Abstract
A search for dominant-negative mutant polypeptides hampering infectious bursal disease virus (IBDV) replication has been undertaken. We have found that expression of a mutant version of the VP3 structural polypeptide known as VP3/M3, partially lacking the domain responsible for the interaction with the virus-encoded RNA polymerase, efficiently interferes with the IBDV replication cycle. Transformed cells stably expressing VP3/M3 show a significant reduction (up to 96%) in their ability to support IBDV growth. Our findings provide a new tool for the characterization of the IBDV replication cycle and might facilitate the generation of genetically modified chicken lines with a reduced susceptibility to IBDV infection.
Infectious bursal disease virus (IBDV), the best-characterized member of the Birnaviridae family, grouping viruses with bipartite double-stranded RNA genomes (16), is responsible for an immunosuppressive disease that affects juvenile chickens (Gallus gallus) and causes substantial economic losses to the poultry industry (12). Although the susceptibility to IBDV infection varies among different chicken breeds, all tested chicken lines undergo bursal damage after experimental infection with very virulent IBDV strains (2). The implementation of intensive IBDV vaccination programs has failed to prevent both the dissemination of the disease and the emergence of very virulent virus strains (15). Hence, there is a need for alternative strategies that might improve the effectiveness of current IBDV control measures.
We sought to find dominant-negative polypeptides interfering with IBDV replication. Previous information suggested that the inner capsid protein VP3 might be a suitable candidate in our search for an effective dominant-negative polypeptide. VP3 (28.8 kDa) controls the assembly of the major capsid polypeptide VP2 (7, 8, 10) and interacts with both the viral genome (4, 14) and the virus-encoded RNA polymerase VP1 (5). Additionally, several VP3 functional domains have been mapped and characterized (4, 5, 7, 14).
We initiated our study by assessing the effect of three previously described (4, 7) VP3 mutant polypeptides, M1, M2, and M3, lacking specific functional domains (Fig. 1A). M1 (29.37 kDa) contains a deletion of 25 residues that abrogates VP3-RNA interactions (4). M2 (27.19 kDa) has a 42-residue C-terminal deletion that eliminates both the oligomerization and the VP1-binding domains (7). M3 (30.94 kDa) lacks the 10 C-terminal residues affecting the VP1-binding domain (7). In order to allow their detection, a 2.5-kDa histidine tag was fused to the N termini of the mutant polypeptides.
FIG. 1.
Expression of VP3 deletion mutant polypeptides reduces IBDV replication. (A) The diagram depicts the VP3 deletion mutant polypeptides M1, M2, and M3, expressed by different rBVs used in our analyses. Gray rectangles indicate the presence of six-histidine tags fused to the N termini of the different VP3-derived constructs. The upper part of the diagram represents the wild-type VP3 polypeptide (257 residues), indicating the positions of the RNA-binding (RNA-BD), oligomerization (OlD), and VP1-binding (VP1-BD) domains. (B) BSC1 cultures were transduced with recombinant baculoviruses expressing M1, M2, and M3. At 48 h posttransduction, cells were harvested and the corresponding extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using an anti-histidine tag monoclonal antibody. The positions of molecular mass markers are shown in kilodaltons (lane MW). (C) Monolayers of BSC1 cells were transduced with recombinant baculoviruses expressing EFGP (•), M1 (▪), M2 (□), and M3 (▴). Mock-transduced monolayers (○) were used as controls for this experiment. Cells were infected at 12 h after transduction with IBDV at a multiplicity of infection of 0.1 PFU/cell. At the indicated times p.i., cultures were harvested and total virus titers were determined by plaque assay on BSC1 cells.
