Abstract
Resting CD4+ T cells are the best-defined reservoir of latent human immunodeficiency virus type 1 (HIV-1) infection, but how the reservoir is formed is unclear. Understanding how the reservoir of latently infected cells forms is critical because it is a major barrier to curing HIV infection. The system described here may provide an in vitro model of latent HIV-1 infection in resting CD4+ T cells. We demonstrated that HIV-1 integrates into the genomes of in vitro-inoculated resting CD4+ T cells that have not received activating stimuli and have not entered cell cycle stage G1b. A percentage of the resting CD4+ T cells that contain integrated DNA produce virus upon stimulation, i.e., are latently infected. Our results show that latent HIV-1 infection occurs in unstimulated resting CD4+ T cells and suggest a new route for HIV-1 reservoir formation.
The introduction of highly active antiretroviral therapy (HAART) in the United States led to impressive declines in human immunodeficiency virus (HIV)-related morbidity and mortality (20, 23, 55). The two-phase viral decay kinetics observed in patients receiving HAART suggested that eradication of HIV might be possible (56). Indeed, patients treated successfully with HAART achieved undetectable levels of viremia. However, in almost every patient, when successful drug therapy was stopped, viremia recurred (3, 57).
HIV type 1 (HIV-1) infection remains incurable because reservoirs of latently infected cells exist. The latent virus in reservoirs is not susceptible to antiretroviral therapy or host immune responses (3, 57). Resting CD4+ T cells are the best-defined reservoir of latently infected cells. In HIV-1-infected individuals, a small percentage (0.01%) of resting CD4+ T cells isolated from blood contain integrated DNA (3, 57). However, these cells do not produce new virions constitutively (42, 43), and only a very small percentage (≤0.0001%) are latently infected, i.e., produce new virions when stimulated (3, 57). Because of the low percentage of latently infected resting CD4+ T cells in vivo, it has been difficult to study HIV-1 reservoir formation.
It is unclear how HIV-1 reservoirs form in resting CD4+ T cells. A central question is what role does T-cell stimulation play in the establishment of latent HIV-1 infection and reservoir formation? One hypothesis is that reservoirs form when HIV-1-infected activated T cells return to a resting state (3, 57). A related hypothesis is that HIV-1-infected resting CD4+ T cells receive transient activating stimuli that allow integration to occur (66, 70, 72).
The prevailing belief is that HIV-1 does not integrate into unstimulated resting CD4+ T cells (3, 57, 66). This belief is based on results from several early experiments. First, reverse transcription is very inefficient in resting CD4+ T cells (63, 76, 77). Furthermore, nuclear import (68) and integration (67) are not detected in HIV-1-inoculated resting CD4+ T cells unless the cells are activated to enter the cell cycle. Progression to cell cycle stage G1b enhances the efficiency of reverse transcription (39) and results in productive infection (16), suggesting that entry into G1b is required for integration to occur. Finally, in HIV-1-infected individuals, proviruses are enriched among resting CD4+ T cells with a memory phenotype (11, 53), implying that prior activation enables integration to occur.
Our hypothesis is that HIV-1 can integrate into resting CD4+ T cells in the absence of activating stimuli. Previously, we measured the kinetics of reverse transcription in HIV-1-inoculated resting CD4+ T cells (69) and found, as have others (58, 63), that reverse transcription occurs inefficiently in resting T cells; however, we also found that the long reverse transcripts in resting T cells are more stable than those in activated T cells (69). The presence (58, 63) and stability (69) of long reverse transcripts in resting CD4+ T cells led us to hypothesize that HIV-1 could integrate into resting CD4+ T cells. Consistent with our hypothesis, HIV-1 RNA production is detected in CD45RA+ CD4+ T cells in HIV-1-infected individuals (78) and in HIV-infected lymphoid organ cultures (17). The expression of CD45RA, a marker for naïve cells (14), suggests that cellular activation may not be necessary for viral production and hence for proviral integration.
Here we showed that HIV can integrate into resting CD4+ T cells in the absence of activating stimuli. A percentage of the resting T cells that contain integrated DNA produce HIV-1 when stimulated. Therefore, latent HIV-1 infection occurs in resting (G0/1a) CD4+ T cells. These results suggest that our in vitro system may provide a model for HIV-1 latency.
MATERIALS AND METHODS
Cells.
CEMss cells were cultured in 10% fetal calf serum in RPMI plus 1% penicillin-streptomycin. CD4+ T cells were cultured in 10% autologous serum in RPMI with 1% penicillin-streptomycin at 5 × 106/ml after spinoculation. The integration standard was prepared as described elsewhere (51), except the cultures were maintained in efavirenz to prevent wild-type recombinant virus from replicating. The integration standard cell line was cultured for only 4 weeks after transduction before preparing genomic DNA to minimize the amount of linear DNA present (confirmed by Southern blotting) and to maximize the diversity of integration sites.
Peripheral blood mononuclear cells (PBMCs) were prepared as described previously (69) and stained with saturating concentrations of the following fluorescein isothiocyanate (FITC)-labeled antilineage (CD8, CD14, CD16, CD20, and CD56) monoclonal antibodies and phycoerythrin-labeled antiactivation (CD69 and HLA-DR, as well as CD25 in indicated experiments) markers (BDIS, San Diego, Calif.). CD4+ T cells were sorted into two populations based on the presence or absence of activation markers on a Becton Dickinson FACSDiVa SE ultra-high-speed digital flow cytometer. A lineage-negative population that expressed no activation markers was labeled resting CD4+ T cells, and a lineage-negative population that expressed high levels of activation markers was labeled endogenous activated CD4+ T cells.
Monitoring cells for activation.
Sort-purified resting CD4+ T cells were stained with saturating quantities of anti-CD69 and HLA-DR (and CD25 in specified experiments) before and after culture and spinoculation. Cells were monitored for cell cycle status before and after culture and spinoculation by DNA and RNA staining as described elsewhere (69). In vitro-activated T cells were prepared by stimulating PBMCs for 4 days in the presence of 50 μg/ml staphylococcal enterotoxin B (SEB; Sigma). These cells were used to define G2 of the cell cycle. The same cells treated with 10 μM aphidicolin defined G1b. The same cells stimulated for 1 day with SEB and 2.5 mM sodium butyrate defined G1a. Aphidicolin prevents entry into S phase. Sodium butyrate inhibits entry into G1b.
Viruses and vectors.
