609 to 127,807) (35) or constitutive LTc (pLTc-luc, nucleotides 127,816 to 129,375) promoters. Full-length RTA was supplied from a cotransfected cytomegalovirus (CMV) enhancer-driven expression plasmid (pCMV-RTA). When the LTd fragment was inserted in the correct orientation with respect to luciferase (pLTd-luc), it gave a significant increase in promoter activity over that of the empty vector (pGL3-luc), and this was increased further (2.8-fold) in the presence of RTA. Induction was modest compared to LTi, which displays very little constitutive activity but was induced by more than 100-fold by addition of RTA. In contrast, the activity of the constitutive promoter pLTc-luc was reduced by RTA. To ask whether induction of pLTd-luc was specific, the assay was repeated using a version of RTA lacking the activation domain (pCMV-RTAΔAD, Fig. 4C). Again pLTd-luc displayed significant constitutive activity, and this was boosted 5.1-fold by expression of the wild-type but not truncated RTA. As controls, we also tested the RTA-responsive LTi and K14 (pK14-luc, nucleotides 127,298 to 128,114) promoters and both were induced strongly by wild type RTA but not the truncation. Thus the small but significant induction of the LTd promoter was dependent on the RTA activation domain.
The same reporter set was tested in TRExBCBL1-Rta cells (Fig. 4D). A total of 1 × 107 cells were transfected with 2 μg of each reporter plasmid using Lipofectamine 2000, split into six separate dishes, and treated with either 1 μg/ml Dox or water for 24 h before being harvested and before luciferase activity was measured. With the exception of pdLT-luc, each reporter showed a significant activity compared to that of the promoterless plasmid but differed in terms of responsiveness to RTA. K14 was induced 49.7-fold, LTi by 40.5-fold, and LTd by 21.8-fold. Note that the apparent 19.3-fold increase observed with the reversed-orientation construct pdLT-luc is most likely an artifact of the barely measurable activity in the uninduced state. These results confirm that the 700-bp LTd fragment contains a constitutively active promoter that can be further induced by forced expression of RTA.
Finally, we asked if the deacetylase inhibitor sodium butyrate (NaB) could augment the response to RTA (Fig. 4E). Previously, we had found that treatment of TRExBCBL1-Rta cells with Dox and NaB for periods of less than 24 h leads to an elevated response from RTA-inducible promoters (35). As shown in Fig. 4E, NaB had no effect on the constitutive activity of LTd but increased the response to Dox from four- to eightfold. By comparison, the Dox-mediated induction of LTi went from 11-fold to 35-fold. This result confirms that the moderate stimulation of LTd in TRExBCBL1-Rta cells is most likely due to expression of RTA rather than a nonspecific effect of Dox.
In summary, our studies show that the major cluster of KSHV latency-associated genes (ORFs K12 to 73 and all 11 miRNAs) contains at least three separate promoters (LTc, LTi, and LTd), giving rise to a complex and interwoven network of mono-, di-, and tricistronic mRNAs. The LTd promoter described here is constitutively active, mirroring LTc, but transcription can be further boosted by expression of RTA, reminiscent of LTi. Induction may occur as a direct result of RTA transactivation or through indirect effects on the cellular transcription machinery. Several RTA-responsive promoters, including LTi, contain binding sites for the Notch-regulated transcription factor CSL (CBF1/RBP-Jκ) (29, 30, 35), and two candidate binding sites (123,704-GTGAGAA-123,710 and 124,167-TTTCCCA-124,173) are found in the LTd promoter fragment tested here.
Although initially characterized as a short 0.7-kb transcript, subsequent studies have shown that ORF K12 is expressed as an ∼2.5-kb spliced mRNA that initiates within the region designated LTd (27, 45). These K12 transcripts are very abundant in KS tumor samples and PEL lines but can be induced further by treatment with tetradecanoyl-phorbol-13-acetate, a potent inducer of RTA expression and lytic reactivation. Kaye and colleagues have reported that a genomic fragment spanning nucleotides 123,526 to 124,242 displays robust promoter activity in uninfected BJAB cells indicative of a constitutive promoter (27). Their fragment overlaps extensively with the LTd promoter fragment (nucleotides 123,568 to 124,267) characterized here, and it is likely the two studies characterize the same promoter, albeit one with an unusually broad spread of initiation sites. Primer 2a (coordinates 123,544 to 123, 567), used here for both primer extension and 5′-RLM-RACE, lies within the intron (118,779 to 123,594) of the K12 transcript described previously by Kaye and colleagues and highlights the fact that the LTd promoter gives rise to multiple mRNAs through alternative splicing as well as multiple initiation points. We favor the hypothesis that transcripts starting within LTd can give rise to two major mRNA species: the unspliced 1.5-kb transcript encoding ORFs 71 and 72 and the spliced 2.5-kb K12 transcript described by Kaye. The model requires that the polyadenylation signals immediately downstream of ORF 71, visualized by RNase protection in Fig. 3B, be ignored by a significant proportion of the transcripts. Consequently, read-through transcripts would transverse the miRNA cluster and terminate downstream of K12. Removal of the large intron that includes ORFs 71 and 72 would give rise to the known 2.5-kb mRNA encoding kaposin. This intron might serve as a source for the 10 of the 11 miRNAs. A less appealing possibility is that the K12 and ORF 71/72 transcripts might initiate from closely spaced but functionally distinct promoters, and only those transcripts initiating at the more distal promoter initiate splicing and terminate after the K12 coding sequences. Regardless of the precise details, our findings provide compelling evidence that levels of expression of ORFs 71, 72, and K12 are linked by usage of a common promoter region.