Intracellular expression of the VP3 mutant polypeptides was achieved by transducing IBDV-susceptible BSC1 cells (American Type Culture Collection) with recombinant baculoviruses (rBVs) containing the corresponding genes under the control of a mammalian transcriptional cassette (1). For this, a hybrid vector, pFBCI, was constructed, containing the transcriptional regulatory sequences from the expression vector pCI-neo (Promega) and sequences required for the generation of rBVs from pFBac1 (Invitrogen). This plasmid was generated by cloning a BglII/BsaBI fragment from pCI-neo into pFBac1, which had been previously digested with BclI and SnaBI. DNA fragments containing the M1 and M3 mutant genes were obtained from pFBHTc/VP3ΔM10 (4) and pFBHTc/VP3Δ1002 (7), respectively. These plasmids were digested with RsrII and then subjected to Klenow treatment and KpnI digestion, and the corresponding fragments were cloned into pFBCI treated with EcoRI, Klenow fragment, and KpnI. M2 cloning was carried out by inserting into SmaI/NotI-digested pFBCI a DNA fragment isolated from pFBHTc/VP3Δ971-1012 (4) by digestion with RsrII, followed by Klenow treatment and NotI digestion. rBVs were generated using the Bac-to-Bac system (Gibco BRL). An rBV expressing the green fluorescent protein (EGFP) was also generated. For this, a DNA fragment obtained by restriction of pFB/EGFP with EcoRI and NotI was inserted into pFBCI digested with the same enzymes. rBVs were purified as previously described (11) and used to transduce BSC1 monolayers. Expression of the different VP3 deletion mutants was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting of extracts from BSC1-transduced cells (Fig. 1B). The transduction efficiencies were determined by immunofluorescence and found to be, in all cases, above 95% (data not shown). At 12 h after transduction, cultures were infected (0.1 PFU/cell) with the IBDV Soroa strain adapted to grow in BSC1 cells (3). At different times postinfection (p.i.), cultures were harvested and total infectious virus concentrations (intra- and extracellular) were titrated on fresh BSC1 monolayers. The experiment was repeated three times with similar results. Figure 1B shows the results of a representative experiment. As expected, EGFP expression does not affect virus replication. In contrast, all three VP3 mutant polypeptides appear to disturb virus growth. The most conspicuous effect, a drop of almost 2 logarithmic titer units, was consistently observed in M3-expressing cells.
The results described above suggested that M3 might be an efficient dominant-negative polypeptide. It was therefore important to determine whether a similar effect could be observed in cells constitutively expressing this polypeptide. Accordingly, the M3 gene was inserted into the mammalian expression vector pCI-neo by cloning a NheI/NotI fragment containing the M3 coding sequence from pFBCI/M3 into pCI-neo digested with the same enzymes. The resulting plasmid, pCI-neo/M3, was transfected into BSC1 cell cultures. Stably transformed cells were selected and cloned, using medium supplemented with G418 as previously described (13). G418-resistant clones were expanded and used to analyze the expression of the M3 polypeptide by immunofluorescence using a VP3-specific antiserum (5). In order to assess the relative expression level in each clone, cell extracts were analyzed by Western blotting using a mixture of rabbit anti-human β-actin (Sigma) and anti-VP3 sera (5). Three clones, showing high (BSC1/M3H), medium (BSC1/M3M), and low (BSC1/M3L) M3 expression levels (Fig. 2), were selected for further analyses. A comparative analysis of the relative M3 accumulation levels in the three described lines (BSC1/M3L, BSC1/M3M, and BSC1/M3H) was carried out. Serial dilutions of the corresponding cell extracts were analyzed by Western blotting using anti-VP3 serum. Dilutions of affinity-purified M3, produced as previously described (4), were used as an internal standard for this analysis. After being developed, the filters were scanned and the integrated density of M3 bands in each sample was determined using the NIH image software (http://rsb.info.nih.gov/nih-image/). The results of this analysis showed that M3 accumulation in BSC1/M3H is approximately 20- and 50-fold higher than in BSC1/M3M and BSC1/M3L, respectively (data not shown). A BSC1-derived line expressing the EGFP polypeptide, BSC1/EGFP, was also generated by transfecting BSC1 cells with the pCI-neo/EGFP vector. This plasmid was built by cloning a NotI/EcoRI DNA fragment containing the EGFP open reading frame from pFBCI/EGFP into pCI-neo digested with the same enzymes.