293T cells were transfected with pIIIB (NG38). The transfection supernatants were passaged in CEMss cells to diminish the amount of input plasmid DNA. Viral supernatants with peak reverse transcriptase activity were treated with DNase at 30 μg/ml (Roche Molecular Biochemicals, Indianapolis, Ind.) for 2 h at 25°C. One stock of virus with 1 μg/ml p24 was used in the experiments shown in all figures except Fig. 6, below. In Fig. 6 a different viral stock was used, but it was diluted to match the potency of the viral stock used in the experiments shown in Fig. 1 to 5, below, resulting in an integration level of 0.1 proviruses/cell. Similar results were found with other viral stocks, including pNL4-3 and pYU-2.
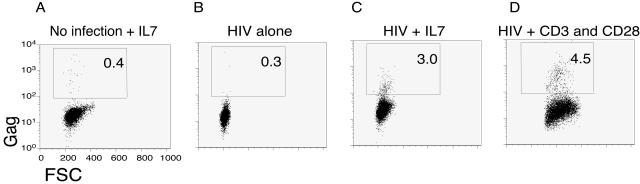
FIG. 6.
A fraction of resting CD4+ T cells bearing integrated HIV-1 DNA was latently infected. In each plot, the percentage of events in the boxed region is shown. The experiments were performed in the presence of the protease inhibitor saquinavir to prevent spreading infection. (A) IL-7 alone. Uninfected cells treated with IL-7 showed minimal background staining with anti-Gag antibodies. Uninfected resting CD4+ T cells were cultured for 3 days and then stimulated with IL-7 for an additional 3 days. This was a control for background staining with the anti-Gag monoclonal antibody. Isotype control antibodies gave less background staining than anti-Gag antibodies. (B) HIV alone. Unstimulated HIV-inoculated cells stained minimally with anti-Gag antibodies. Resting cells were spinoculated with HIV and 6 days later were permeabilized and stained for intracellular Gag. (C) HIV plus IL-7. IL-7 stimulated 3.0% of HIV-inoculated cells to produce Gag. Resting cells were spinoculated with HIV, 3 days later they were stimulated with IL-7, and 3 days after IL-7 addition they were permeabilized and stained for intracellular Gag. Gag was measured 3 days after IL-7 stimulation because Gag production peaked at that time. (D) HIV plus CD3 and CD28 beads. Anti-CD3 and -CD28 beads stimulated 4.5% of HIV-inoculated cells to produce Gag. Resting cells were spinoculated with HIV, 3 days later they were stimulated with anti-CD3 and -CD28 beads in the presence of the integrase inhibitor L870,810, and 24 h later they were permeabilized and stained for intracellular Gag. Gag was measured 24 h after bead stimulation because Gag production peaked at that time. We demonstrated that our integrase inhibitor was effective by inoculating resting CD4+ T cells and culturing them in the presence of the integrase inhibitor L870,810 at 100 nM. Under these conditions, we saw no reduction in the frequency of reverse transcripts, but we found that the frequency of proviruses/cell was reduced by 96.7%.
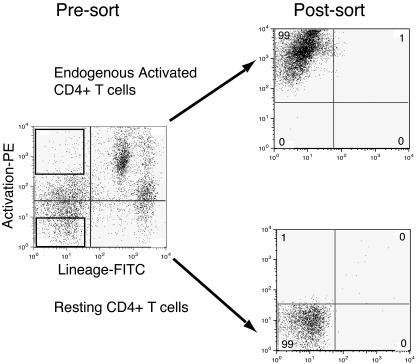
FIG. 1.
Purification of resting and endogenous activated CD4+ T cells. Mononuclear cells were stained with a cocktail of monoclonal antibodies against lineage-specific antigens, including FITC-labeled anti-CD8, -CD14, -CD16, -CD20, and -CD56. Simultaneously, the cells were stained with phycoerythrin-labeled monoclonal antibodies against the activation markers CD69 and HLA-DR. The gates used to define the two populations are shown on the presort panel. Postsort analysis showed that endogenous activated CD4+ T cells expressed high levels of activation markers and resting CD4+ T cells did not express detectable levels of activation markers. To determine the purity of the resting CD4+ T cells, the quadrants were set so that 99% of unstained lymphocytes were in the lower left quadrant. The sorting gates were set conservatively to ensure high purity. We consistently found that only 1% of the resting T cells were contaminants and 1% of the activated cells were contaminants. Dendritic cells copurified with the endogenous activated T cells. We chose not to remove dendritic cells from the activated T cells. We reasoned that the activated CD4+ T cells were more likely to remain activated in the presence of dendritic cells. The resting CD4+ T cells did not contain dendritic cells.
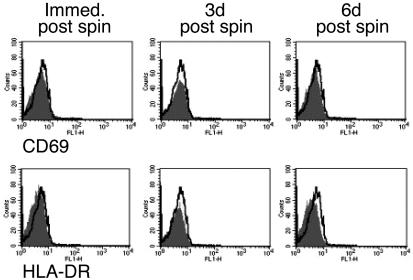
FIG. 5.
Spinoculation and culture did not induce resting CD4+ T cells to express HLA-DR or CD69. Sort-purified resting CD4+ T cells were stained with anti-CD69 or anti-HLA-DR-FITC immediately after spinoculation (immed. post spin), 3 days after spinoculation (3d post spin), and 6 days after spinoculation (6d post spin). The gray boxes represent the staining seen with anti-HLA-DR and anti-CD69 antibodies. The black line represents the staining obtained with an IgG1 isotype control antibody immediately after sorting. CD69 expression was also not detected at 2, 6, and 20 h after spinoculation (data not shown). The data are representative of three experiments.
Inoculations.
Spinoculation was performed as described previously (52), except resting CD4+ T cells were resuspended at 2 × 107cells/ml in viral supernatant. Viral cell suspensions of 0.4 ml were spun in a 24-well plate at 1,200 × g for 2 h at 25°C. After spinoculation the cells were washed three times and resuspended in 10% autologous serum in the presence of 1.25 μM saquinavir (Roche US Pharmaceuticals). For comparison, cells were inoculated with the same viral supernatant at the same cell concentration at atmospheric pressure (1 g).
Assays for viral DNA intermediates.
DNA was prepared immediately after inoculation and at multiple time points after culture using a QIAmp blood and cell culture kit (QIAGEN, Valencia, Calif.). Zidovudine at 10 μg/ml and L870,810 at 100 nM (Merck Research Laboratories, West Point, Pa.) were used to inhibit reverse transcription and integration (26, 27) where indicated.