Although two or more major polyadenylation sites have been identified, it is not known where each of the primary transcripts initiating from LTd, LTi, or LTc terminate. In principle, all three promoters could contribute substrate RNAs for the production of the 11 known miRNAs (9, 40, 46). miRNAs are usually transcribed by RNA polymerase II and are often derived from capped and polyadenylated mRNAs. LTd lies immediately upstream of the miRNA cluster and would provide transcripts that can be processed into pre-miRNA hairpins. Estimates of the relative abundance of the mature miRNAs suggest that they are derived from one or more moderately active promoters and can be induced slightly by treatment with TPA (9). This behavior is also consistent with the observed properties of LTd.
Work by others has found that RNA interference (RNAi) knockdown of v-FLIP depletes v-Cyclin but not LANA and vice versa (21, 23). Although unusual RNA structures that insulate the RNAi effects or the existence of unknown RNA processing events there that convert the tricistronic precursor into monocistronic ORF 73 and dicistronic ORF 71/72 mRNAs have been proposed, a simpler explanation is that the LTd promoter generates sufficient levels of ORF 71/72 mRNA to compensate for destruction of the tricistronic precursor. Likewise, it has been shown that treatment of PEL cells with the triterpenoid glycyrrhizic acid leads to a marked down-regulation of the 5.4-kb transcript encoding LANA accompanied by significant up-regulation in the expression of 1.4- to 1.5-kb transcripts encoding v-FLIP or v-Cyclin transcripts (13). While this may reflect changes in splice site usage, it is equally possible that the drug selectively inhibits activity of the LTc promoter without affecting the LTd promoter. Reduction in promoter occlusion due to read-through transcripts from LTc may account for the increase in abundance of 1.4- to 1.5-kb transcripts derived from LTd.
Usage of multiple LT promoters provides the KSHV with a mechanism to adjust the relative levels of v-FLIP, v-Cyclin, and LANA. This may allow the virus to fine-tune the expression of each protein to fit the specific environment of a host cell or adapt to changing growth properties of a developing tumor. In situ hybridization studies of late-stage nodular KS lesions reveal significantly higher levels of bicistronic transcripts encoding ORF 71/72 compared to those encoding LANA (48). This can be explained by preferential usage of LTd in the tumor cells and is also consistent with the abundant expression of K12 RNA in the same cells. KS lesions are continually exposed to inflammatory cytokines such as tumor necrosis factor alpha and gamma interferon and are infiltrated by activated T cells, both of which provide strong proapoptotic signals (16). By selectively elevating the expression of antiapoptotic factors v-FLIP and v-Cyclin, KSHV is able to prevent the host cell from initiating programmed cell death without altering the proliferation and episome maintenance functions provided by LANA.
Acknowledgments
We thank John Souvlis and Jae Jung for sharing their TRExBCBL1-Rta cell line, Rolf Renne for sharing unpublished data and helpful discussions, and Francesca Curreli and Ornella Flore for providing BC-1 and BC3 cells. Naoko Tanese and Ian Mohr provided valuable comments on the manuscript.
This work was supported by NIH grant GM61139-04. S.M. is a postdoctoral fellow of the Rett Syndrome Research Foundation (RSRF).