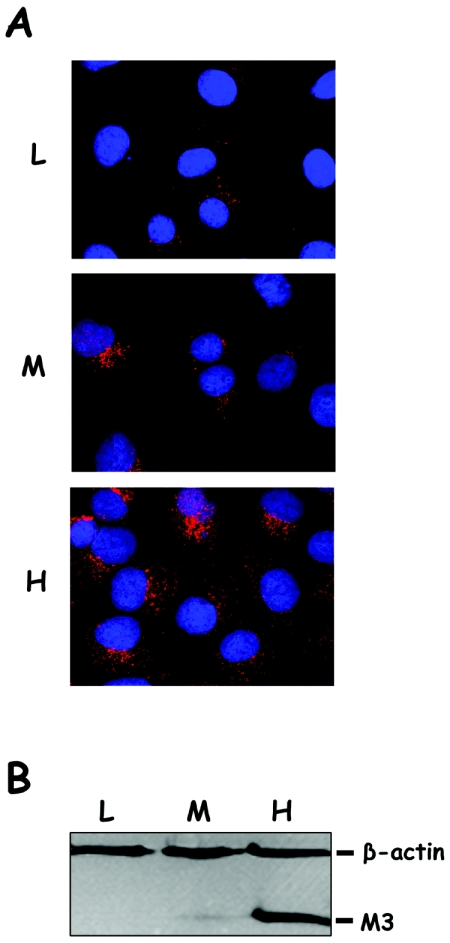
FIG. 2.
Characterization of BSC1 cells stably expressing the VP3 M3 deletion mutant polypeptide. BSC1 cells expressing the M3 polypeptide were generated by transfection with plasmid pCI-neo/M3 and subsequent selection in the presence of G418. (A) Immunofluorescence analysis of three selected clones, BSC1/M3L (L), BSC1/M3M (M), and BSC1/M3H (H). Cells were grown on coverslips, fixed, and incubated with rabbit anti-VP3 serum, followed by incubation with goat anti-rabbit immunoglobulin coupled to Alexa 594 (red). Cell nuclei were stained with ToPro-3 (blue). Samples were visualized by epifluorescence using a Zeiss Axiovert 200 microscope equipped with a Bio-Rad Radiance 2100 confocal system. Images were captured using the Laser Sharp software package (Bio-Rad). (B) Cultures of BSC1/M3L (L), BSC1/M3M (M), and BSC1/M3H (H) were harvested and analyzed by Western blotting using a mixture of mouse anti-human β-actin and rabbit anti-VP3 sera, followed by addition of a mixture of horseradish peroxidase-conjugated goat anti-mouse and rabbit anti-rabbit immunoglobulins. Signals were detected by incubation with 1-chloro-4-naphthol in the presence of hydrogen peroxide. The positions of the β-actin and M3 polypeptides are indicated.
Cultures of BSC1, BSC1/EGFP, and the three selected BSC1/M3 cell lines were infected with 0.1 PFU/cell of IBDV. At different times p.i., cultures were harvested and the total virus titers determined. As illustrated in Fig. 3A, the three M3-expressing BSC1 clones showed a decreased ability to support IBDV growth. A reduction close to 2 logarithmic units was detected with BSC1/M3H. In order to determine the effect of M3 expression on IBDV yield, five independent infections were performed. Virus yields at 72 h p.i. were calculated using the value obtained in the control BSC1 cell line as the 100% standard. The results of this analysis are presented in Fig. 3B. Again, the highest reduction, over 96%, was observed with the BSC1/M3H line. BSC1/M3M and BSC1/M3L lines showed reductions of 84 and 60%, respectively. As expected, the BSC1/EGFP cell line did not show detectable effects on IBDV yields. To further confirm that the interference in IBDV replication observed with the BSC1/M3H cell line is genuinely due to M3 expression, the ability of IBDV to replicate in cultures of additional lines expressing M3 at high levels was tested. The results obtained were almost identical (less than 2% variation in IBDV yields) to those obtained with BSC1/M3H (data not shown). As shown in Fig. 3C, the reduction in IBDV yields in the M3-expressing cell lines is accompanied by a decrease in the accumulation of the major structural IBDV polypeptides pVP2, VP2, and VP3. As expected, a conspicuous lessening of IBDV-induced cytopathic effect was observed in BSC1/M3H cells (Fig. 4A). In order to test the specificity of the M3 effect, BSC1 and BSC1/M3H cells were infected with two unrelated viruses, vaccinia virus (WR strain) and influenza virus (A/WSN/33 strain). The yields of these two viruses were nearly identical in both cell lines (data not shown).