Kinetic PCR was performed for strong stop (RU5) to detect short reverse transcripts and β-globin as described previously (69). Second strand transfer to detect long reverse transcripts was measured using the following forward and reverse primers and molecular beacon: 5′GCTAGCTAGGAAACCCACTGC TTA3′, 5′-CTGCGTCGAGAGAGCTCCTCTGGTT-3′, and 5′-6-carboxyfluorescein-GCGAGTCACACAACAGACGGGCACACACTACTCGC-4-(4′-di- methylamino-phenylazo)-benzene-3′(Midland Certified Reagent Co., Midland, Tex.). Reactions were carried out in 50-μl volumes containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 5.0 mM MgCl2, 250 μM (each) dATP, dCTP, dGTP, and dTTP (Promega, Madison, Wis.), 1,000 nM forward and reverse primers, 200 nM probe, 0.025 U of Platinum Taq (Invitrogen, Carlsbad, Calif.), and 500 nM carboxy-X-rhodamine as a passive reference (Molecular Probes, Eugene, Oreg.). Cycling conditions were 95°C for 2 min and then 40 cycles of 95°C for 15 s, 56°C for 30 s, and 72°C for 1 min.
Integrated DNA was measured by Alu-PCR as described elsewhere with minor modifications (51). The assay included two nested PCR amplification steps. The first step used primers to Alu and gag. The second step used internal primers to RU5 and a molecular beacon to detect the amplicons kinetically. Controls included performing the first PCR step with only the gag primer (linear preamplification of DNA) as well as performing the first step without any primers (no preamplification of DNA) as described elsewhere (51). The gag primer control monitors the signal contributed from unintegrated DNA. The integration standard was diluted in 2 μg/ml uninfected PBMC DNA to keep the number of Alu sites per reaction mixture constant. Samples were always assayed at two concentrations in triplicate to demonstrate that PCR inhibitors were absent. Dilutions were performed in a constant amount of uninfected PBMC DNA (2 μg/ml) again to keep the number of Alu sites per reaction mixture constant.
Assay for viral binding.
The amount of p24gag associated with the cells at the end of inoculation was measured by enzyme-linked immunosorbent assay (ELISA; Coulter Corporation, Miami, FL). The p24 ELISA does not detect p55. The amount of p24 capsids associated with the cells at the end of the inoculation was used to estimate the number of virions bound, as previously described (52). The calculation assumes ∼1,500 p24 capsids present per viral particle. The number of virions bound at the end of the inoculation may be an overestimate, as the viral supernatant may contain virus-like particles that lack viral RNA.
Resting CD4+ T cells were stimulated to produce new virions with 20 ng/ml interleukin-7 (IL-7; Sigma, St. Louis, Mo.) or with artificial beads coated with anti-CD3 and anti-CD28 as described elsewhere (8, 9, 59). The percentage of cells expressing Gag was measured by intracellular staining for Gag. Cells were fixed with 1% paraformaldehyde for 10 min, washed, and stained in permeabilization buffer (2% fetal calf serum, 25 μg/ml murine immunoglobulin G [IgG; BDIS], 0.5% Saponin [Sigma]) with phycoerythrin-labeled KC57 (Beckman Coulter Immunology Systems, Miami, Fla.). This IgG1 antibody recognizes p55, p39, p33, and p24 proteins of the core antigens of HIV-1.
RESULTS
Our sort strategy.
We set out to develop a strategy that would (i) purify resting CD4+ T cells without activating them during selection and (ii) control for the percentage of contaminating activated cells in our resting population. CD4+ T cells were sorted to be negative for lineage markers CD8, -14, -16, -20, and -56. In addition, the cells were sorted into two populations based on the presence or absence of activation markers HLA-DR and CD69.
The two sorted populations were called endogenous activated CD4+ T cells and resting CD4+ T cells. The endogenous activated cells expressed high levels of activation markers, while the resting cells did not express detectable levels of activation markers (Fig. 1, presort). In order to measure the percentage of contaminating cells, we analyzed the purity of the two populations postsort. To determine the percentage of cells that lacked activation and lineage markers, we used unstained lymphocytes to set the quadrants in Fig. 1. Postsort analysis showed that 1% of the endogenous activated population were contaminants and that 1% of the resting population were contaminants (Fig. 1, postsort). Most of the contaminants in the resting population were not strongly activated cells but were intermediate cells that expressed low levels of activation markers (Fig. 1, postsort). Since both resting and endogenous activated populations were contaminated with intermediate cells, in some experiments we also sorted for cells that expressed low levels of activation markers.
The postsort data demonstrated that our sort strategy largely removed contaminating cells from the resting CD4+ T-cell population. As shown in later experiments, our sort strategy allowed us to measure the level of HIV-1 integration in endogenous activated T cells. Therefore, we could measure the signal contributed by contaminating activated cells in the resting cells.
In early experiments CD4+ T cells were negatively selected against the activation marker CD25, in addition to HLA-DR and CD69. We compared HIV-1 integration frequency in CD25− HLA-DR− CD69− CD4+ T cells to that in HLA-DR− CD69− CD4+ T cells and found no difference. As a result, we performed the majority of experiments with HLA-DR− CD69− CD4+ T cells. When we used CD25− HLA-DR−, and CD69− CD4+ T cells, that fact is indicated in the text and/or figure legend.
Reverse transcription in CD4+ T cells.
We compared the extent of reverse transcription in resting and endogenous activated CD4+ T cells. The two populations of primary CD4+ T cells were spinoculated with HIV-1IIIB and washed and cultured for 3 days, and DNA was prepared for reverse transcription assays. Reverse transcription was measured on postinoculation day 3 because reverse transcription plateaus at this time in resting CD4+ T cells (58, 69).
Reverse transcripts (Fig. 2) were detected at slightly different levels in the two primary T-cell populations. The activated population contained twofold more reverse transcripts per cell than the resting population (Fig. 2). Consistent with prior work (69), 100-fold more reverse transcripts were detected in an activated T-cell line, CEMss, at 18 h postinoculation (peak of reverse transcription in activated CEMss T-cell line) compared to resting T cells at 3 days postinoculation (peak of reverse transcription in resting CD4+ T cells). Reverse transcription, entry, or a combination of both occurred much more efficiently in the activated T-cell line CEMss than in endogenous activated T cells. For example, we measured 50 reverse transcripts per cell in CEMss cells compared to 0.8 reverse transcripts per cell in endogenous activated T cells (Fig. 2). We found that the number of reverse transcripts per cell in the CEMss cell line is similar to the number in artificially (e.g., phytohemagglutinin, CD3+ CD28 beads, SEB) activated T-cell blasts (69).