REFERENCES
- 1.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol. 4:62-73. [PubMed] [Google Scholar]
- 5.Bieleski, L., C. Hindley, and S. J. Talbot. 2004. A polypyrimidine tract facilitates the expression of Kaposi's sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J. Gen. Virol. 85:615-620. [DOI] [PubMed] [Google Scholar]
- 6.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshoff, C., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52:453-470. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff, C., and R. Weiss. 2002. AIDS-related malignancies. Nat. Rev. Cancer 2:373-382. [DOI] [PubMed] [Google Scholar]
- 9.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 11.Chiou, C.-J., L. J. Poole, P. S. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J.-C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloutier, N., A. Gravel, and L. Flamand. 2004. Multiplex detection and quantitation of latent and lytic transcripts of human herpesvirus-8 using RNase protection assay. J. Virol. Methods 122:1-7. [DOI] [PubMed] [Google Scholar]
- 13.Curreli, F., A. E. Friedman-Kien, and O. Flore. 2005. Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. J. Clin. Investig. 115:642-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiorelli, V., R. Gendelman, M. C. Sirianni, H. K. Chang, S. Colombini, P. D. Markham, P. Monini, J. Sonnabend, A. Pintus, R. C. Gallo, and B. Ensoli. 1998. Gamma-interferon produced by CD8+ T cells infiltrating Kaposi's sarcoma induces spindle cells with angiogenic phenotype and synergy with human immunodeficiency virus-1 Tat protein: an immune response to human herpesvirus-8 infection? Blood 91:956-967. [PubMed] [Google Scholar]
- 17.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 18.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 19.Gillery, P., N. Georges, A. Randoux, F. Lefevre, F. X. Maquart, and J. P. Borel. 1996. Modulation of protein synthesis by extracellular matrix: potential involvement of two nucleolar proteins, nucleolin and fibrillarin. Biochem. Biophys. Res. Commun. 228:94-99. [DOI] [PubMed] [Google Scholar]
- 20.Godden-Kent, D., S. J. Talbot, C. Boshoff, Y. Chang, P. Moore, R. A. Weiss, and S. Mittnacht. 1997. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J. Virol. 71:4193-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey, A., J. Anderson, A. Papanastasiou, Y. Takeuchi, and C. Boshoff. 2004. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood 105:2510-2518. [DOI] [PubMed] [Google Scholar]
- 22.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199:993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Herndier, B., and D. Ganem. 2001. The biology of Kaposi's sarcoma. Cancer Treat. Res. 104:89-126. [DOI] [PubMed] [Google Scholar]
- 25.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliche, S., W. Nagel, E. Kremmer, C. Atzler, A. Ege, T. Knorr, U. Koszinowski, W. Kolanus, and J. Haas. 2001. Signaling by human herpesvirus 8 kaposin A through direct membrane recruitment of cytohesin-1. Mol. Cell 7:833-843. [DOI] [PubMed] [Google Scholar]
- 27.Li, H., T. Komatsu, B. J. Dezube, and K. M. Kaye. 2002. The Kaposi's sarcoma-associated herpesvirus K12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J. Virol. 76:11880-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M., H. Lee, D.-W. Yoon, J. C. Albrecht, B. Fleckenstein, F. Neipel, and J. U. Jung. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 71:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, Y., and D. Ganem. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 78:6818-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, L., M. T. Eby, N. Rathore, S. K. Sinha, A. Kumar, and P. M. Chaudhary. 2002. The human herpes virus 8 encoded viral FLICE inhibitory protein physically associates with and persistently activates the IkappaB kinase complex. J. Biol. Chem. 277:13745-13751. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., and M. A. Gorovsky. 1993. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 21:4954-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low, W., M. Harries, H. Ye, M.-Q. Du, C. Boshoff, and M. Collins. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 75:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumura, S., Y. Fujita, E. Gomez, N. Tanese, and A. C. Wilson. 2004. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50). J. Virol. 79:8493-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, N. Azumi, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muralidhar, S., G. Veytsmann, B. Chandran, D. Ablashi, J. Doniger, and L. J. Rosenthal. 2000. Characterization of the human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) oncogene, kaposin (ORF K12). J. Clin. Virol. 16:203-213. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ojala, P. M., K. Yamamoto, E. Castanos-Velez, P. Biberfeld, S. J. Korsmeyer, and T. P. Makela. 2000. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates Bcl-2. Nat. Cell Biol. 2:819-825. [DOI] [PubMed] [Google Scholar]
- 40.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 41.Platt, G. M., E. Cannell, M. E. Cuomo, S. Singh, and S. Mittnacht. 2000. Detection of the human herpesvirus 8-encoded cyclin protein in primary effusion lymphoma-derived cell lines. Virology 272:257-266. [DOI] [PubMed] [Google Scholar]
- 42.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 43.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sturzl, M., C. Hohenadl, C. Zietz, E. Castanos-Velez, A. Wunderlich, G. Ascherl, P. Biberfeld, P. Monini, P. J. Browning, and B. Ensoli. 1999. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 91:1725-1733. [DOI] [PubMed] [Google Scholar]
- 49.Sun, Q., H. Matta, and P. M. Chaudhary. 2003. The human herpes virus 8-encoded viral FLICE inhibitory protein protects against growth factor withdrawal-induced apoptosis via NF-kappa B activation. Blood 101:1956-1961. [DOI] [PubMed] [Google Scholar]
- 50.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, T., M. Sugaya, A. M. Atkins, E. A. Aquilino, A. Yang, D. L. Borris, J. Brady, and A. Blauvelt. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J. Virol. 77:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]