Abstract
Previous evaluation of HWP1 in systemic candidiasis in CBA/J mice was done with Candida albicans strains with differing genetic locations of URA3 as a result of Ura-blaster mutagenesis. In this study, the presence of HWP1 and the location of URA3 contributed to the severity of murine systemic candidiasis in BALB/c mice.
In previous work (8), the HWP1 gene was required for virulence in systemic candidiasis. A perplexing observation from this study was the suggestion of a trend of increased survival, or slight loss of virulence, of mice given CAH7 (HWP1/hwp1) compared to that of mice given CAHR3 (HWP1/hwp1 revertant). The survival differences were apparently not related to differences in HWP1 gene expression as the amounts of HWP1 mRNA and protein were equivalent for the two strains. Moreover, the strains did not differ in growth rates. A difference between strains CAH7 and CAHR3 is the location of the selectable marker gene URA3. The URA3 gene is within the open reading frame of HWP1 in CAH7 but within ENO1 in CAHR3.
The use of the Ura-blaster technique in Candida albicans (2) results in both inactivation of a gene of interest and ectopic placement of URA3 within the gene. The observation that C. albicans auxotrophic mutants that cannot produce orotidine 5′-monophosphate (OMP) decarboxylase because they lack the URA3 gene are not pathogenic (4) suggests that if reduced levels of OMP decarboxylase were to be produced in vivo, intermediate virulence might result. Placement of the C. albicans URA3 gene at a locus other than the URA3 locus during implementation of the Ura-blaster technique has been shown to result in reduced OMP decarboxylase activities in vitro, and the levels of reduction were different for different loci. However, a definitive relationship between OMP decarboxylase enzyme activity in in vitro cultures and virulence was not revealed (6). These studies raised the possibility that the genetic location of the URA3 gene affects differential in vivo growth rates of strains CAH7 and CAHR3, leading to a trend of decreased virulence of CAH7.
The goals of the present study were twofold. The first was to make the expression of the URA3 gene independent of positional effects arising from placement at the HWP1 locus. The second goal was to be able to compare C. albicans strains that are identical with regard to the location of the URA3 gene. Two new C. albicans strains with disruptions at the HWP1 locus were created by introducing a DNA fragment with eno1::URA3 (7) into the Ura− strains CAH7-1 (HWP1/hwp1) and CAH7-1A1 (hwp1/hwp1) (8). Previous studies with a strain bearing a URA3 disruption of one of four ENO1 homologues did not reveal reduced growth on pyruvate or glucose (7) or a noticeable reduction in ENO1 mRNA levels (7, 8), and germ tube formation was unaffected (1, 7).
Ura+ transformants were initially screened by PCR for homologous recombination of the eno1::URA3 fragment at ENO1, and positive transformants were confirmed by Southern blot analysis (data not shown). These strains, CAH7-1E1 (HWP1/hwp1) and CAH7-1A1E2 (hwp1/hwp1), are comparable to the strains, CAH7 and CAH7-1A, used in the previous experiment. However, URA3 was targeted to the ENO1 locus in the newly constructed strains as in the revertant strain CAHR3 used in the previous study (Table 1). No differences in growth rates between strains were found in yeast nitrogen base medium without uridine. In addition to the new strains, CAH49, an additional HWP1/hwp1 heterozygote with URA3 integrated into the HWP1 locus, was included along with CAH7.
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| SC5314 | Wild type, parent of CAI4 | 5 |
| CAH7a | HWP1/hwp1::hisG-URA3-hisG | 8 |
| CAH49 | HWP1/hwp1::hisG-URA3-hisG | 8 |
| CAH7-1 | HWP1/hwp1::hisG | 8 |
| CAH7-1A | hwp1::hisG-URA3-hisG/hwp1::hisG | 8 |
| CAH7-1A1 | hwp1::hisG/hwp1::hisG | 8 |
| CAHR3 | hwp1::hisG/HWP1 eno1::URA3 (revertant) | 8 |
| CAH7-1E1 | HWP1/hwp1::hisG eno1::URA3 | This study |
| CAH7-1A1E2 | hwp1::hisG/hwp1::hisG eno1::URA3 | This study |
All CAH strains were derived from the Ura− strain CAI4 (2).