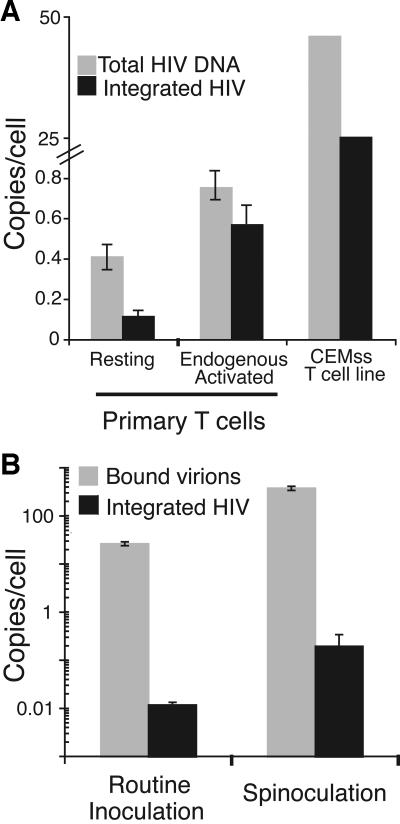
FIG. 2.
HIV-1 integrated into resting CD4+ T cells. (A) The relative levels of total DNA and integrated DNA in the two populations of primary T cells at 3 days postinoculation are shown. Total DNA was measured by primers that detect second strand transfer. Integrated DNA was measured by Alu-PCR (51). The relative levels of total DNA and integrated DNA for the activated T-cell line, CEMss, at 18 h postinoculation are also shown. Immediately after spinoculation only very low levels of total DNA (<0.001 copies/cell) and no integrated DNA were detected (not shown), indicating that our viral supernatants contained minimal amounts of contaminating viral DNA from the initial transfection. Comparing the resting and endogenous activated CD4+ T cells, there was a significant difference in the amount of total DNA (P < 0.02) and integrated DNA (P ≪ 0.01) by Student's t test. Similar results were obtained with HIV-1YU2, a CCR5-tropic virus, and pNL4-3, another CXCR4-tropic virus (not shown). The data are representative of four experiments. (B) Spinoculation did not induce integration. Binding and integration were increased proportionally by spinoculation. Resting CD4+ T cells were inoculated under routine and spinoculation conditions and washed to remove unbound virions, and the number of bound virions per cell was measured as described elsewhere (52). The number of integrated proviruses per cell was measured 3 days after inoculation as described previously (51). The data are representative of three experiments.
HIV-1 integrated into resting CD4+ T cells.
We compared the extent of integration between resting and endogenous activated CD4+ T cells. In the same experiment as above, the cells were spinoculated with HIV-1IIIB and washed and cultured for 3 days, and DNA was prepared to measure integration. On average, 1 in 10 cells contained integrated DNA. Our HIV-1 integration assay included two nested PCR amplification steps (51). The first step used primers specific for the repetitive element Alu and HIV-1 gag. The second step was kinetic and HIV-1 specific (51). We previously demonstrated that our assay is highly sensitive and specific for integration (51).
Surprisingly, integrated DNA (Fig. 2) was detected in resting CD4+ T cells 3 days after inoculation. The endogenous activated population contained fivefold more integrated DNA than the resting population (Fig. 2). Fifty-fold more proviruses were detected in CEMss cells than in endogenous activated T cells (Fig. 2). In both CEMss and endogenous activated T cells, the majority of reverse transcripts are integrated, while in resting T cells a minority (∼1 in 5) are integrated.
Contaminating activated cells could not account for the integration signal in resting cells. In order for contaminating activated cells to account for the integration signal in the resting population, 20% of the “resting” cells would need to be strongly activated CD4+ T cells. However, in the sort strategy we set conservative gates for the resting and activated populations so that there would be little cross-contamination between the populations. Indeed, postsort analysis confirmed that there was very little cross-contamination—the resting population contained only 1% activated cells (Fig. 1, postsort).
It was possible that intermediate CD4+ T cells, cells that expressed low levels of activation markers, contributed to the integration signal in resting cells. To test this hypothesis, we sorted the intermediate cells, inoculated them with HIV-1, measured the level of integration, and compared it to the level of integration in resting cells. Intermediate and resting cells had similar integration levels (0.11 ± 0.04 proviruses/cell in intermediate cells and 0.11 ± 0.06 proviruses/cell in resting cells in one representative experiment). Therefore, intermediate cells could not account for the integration signal in resting cells. In addition, depleting CD25+ cells from the resting population did not change the level of integration. CD25− HLA-DR− CD69− CD4+ T cells had the same integration level as HLA-DR− CD69− CD4+ T cells (0.10 ± 0.06 provirus/cell for CD25− HLA-DR− CD69− versus 0.13 ± 0.02 for HLA-DR− CD69−).
Effect of spinoculation on T-cell susceptibility to integration.
We previously demonstrated that, compared to routine inoculation (1 g), spinoculation (1,200 × g) increased viral binding to resting CD4+ T cells by ∼20-fold and increased binding and reverse transcription to similar extents in activated T-cell lines (52). Here we wanted to determine whether spinoculation made cells more susceptible to HIV-1 integration. We inoculated resting CD4+ T cells under routine and spinoculation conditions and then measured binding and integration frequencies. If spinoculation made the cells more susceptible to integration, it would increase integration to a greater extent than binding. We found that spinoculation increased binding and integration to similar extents (Fig. 2B). Thus, spinoculation did not appear to induce HIV-1 integration.
Kinetics of reverse transcription and integration were delayed in resting CD4+ T cell.
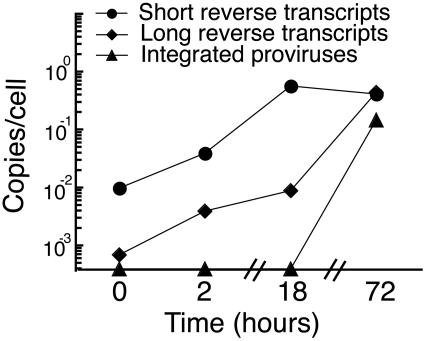
We and others have shown that the kinetics of reverse transcription are delayed in resting CD4+ T cells (10, 58, 63, 69). Here we measured the kinetics of reverse transcription again in order to compare the kinetics of integration relative to reverse transcription. To do this, we spinoculated resting CD4+ T cells, prepared DNA at multiple time points, and measured both the number of reverse transcripts and proviruses per cell.
Short reverse transcripts (Fig. 3) rapidly accumulated in resting CD4+ T cells, while long reverse transcripts (Fig. 3) accumulated over the 3-day period. In contrast, reverse transcription in endogenous activated T cells (not shown) reached a plateau by 18 h postinoculation. The difference in reverse transcription kinetics was one parameter that distinguished resting CD4+ T cells from endogenous activated CD4+ T cells.