FIG. 3.
Stable expression of VP3/M3 interferes with IBDV replication. (A) Cultures of BSC1 (○), BSC1/EGFP (•), BSC1/M3L (▪), BSC1/M3M (□), and BSC1/M3H (▴) were infected with IBDV at a multiplicity of infection (MOI) of 0.1 PFU/cell. At the indicated times p.i., cultures were harvested and total virus titers were determined by plaque assay on fresh BSC1 cells. (B) Comparison of the IBDV yields in BSC1/EGFP and the different M3-expressing cells. Cultures infected at an MOI of 0.1 PFU/cell were harvested at 72 h p.i., and virus titers were determined as described for panel A. Titers obtained with BSC1 cells were used as the standard 100% value. The graph was generated with data from five independent experiments. EGFP, BSC1/EGFP; M3L, BSC1/M3L; M3M, BSC1/M3M; M3H, BSC1/M3H. (C) Accumulation of IBDV structural polypeptides in M3-expressing BSC1 cells. Cultures of BSC1, BSC1/EGFP (EGFP), BSC1/M3L (L), BSC1/M3M (M), and BSC1/M3H (H) infected at an MOI of 0.1 PFU/cell were harvested at 72 h p.i. Cell extracts were analyzed by Western blotting using rabbit anti-VP2 and anti-VP3 sera, followed by addition of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. Signals were detected by incubation with 1-chloro-4-naphthol in the presence of hydrogen peroxide. Lanes I and MW correspond to purified virus and prestained molecular weight markers, respectively. The positions of pVP2, VP2, VP3, and M3 polypeptides are indicated.
FIG. 4.
(A) Stable expression of VP3/M3 reduces IBDV-induced cytopathic effects. Monolayers of BSC1 and BSC1/M3H were either mock infected or infected with IBDV at a multiplicity of infection of 0.1 PFU/cell. Phase-contrast images at a magnification of ×200 were recorded at 72 h p.i. (B) Effects of IBDV infection on the subcellular distribution of the M3 polypeptide. Mock- and IBDV-infected BSC1/M3H cells were processed for confocal laser scanning microscopy using specific antisera. Green (Alexa 488) corresponds to VP2, and red (Alexa 594) corresponds to the His tag of the M3 polypeptide. The blue signal (ToPro-3) corresponds to nuclear staining. The merge panels show the overlays of the three fluorescent signals.
Data presented here demonstrate that the M3 polypeptide is a specific dominant-negative polypeptide that interferes with IBDV replication, thus suggesting that it might affect the assembly process. To investigate this possibility, an immunofluorescence analysis was performed on cultures of mock- and IBDV-infected BSC1/M3H cells. At 48 h p.i., cell monolayers were fixed, incubated with anti-VP2 and anti-His sera, and processed for confocal laser scanning microscopy as previously described (10). The results of this analysis are shown in Fig. 4B. While in mock-infected cells the M3 polypeptide is distributed throughout the cell cytoplasm, in IBDV-infected cells it accumulates in large viroplasm-like aggregates, colocalizing with the major structural polypeptide VP2. This dramatic alteration in the M3 subcellular distribution strongly suggests that it participates in the assembly of the virus progeny. Previous studies have shown that although M3 is unable to form complexes with the virus-encoded RNA-dependent RNA polymerase (6), it retains the ability to oligomerize (7) and to interact with the virus genome (4). The presence of M3 at the IBDV assembly factories might hamper the correct incorporation of the VP1 polypeptide into the newly formed particles. This hypothesis is supported by the previous observation that treatment of IBDV-infected cells with Trojan peptides containing the VP1-binding motif found in VP3 causes a significant reduction in infective-virus production (6).