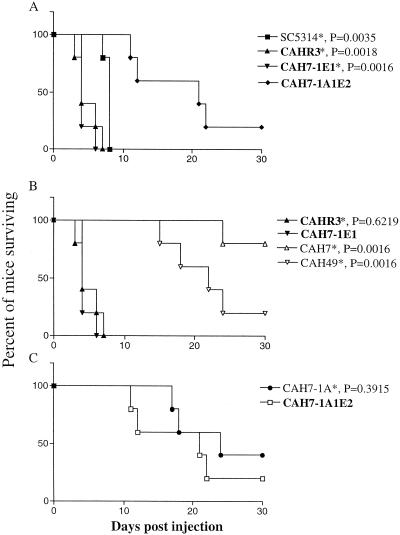
The effect of HWP1 in systemic candidiasis was reevaluated by comparing the survival curves of mice given isogenic strains differing in HWP1 copy number but identical in the location of the URA3 gene. Survival curves of mice given the homozygous hwp1/hwp1 mutant CAH7-1A1E2 were compared to those of mice given the heterozygous HWP1/hwp1 and revertant strains CAH7-1E1 and CAHR3, respectively. Each BALB/c mouse (female, 8 weeks old, n = 5) was given 0.1 ml of yeast cell suspension (5 × 106 cells/ml) (as per C. albicans strain CA-1 in reference 3) in phosphate-buffered saline and observed daily over a 30-day period. The animals used in this study were housed at the Association for Assessment and Accreditation of Laboratory Animal Care-certified Resources Center at Mon-tana State University under the National Institutes of Health guidelines for the care and use of laboratory animals. The survival curves showed that virulence of the hwp1/hwp1 mutant, CAH7-1A1E2, was reduced compared to that of the other strains (Fig. 1A). The differences in survival between CAH7-1A1E2 (hwp1/hwp1) and CAH7-1E1 or CAHR3 (both HWP1/hwp1) were statistically significant (P = 0.0018 and P = 0.0016, respectively, as calculated by the log rank test as before [3]). This is in agreement with the result of the previous study and supports the conclusion that HWP1 is important for systemic candidiasis.
FIG. 1.
Survival of BALB/c mice injected intravenously with C. albicans strains. (A) Survival of mice injected with HWP1 mutant strains with URA3 at the ENO1 locus (in bold) and with wild-type strain SC5314. The significance of survival differences relative to the hwp1/hwp1 strain CAH7-1A1E2 by the log rank test is indicated by an asterisk. (B) Survival of mice injected with heterozygous HWP1/hwp1 strains with URA3 at the HWP1 or ENO1 locus (in bold). The significance of survival differences relative to CAH7-1E1 is indicated by an asterisk. (C) Survival of mice injected with homozygous hwp1/hwp1 strains with URA3 at the HWP1 or ENO1 locus (in bold). The significance of survival relative to CAH7-1A1E2 is indicated by an asterisk.
In contrast to results with strain CAH7, the survival curve of the new HWP1/hwp1 heterozygote, CAH7-1E1, was equivalent to that of the revertant strain CAHR3 (Fig. 1A) and both of these strains were as virulent as wild-type strain SC5314. A trend of reduced virulence relative to CAHR3 was not found. The results strongly suggested that the location of the URA3 gene at the HWP1 locus in strain CAH7 contributed to its reduced virulence relative to CAHR3 in the previous study. A role for the genetic location of URA3 was also shown by differences in survival curves between the HWP1/hwp1 heterozygote strains that differed in the location of the URA3 gene. Strains CAH49 and CAH7 (HWP1/hwp1), with URA3 interrupting HWP1, were each reduced in virulence compared to CAH7-1E1 (HWP1/hwp1), with URA3 disrupting ENO1 (P = 0.0016 for both strains versus CAH7-1E1) (Fig. 1B). In contrast to the HWP1/hwp1 heterozygotes, targeting the URA3 gene to the enolase locus did not increase the virulence of the homozygous hwp1/hwp1 mutant. Survival curves of mice injected with strains CAH7-1A1E2 and CAH7-1A (both hwp1/hwp1) were not different (P = 0.3915) (Fig. 1C). The absence of HWP1 led to reduced virulence that was not enhanced by placing the URA3 gene at ENO1. This result strengthens the conclusion that HWP1 is important for virulence.
By comparing the virulence of strains with the selectable marker URA3 positioned identically, we were able to confirm that HWP1 is important for systemic candidiasis in mice. However, we also showed that the location of the URA3 selectable marker may influence the in vivo performance of C. albicans strains. Although measurement of URA3 gene activity in vivo is difficult and impractical, the results presented in this study suggest that placing URA3 at the HWP1 locus may lead to decreased URA3 expression in vivo, thereby causing a reduction in virulence compared to strains with URA3 at the ENO1 locus. The mechanism leading to the differences in expression based on genetic location are unknown; however, epigenetic regulation of URA3 at the HWP1 locus may lead to diminished URA3 expression. In addition, the induction of HWP1 during hyphae production may further limit URA3 levels in vivo. Perhaps these features compromise the cells for Ura3p.
Acknowledgments
Support for this research was provided from grant RO1 DE11375-05A2 from the National Institute of Dental and Craniofacial Research and from the Burroughs Wellcome fund to P.S. and by grants RO1AI24912 and PO1 AI37194 to J.E.C.
Editor: T. R. Kozel
REFERENCES
- 1.Bahn, Y. S., and P. Sundstrom. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183:3211-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han, Y., T. R. Kozel, M. X. Zhang, R. S. MacGill, M. C. Carroll, and J. E. Cutler. 2001. Complement is essential for protection by an IgM and an IgG3 monoclonal antibody against experimental, hematogenously disseminated candidiasis. J. Immunol. 167:1550-1557. [DOI] [PubMed] [Google Scholar]
- 4.Kirsch, D. R., and R. R. Whitney. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtz, M. B., M. W. Cortelyou, and D. R. Kirsch. 1986. Integrative transformation of Candida albicans, using a cloned Candida ADE2 gene. Mol. Cell. Biol. 6:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postlethwait, P., and P. Sundstrom. 1995. Genetic organization and mRNA expression of enolase genes of Candida albicans. J. Bacteriol. 177:1772-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]