FIG. 3.
Reverse transcription and integration have delayed kinetics in resting CD4+ T cells. The number of copies per cell of short reverse transcripts and long reverse transcripts are shown at several time points after inoculation. Short reverse transcripts were detected using RU5-specific primers. Long reverse transcripts were detected using primers specific for reverse transcripts that form after second strand transfer. Integrated provirus is shown at several time points after inoculation. The cells were cultured in the presence of the protease inhibitor saquinavir to prevent spreading infection. When we inoculated resting CD4+ T cells with HIV and cultured in the presence of the integrase inhibitor L870,810 at 100 nM, the integration level was reduced by 96.7% while the level of reverse transcripts was unchanged, demonstrating the specificity of our integration assay. When we inoculated and cultured the cells in the presence of zidovudine at 10 μg/ml, the number of reverse transcripts was reduced by >99%, demonstrating that our viral supernatant contained minimal amounts of contaminating viral DNA. The error bars (standard deviations) are obscured by the symbols. The coefficient of variation ranged from 3 to 17% in this experiment.
Integration kinetics were also delayed in resting CD4+ T cells. Integrated provirus (Fig. 3) was not detected in resting CD4+ T cells at 18 h postinoculation but was detected at 3 days postinoculation. Low levels of integration in resting CD4+ T cells were detected at 48 h postinoculation (not shown). The delayed integration kinetics in resting CD4+ T cells were consistent with the slower kinetics of reverse transcription in these cells (69) (Fig. 3). By comparison, integration in endogenous activated T cells was detected by 18 h postinoculation (not shown). The difference in integration kinetics was another parameter that distinguished resting CD4+ T cells from endogenous activated CD4+ T cells.
Effects of culture and spinoculation on T-cell activation.
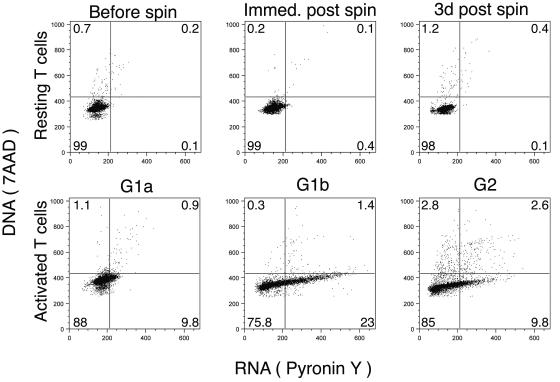
It was possible that culture and spinoculation activated resting CD4+ T cells. We assayed the activation status of cultured, spinoculated cells in several ways. First, using DNA/RNA fluorescence-activated cell sorter (FACS) analysis (Fig. 4), we showed that the cell cycle status did not change before spinoculation, immediately after spinoculation, and 3 days after spinoculation. Second, the cells remained negative for HLA-DR and CD69 expression immediately postspinoculation, at 2, 6, and 18 h postspinoculation, and at 3 and 6 days postspinoculation (Fig. 5 and data not shown). We assayed for CD69 expression at multiple time points because CD69 is one of the first markers to upregulate after cellular activation (13, 44). Third, when CD25− HLA-DR− CD69− cells were spinoculated and cultured, CD25 did not upregulate (not shown). Fourth, spinoculated cells did not take up bromodeoxyuridine for the duration of the experiment (not shown). We concluded that culture and spinoculation did not appear to activate resting CD4+ T cells.
FIG. 4.
Spinoculation and culture did not induce resting CD4+ T cells to enter the cell cycle. Data are plotted as DNA (7AAD) on the y axis versus RNA (pyronin Y) on the x axis. Resting CD4+ T cells were fixed at various time points: immediately before spinoculation (before spin), immediately after spinoculation (immed. post spin), and 3 days after spinoculation (3d post spin). Cell cycle stages were defined by activating T cells by treating PBMCs with SEB in the presence of the inhibitor sodium butyrate for 1 day (G1a), in the presence of aphidicolin for 4 days (G1b), or in the absence of an inhibitor for 4 days (G2). To demonstrate that cells did not enter the cell cycle after spinoculation, quadrants were set using CD4+ T cells before spinoculation. The data are representative of three experiments.
HIV-1-inoculated resting CD4+ T cells were latently infected.
We tested whether resting CD4+ T cells that contained integrated HIV-1 provirus (on average ∼1 in 10 cells) were latently infected, i.e., were capable of producing new virus upon stimulation. We spinoculated resting cells, exposed the cells to different stimuli, and then measured intracellular Gag as a marker for new virus production. When we stimulated HIV-1-inoculated resting CD4+ T cells (CD25− HLA-DR− CD69−) with anti-CD3 anti-CD28 beads, 4.5% of the cells produced Gag, i.e., were latently infected (Fig. 6D). When we stimulated HIV-1-inoculated resting CD4+ T cells with IL-7, approximately 3% of the cells produced Gag (Fig. 6C). We obtained similar results when we treated the cells with IL-15 (similar to IL-7), but treatments with IL-2 and with tumor necrosis factor alpha plus IL-6 were less effective at stimulating new virus production (not shown). We concluded that a small percentage of HIV-1-inoculated resting CD4+ T cells were latently infected.
We wanted to demonstrate that HIV-1 integration occurred in resting cells prior to, not as a result of, cellular stimulation. Therefore, we performed CD3-CD28 costimulation in the presence of an integrase inhibitor (diketo-acid L870,810) (26). Under these conditions, new virus production was still detected, demonstrating that integration occurred in resting CD4+ T cells prior to cellular stimulation (Fig. 6D). The concentration of L870,810 used (100 nM) reduced the level of integration by 99% in activated T cells and by 97% in resting CD4+ T cells. The same concentration of the integrase inhibitor decreased productive infection of CD3-CD28-activated T cells, as measured by intracellular p24 FACS, from 10% to 0.2% production, effectively blocking productive infection (data not shown).
DISCUSSION
Our results support two major conclusions: (i) HIV can integrate into primary, resting (G0/1a) CD4+ T cells, and (ii) a percentage of resting T cells that contain integrated DNA are latently infected. The prevailing belief is that entry into cell cycle stage G1b is necessary for HIV-1 integration. This belief comes from the observation that HIV-inoculated resting T cells produce new virions when stimulated, through various T-cell activating pathways, to enter cell cycle stage G1b (2, 5-7, 12, 16, 19, 36, 54, 72). Here, we demonstrated, using a sensitive kinetic PCR assay, that HIV-1 can integrate into G0/1a resting CD4+ T cells and that upon stimulation a fraction of the cells containing integrated DNA produce HIV. Thus, we showed that HIV-1 can establish latent infection in resting (G0/1a) CD4+ T cells. These results suggest that our in vitro system provides a new model of latent HIV-1 infection. We speculate that in vivo reservoirs may form in the absence of T-cell activation.