The BSC1/M3H cell line provides an excellent tool to gain information about the as yet poorly characterized IBDV replication cycle. Hopefully, recent developments in the generation of germ line-transgenic chickens (9) will facilitate testing the potential of M3 to establish genetically modified chicken lines with a reduced susceptibility to IBDV infection.
Acknowledgments
This work was supported by grants AGL2003-07189 from Dirección General de Investigación del Ministerio de Educación and 07B/0041/2002 from the Subdirección General de Investigación of the Comunidad Autónoma de Madrid. F.A. was the recipient of a fellowship from the Comunidad Autónoma de Madrid.
We thank Dolores Rodríguez for her guidance on the generation of stable cell lines and Amelia Nieto for the A/WSN/33 influenza virus strain. We are grateful for the technical assistance provided by Sylvia Gutierrez, head of the CNB Confocal Laser Scanning Microscopy Service.
REFERENCES
- 1.Boyce, F. M., and N. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 93:2348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bumstead, N., R. L. Reece, and J. K. Cook. 1993. Genetic differences in susceptibility of chicken lines to infection with infectious bursal disease virus. Poult. Sci. 72:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Arias, A., S. Martínez, and J. F. Rodríguez. 1997. The major antigenic protein of infectious bursal disease virus, VP2, is an apoptotic inducer. J. Virol. 71:8014-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochan, G., D. González, and J. F. Rodríguez. 2003. Characterization of the RNA-binding activity of VP3, a major structural protein of Infectious bursal disease virus. Arch. Virol. 148:723-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombardo, E., A. Maraver, J. R. Castón, J. Rivera, A. Fernández-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maraver, A., R. Clemente, J. F. Rodríguez, and E. Lombardo. 2003. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J. Virol. 77:2459-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maraver, A., A. Oña, F. Abaitua, D. González, R. Clemente, J. A. Ruiz-Díaz, J. R. Castón, F. Pazos, and J. F. Rodriguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Torrecuadrada, J. L., J. R. Castón, M. Castro, J. L. Carrascosa, J. F. Rodríguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 9.McGrew, M. J., A. Sherman, F. M. Ellard, S. G. Lillico, H. J. Gilhooley, A. J. Kingsman, K. A. Mitrophanous, and H. Sang. 2004. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 5:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oña, A., D. Luque, F. Abaitua, A. Maraver, J. R. Castón, and J. F. Rodríguez. 2003. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322:135-142. [DOI] [PubMed] [Google Scholar]
- 11.Sarkis, C., C. Serguera, S. Petres, D. Buchet, J.-L. Ridet, L. Edelman, and J. Mallet. 2000. Efficient transduction of neural cells in vitro and in vivo by a baculovirus-derived vector. Proc. Natl. Acad. Sci. USA 97:14638-14643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma, J. M., I. J. Kim, S. Rautenschlein, and H. Y. Yeh. 2000. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev. Comp. Immunol. 24:223-235. [DOI] [PubMed] [Google Scholar]
- 13.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 14.Tacken, M. G. J., B. P. H. Peeters, A. A. M. Thomas, P. J. M. Rottier, and H. J. Boot. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J. Virol. 76:11301-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Berg, T. P., N. Eterradossi, D. Toquin, and G. Meulemans. 2000. Infectious bursal disease (Gumboro disease). Rev. Sci. Technol. 19:509-543. [PubMed] [Google Scholar]
- 16.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.