Our in vitro latency model parallels important aspects of in vivo HIV-1 latency. First, in our model resting CD4+ T cells did not enter the cell cycle for the duration of the experiment. Similarly, in vivo resting CD4+ T cells do not enter the cell cycle for long periods of time (with half-lives on the order of months) (46, 48, 64, 71). Second, in our model a minority of reverse transcripts integrated. In vivo a minority (albeit a smaller minority) of reverse transcripts integrate (11, 53). Third, in our model a fraction of HIV-1-inoculated resting CD4+ T cells were latently infected. Similarly, a fraction (albeit a smaller fraction) of in vivo-infected resting T cells are latently infected.
Our in vitro system differs from in vivo infection in that there is a higher level of integration and latent infection. We speculate that the higher level of infection is due to a higher level of input virions and a lower mutation frequency in our system. We achieve a higher level of input virions by using spinoculation. The higher level of input virions may result in a higher level of integration by overcoming a restriction factor (18) that blocks entry, reverse transcription, nuclear import, or integration. Increased levels of fused virions could potentially saturate such a cellular restriction factor. In addition, the enhanced level of latently infected cells may be due to a lower mutation frequency in our system. In vivo fewer proviruses may be capable of production due to a higher rate of defective (mutated) proviruses. The cytidine deaminases APOBEC-3F and APOBEC-3G (1, 25, 62) each play a role in mutation in vivo (29, 35, 73), but these mutations are rare in our system (10), as our virions are prepared from cells that express low levels of APOBEC-3F and APOBEC-3G.
The original studies of HIV infection of primary T cells had some technical limitations that we sought to overcome. First, resting CD4+ T cells were often contaminated with activated T cells and dendritic cells (63, 67, 76) or were positively selected with anti-CD4, which could have activated the T cells and copurified dendritic cells. Second, the infection level of resting T cells was compared to the infection level of T blasts that were artificially activated (e.g., phytohemagglutinin activated) (76). But, in this paper we showed that endogenous activated T cells are less permissive to HIV than artificially activated cells and thus are a more relevant control. Third, because of the low infection frequency in resting cells, integration events may have been so rare that the events were below the assay's detection limit (2, 67). Fourth, endpoint PCR was used to estimate the frequency of integration (67). Although state-of-the-art at the time, endpoint PCR is only semiquantitative.
Our methods differed from previous methods in four ways. First, to purify target cells, we used a negative selection procedure that labeled non-CD4+ T cells in order to avoid CD4+ T-cell activation by cross-linking antibodies. Second, we simultaneously purified resting and endogenous activated CD4+ T cells by adding antibodies to activation markers that were labeled with a second fluorescent marker. This enabled us to control more precisely for the integration signal from contaminating activated cells. This was an important control because endogenous activated T cells were less permissive to HIV-1 reverse transcription and integration than T-cell lines and in vitro-activated T-cell blasts—the cells that were compared to resting CD4+ T cells in previous studies. Third, we used spinoculation to increase the infection frequency by pelleting virions onto cells (52). Because we used spinoculation we could avoid using Polybrene, a potentially activating agent, to enhance infection (38, 39, 76). Fourth, we developed kinetic PCR-based, quantitative assays to measure HIV-1 binding (52), reverse transcription (69), and integration (51).
It could be argued that contaminating activated cells caused the integration signal in resting CD4+ T cells. However, we controlled for the signal contributed by contaminating cells by postsort analysis. For example, for the integration signal in resting cells to be due to contaminating endogenous activated cells, 20% of the resting cells would have had to be activated cells. Yet, postsort analysis showed that only 1% of the resting cells were activated cells. The kinetics of reverse transcription and integration in the resting CD4+ T cells were slower compared to the activated cells, further distinguishing the two cell populations. Therefore, we concluded that the integration signal detected in resting cells was not due to contaminating activated cells.
It could be argued that cell culture, spinoculation, or exposure to high concentrations of virions activated the resting T cells and induced HIV-1 integration. In other work, PBMCs exposed to high concentrations of virions enter G1b and become productively infected (4). It had been assumed that exposure to high concentrations of virions activated the cells. However, the T cells in that study may have been activated by dendritic cell-T-cell interactions in the presence of virus, rather than by exposure to high concentrations of virions alone. Our HIV-spinoculated, HLA-DR− CD69− CD4+ T cells did not become activated in culture as assessed by DNA/RNA FACS, bromodeoxyuridine uptake staining, and activation markers. While some mononuclear cells, such as dendritic cells, become activated in culture (47), highly purified, lymph node CD4+ T cells become less activated in culture (65). In vivo resting CD4+ T cells probably display a spectrum of T-cell activation as a result of cytokine exposure and antigen-independent interactions with dendritic cells. Current literature (65) supports that our resting CD4+ T cells are at the low end of the T-cell activation spectrum compared to in vivo resting CD4+ T cells.
We further tested if the process of spinoculation alters the cells to make them more permissive to infection by comparing routine and spinoculation conditions. Previously we showed that spinoculation increases virus binding and reverse transcription to similar extents and that spinoculation does not enhance fusion (52). In the current study we showed that spinoculation increased binding and integration to similar extents. This suggests that spinoculation does not induce integration or make cells more permissive to integration.
Spinoculation is an important tool in the study of HIV infection in resting CD4+ T cells because it overcomes a significant limitation of routine inoculation. It achieves a higher percentage of HIV-1 integration and latent infection than in vivo infection or routine in vitro inoculation. The higher percentage of latently infected cells in vitro allows us to do experiments that cannot be done using in vivo samples. Spinoculation achieves higher levels of infection because it uses centrifugal force to pellet virions onto cells, thus overcoming the rate-limiting step of virion diffusion to the cell surface. Routine inoculation of T cells results in low levels of infection, because virion diffusion to the cell surface is inefficient. With such low levels of infection it is difficult to detect rare events and difficult to determine whether the rare events are due to contaminating cells. It is possible that spinoculation altered resting T cells and made them more permissive to integration. For example, spinoculation could have induced changes in the expression of stress genes, as occurs in lymphocytes after continuous flow leukopheresis (49). Even if this is true, spinoculation is a useful technique because it achieves high levels of HIV-1 integration and latent infection in our system.
Spinoculation may mimic one aspect of in vivo infection: delivery of multiple virions to a target cell. Similarly, in vivo multiple virions are delivered to one target cell, not through spinoculation but through a virological synapse (30, 31, 45). In vivo infection probably progresses from infected cell to target cell through cell-to-cell spread, because cell-to-cell spread is more efficient than virus-to-cell spread (15, 21, 22, 60). By one account, an activated T cell can produce at least 60 virions at one instant in time (79). Progeny virions budding from an activated infected T cell and HIV-1 receptors on an uninfected target cell cluster in the area of contact between the cells—an area known as the virological synapse (30, 31). Therefore, it is reasonable to assume that in vivo a productively infected activated T cell or dendritic cell bearing multiple HIV-1 virions delivers many virions to an uninfected resting T cell at the virological synapse (30, 31, 45).
Valid in vitro models of HIV-1 latency are needed, because in HIV-1-infected individuals latent reservoirs are a major barrier to cure. Using our model we can ask questions that are difficult to study in vivo due to the low level of latently infected cells (24, 35, 42, 43, 50, 75). For example, what portion of CD4+ T cells are multiply infected (34)? What fraction of the integrated proviruses are mutated and/or defective in the presence of APOBEC-3F and APOBEC-3G (10, 35, 50)? Do the integration sites differ in latently versus productively infected cells (24, 32, 33, 61)? What conditions favor reactivation (28, 37, 40, 41, 74)? What is the relative efficiency of HIV-1 integration in different CD4+ T-cell subsets (11, 53)? In summary, our in vitro system provides a new model of HIV-1 latency that can be used to answer questions about HIV-1 latency and, ultimately, to test therapies aimed at diminishing HIV-1 reservoirs.
Acknowledgments
This study was supported in part by NIH grants K08 HL03984 (U.O.), R01 AI058862-01 (U.O.), and K08 A50458-01 (W.J.S.). Additional support came from the National Blood Foundation (U.O.), the McCabe Award (U.O.), and the W.W. Smith Charitable Trust (W.J.S.).
We thank Liz Colston, Drew Weissman, Rick Bushman, Terri Finkel, and Avinash Bhandoola for critical reading of the manuscript and Michael Malim and Ann Sheehy for helpful discussions. The integrase inhibitor L-870,810 was a generous gift of Merck Research Laboratories, West Point, Pa. We are grateful to Penn CFAR for performing our p24 ELISAs and to the Flow Cytometry Core for excellent technical guidance and cell sorting.
REFERENCES
- 1.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 2.Biancotto, A., J. C. Grivel, F. Gondois-Rey, L. Bettendroffer, R. Vigne, S. Brown, L. B. Margolis, and I. Hirsch. 2004. Dual role of prostratin in inhibition of infection and reactivation of human immunodeficiency virus from latency in primary blood lymphocytes and lymphoid tissue. J. Virol. 78:10507-10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 4.Briant, L., N. Coudronniere, V. Robert-Hebmann, M. Benkirane, and C. Devaux. 1996. Binding of HIV-1 virions or gp120-anti-gp120 immune complexes to HIV-1-infected quiescent peripheral blood mononuclear cells reveals latent infection. J. Immunol. 156:3994-4004. [PubMed] [Google Scholar]
- 5.Brooks, D. G., P. A. Arlen, L. Gao, C. M. Kitchen, and J. A. Zack. 2003. Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc. Natl. Acad. Sci. USA 100:12955-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, D. G., D. H. Hamer, P. A. Arlen, L. Gao, G. Bristol, C. M. Kitchen, E. A. Berger, and J. A. Zack. 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19:413-423. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, D. G., S. G. Kitchen, C. M. R. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459-464. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, R. G., J. L. Riley, B. L. Levine, P. J. Blair, D. C. St Louis, and C. H. June. 1998. The role of co-stimulation in regulation of chemokine receptor expression and HIV-1 infection in primary T lymphocytes. Semin. Immunol. 10:195-202. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, R. G., J. L. Riley, B. L. Levine, Y. Feng, S. Kaushal, D. W. Ritchey, W. Bernstein, O. S. Weislow, C. R. Brown, E. A. Berger, C. H. June, and D. C. St. Louis. 1997. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science 276:273-276. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 11.Chun, T.-W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y.-H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craston, R., M. Koh, A. Mc Dermott, N. Ray, H. G. Prentice, and M. W. Lowdell. 1997. Temporal dynamics of CD69 expression on lymphoid cells. J. Immunol. Methods 209:37-45. [DOI] [PubMed] [Google Scholar]
- 14.De Rosa, S. C., L. A. Herzenberg, and M. Roederer. 2001. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 7:245-248. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrov, D. S., R. L. Willey, H. Sato, L. J. Chang, R. Blumenthal, and M. A. Martin. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67:2182-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducrey-Rundquist, O., M. Guyader, and D. Trono. 2002. Modalities of interleukin-7-induced human immunodeficiency virus permissiveness in quiescent T lymphocytes. J. Virol. 76:9103-9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 18.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849-859. [DOI] [PubMed] [Google Scholar]
- 19.Goletti, D., D. Weissman, R. W. Jackson, F. Collins, A. Kinter, and A. S. Fauci. 1998. The in vitro induction of human immunodeficiency virus (HIV) replication in purified protein derivative-positive HIV-infected persons by recall antigen response to Mycobacterium tuberculosis is the result of a balance of the effects of endogenous interleukin-2 and proinflammatory and antiinflammatory cytokines. J. Infect. Dis. 177:1332-1338. [DOI] [PubMed] [Google Scholar]
- 20.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 21.Gummuluru, S., V. N. KewalRamani, and M. Emerman. 2002. Dendritic cell-mediated viral transfer to T cells is required for human immunodeficiency virus type 1 persistence in the face of rapid cell turnover. J. Virol. 76:10692-10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gummuluru, S., C. M. Kinsey, and M. Emerman. 2000. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J. Virol. 74:10882-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, M. A. Fischl, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 24.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 26.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, Jr., M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. Jin, I. W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 101:11233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stilmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 28.He, G., L. Ylisastigui, and D. M. Margolis. 2002. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 21:697-705. [DOI] [PubMed] [Google Scholar]
- 29.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolly, C., K. Kashefi, M. Hollinshead, and Q. J. Sattentau. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolly, C., and Q. J. Sattentau. 2004. Retroviral spread by induction of virological synapses. Traffic 5:643-650. [DOI] [PubMed] [Google Scholar]
- 32.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22: 1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung, A., R. Maier, J.-P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meryerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 35.Kieffer, T. L., P. Kwon, R. E. Nettles, Y. Han, S. C. Ray, and R. F. Siliciano. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 79:1975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita, S., B. K. Chen, H. Kaneshima, and G. P. Nolan. 1998. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell 95:595-604. [DOI] [PubMed] [Google Scholar]
- 37.Kinter, A. L., C. A. Umscheid, J. Arthos, C. Cicala, Y. Lin, R. Jackson, E. Donoghue, L. Ehler, J. Adelsberger, R. L. Rabin, and A. S. Fauci. 2003. HIV envelope induces virus expression from resting CD4+ T cells isolated from HIV-infected individuals in the absence of markers of cellular activation or apoptosis. J. Immunol. 170:2449-2455. [DOI] [PubMed] [Google Scholar]
- 38.Korin, Y. D., and J. A. Zack. 1999. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J. Virol. 73:6526-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korin, Y. D., and J. A. Zack. 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J. Virol. 72:3161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkosky, J., J. Sullivan, Y. Xu, E. Souder, D. H. Hamer, and R. J. Pomerantz. 2004. Expression of latent HAART-persistent HIV type 1 induced by novel cellular activating agents. AIDS Res. Hum. Retrovir. 20:497-505. [DOI] [PubMed] [Google Scholar]
- 41.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525-531. [DOI] [PubMed] [Google Scholar]
- 43.Lassen, K. G., J. R. Bailey, and R. F. Siliciano. 2004. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 78:9105-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Cabrera, M., A. G. Santis, E. Fernandez-Ruiz, R. Blacher, F. Esch, P. Sanchez-Mateos, and F. Sanchez-Madrid. 1993. Molecular cloning, expression, and chromosomal localization of the human earliest lymphocyte activation antigen AIM/CD69, a new member of the C-type animal lectin superfamily of signal-transmitting receptors. J. Exp. Med. 178:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 46.McLean, A. R., and C. A. Michie. 1995. In vivo estimates of division and death rates of human T lymphocytes. Proc. Natl. Acad. Sci. USA 92:3707-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellman, I., and R. M. Steinman. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255-258. [DOI] [PubMed] [Google Scholar]
- 48.Michie, C. A., A. McLean, C. Alcock, and P. C. L. Beverley. 1992. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360:264-265. [DOI] [PubMed] [Google Scholar]
- 49.Moir, S., E. T. Donoghue, O. K. Pickeral, A. Malaspina, M. A. Planta, T. W. Chun, S. R. Krishnan, S. Kottilil, C. E. Birse, S. F. Leitman, and A. S. Fauci. 2003. Continuous flow leukapheresis induces expression of stress genes in lymphocytes: impact on microarray analyses. Blood 102:3852-3853. [DOI] [PubMed] [Google Scholar]
- 50.Monie, D., R. P. Simmons, R. E. Nettles, T. L. Kieffer, Y. Zhou, H. Zhang, S. Karmon, R. Ingersoll, K. Chadwick, J. B. Margolick, T. C. Quinn, S. C. Ray, M. Wind-Rotolo, M. Miller, D. Persaud, and R. F. Siliciano. 2005. A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients. J. Virol. 79:5185-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative, assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostrowski, M. A., T.-W. Chun, S. J. Justement, I. Motola, M. A. Spinelli, J. Adelsberger, L. A. Ehler, S. B. Mizell, C. W. Hallahan, and A. S. Fauci. 1999. Both memory and CD45RA+/CD62L+ naive CD4+ T cells are infected in human immunodeficiency type 1-infected individuals. J. Virol. 73:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oswald-Richter, K., S. M. Grill, M. Leelawong, and D. Unutmaz. 2004. HIV infection of primary human T cells is determined by tunable thresholds of T cell activation. Eur. J. Immunol. 34:1705-1714. [DOI] [PubMed] [Google Scholar]
- 55.Palella, F. J., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 56.Perelson, A. S., P. Essunger, and Y. Cao. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387: 188-191. [DOI] [PubMed] [Google Scholar]
- 57.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 58.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley, J. L., K. Schlienger, P. J. Blair, B. Carreno, N. Craighead, D. Kim, R. G. Carroll, and C. H. June. 2000. Modulation of susceptibility to HIV-1 infection by the cytotoxic T lymphocyte antigen 4 costimulatory molecule. J. Exp. Med. 191:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato, H., J. Orenstein, D. Dimitrov, and M. Martin. 1992. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology 186:712-724. [DOI] [PubMed] [Google Scholar]
- 61.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 62.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 63.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2877-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sprent, J., and D. F. Tough. 1994. Lymphocyte life-span and memory. Science 265:1395-1400. [DOI] [PubMed] [Google Scholar]
- 65.Stefanova, I., J. R. Dorfman, and R. N. Germain. 2002. Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420:429-434. [DOI] [PubMed] [Google Scholar]
- 66.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853-860. [DOI] [PubMed] [Google Scholar]
- 67.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun, Y., L. M. Pinchuk, M. B. Agy, and E. A. Clark. 1997. Nuclear import of HIV-1 DNA in resting CD4+ T cells requires a cyclosporin A-sensitive pathway. J. Immunol. 158:512-517. [PubMed] [Google Scholar]
- 69.Swiggard, W. J., U. O'Doherty, D. McGain, D. Jeyakumar, and M. H. Malim. 2004. Long HIV type 1 reverse transcripts can accumulate stably within resting CD4+ T cells while short ones are degraded. AIDS Res. Hum. Retrovir. 20:285-295. [DOI] [PubMed] [Google Scholar]
- 70.Swingler, S., B. Brichacek, J. M. Jacque, C. Ulich, J. Zhou, and M. Stevenson. 2003. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature 424:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tough, D. F., and J. Sprent. 1994. Turnover of naive- and memory-phenotype T cells. J. Exp. Med. 179:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams, S. A., and W. C. Greene. 2005. Host factors regulating post-integration latency of HIV. Trends Microbiol. 13:137-139. [DOI] [PubMed] [Google Scholar]
- 75.Ylisastigui, L., J. J. Coull, V. C. Rucker, C. Melander, R. J. Bosch, S. J. Brodie, L. Corey, D. L. Sodora, P. B. Dervan, and D. M. Margolis. 2004. Polyamides reveal a role for repression in latency within resting T cells of HIV-infected donors. J. Infect. Dis. 190:1429-1437. [DOI] [PubMed] [Google Scholar]
- 76.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 77.Zack, J. A., A. M. Haislip, P. Krogstad, and I. S. Y. Chen. 1992. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J. Virol. 66:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, Z.-Q., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]
- 79.Zhang, Z. Q., S. W. Wietgrefe, Q. Li, M. D. Shore, L. Duan, C. Reilly, J. D. Lifson, and A. T. Haase. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 101:5640-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]