Abstract
Although parainfluenza virus 5 (simian virus 5 [SV5]) circumvents the interferon (IFN) response by blocking IFN signaling and by reducing the amount of IFN released by infected cells, its ability to circumvent the IFN response is not absolute. The effects of IFN on SV5 infection were examined in Vero cells, which do not produce but can respond to IFN, using a strain of SV5 (CPI−) which does not block IFN signaling. Thus, by infecting Vero cells with CPI− and subsequently treating the cells with exogenous IFN, it was possible to observe the effects that IFN had on SV5 infection in the absence of virus countermeasures. IFN rapidly (within 6 h) induced alterations in the relative levels of virus mRNA and protein synthesis and caused a redistribution of virus proteins within infected cells that led to the enhanced formation of virus cytoplasmic inclusion bodies. IFN induced a steeper gradient of mRNA transcription from the 3′ to the 5′ end of the genome and the production of virus mRNAs with longer poly(A) tails, suggesting that the processivity of the virus polymerase was altered in cells in an IFN-induced antiviral state. Additional evidence is presented which suggests that these findings also apply to the replication of strains of SV5, parainfluenza virus type 2, and mumps virus that block IFN signaling when they infect cells that are already in an IFN-induced antiviral state.
The family Paramyxoviridae is a large group of enveloped, nonsegmented negative-stranded RNA viruses. The Paramyxoviridae family is divided into two subfamilies, Paramyxovirinae and Pneumovirinae, which are further subdivided into genera according to characteristics such as genome organization, virus morphology, protein characteristics, and relatedness of protein sequences. Studies on parainfluenza virus 5, more commonly referred to as simian virus 5 (SV5) (7), a prototype rubulavirus within the Paramyxovirinae subfamily, have led to important general findings regarding the molecular pathogenesis of this group of viruses. The envelope of SV5 contains three integral membrane proteins, the hemagglutinin-neuraminidase (HN), fusion (F), and small hydrophobic (SH; which is a minor component) proteins; the matrix (M) protein, which is required for the integrity of the virus particle, is located on the inner surface of the envelope. Within the envelope is the internal helical nucleocapsid core. This core structure consists of genomic RNA which is encapsidated by the nucleoprotein (NP). Associated with the nucleocapsid is the virus polymerase complex, consisting of the phospho (P) and large (L) proteins, and the V protein, which is an interferon antagonist (see below). These eight proteins are encoded by seven genes found within the SV5 genome of 15,246 nucleotides (reviewed by Lamb and Kolakofsky [23]). The genome has a 55-nucleotide (nt) 3′ leader sequence and a 30-nt 5′ trailer sequence, which are essential for virus RNA synthesis. Like other paramyxoviruses, SV5 has to generate three different RNA products during its infectious cycle, namely mRNAs, full-length antigenome RNA, and genome RNA. The viral polymerase responsible for transcription enters the template at the 3′ end in the noncoding leader sequence and sequentially synthesizes the NP, P/V, M, F, SH, HN, and L mRNAs, which are capped and polyadenylated, by terminating and restarting at each of the gene junctions (reviewed in reference 36). Between the gene stop and start sequences are highly diverse intergenic regions, which vary in length from 1 to 22 nt (19, 30, 31). Distinct gradient effects on transcription have been observed, with the NP mRNA being the most abundant species and the L mRNA being the least abundant. It is thought that this occurs because the polymerase only binds to the genomic RNA at a position within the 3′ leader sequence but has an increasing chance of disengagement the further it proceeds along the genome during transcription (1, 4, 14, 22). Occasionally, the polymerase fails to terminate at gene end sequences and proceeds to transcribe mRNA across the intergenic regions, generating both bi- and tricistronic mRNA species (26). However, due to ribosomal scanning, only the first cistron is translated into protein and hence transcriptional readthrough results in a decreased expression of the downstream genes.
Interferons (IFNs) are a group of secreted cell signaling glycoproteins that can induce an antiviral state within cells by upregulating the expression of many cellular genes, some of which, such as protein kinase R (PKR), oligo(A) synthetase, and Mx proteins, inhibit virus replication. There are two main subtypes of IFN: alpha/beta IFN (IFN-α/β), produced as a direct consequence of virus infection, and IFN-γ, which is produced by subsets of activated T lymphocytes and NK cells. All members of the Paramyxovirinae so far examined block IFN signaling, thereby inhibiting the expression of IFN-stimulated genes. Interestingly, the molecular mechanisms by which different viruses within this subfamily achieve this are very distinct, and it is likely that the mechanisms employed affect their biological characteristics (for reviews of how paramyxoviruses block the interferon response, see references 10, 17, 21, 25, and 33). However, it is unlikely that blocking IFN signaling alone is sufficient to allow these viruses to fully circumvent the IFN response because any IFN released by infected cells would still induce an antiviral state in surrounding uninfected cells, thereby making it difficult for the virus to spread from the initial foci of infection (3). Consequently, as well as blocking IFN signaling, most paramyxoviruses also limit the production of IFN by infected cells (20, 27, 35). Critical to their ability to interfere with IFN production, members of the Paramyxovirinae subfamily interact with, and inhibit the action of, mda-5, an intracellular signaling molecule that plays a key role in at least one intracellular signaling pathway that can lead to the induction of IFN (2). However, much can still be learned about the molecular pathogenesis of these viruses by further defining exactly how viruses interact with the antiviral effector molecules of the IFN response. For example, although SV5 blocks IFN signaling by targeting STAT1 for degradation and reduces the amount of IFN released by infected cells, its ability to circumvent the IFN response is not absolute, since infected cells still release small amounts of IFN and the replication of SV5 is restricted in cells that have entered an IFN-induced antiviral state prior to infection (3, 8, 12, 34). Indeed, SV5 forms larger plaques in MRC-5 (human diploid) cells that have been engineered to be nonresponsive to IFN (37). Furthermore, using SV5 as a model to study paramyxovirus persistence, we showed that a strain of SV5, termed CPI−, which was isolated from a dog infected with SV5 (strain CPI+), surprisingly failed to target STAT1 for degradation and thus did not block IFN signaling, even though CPI+, used to experimentally infect the dog, blocks IFN signaling. Further studies showed that three amino acid differences in the P/V N-terminal common domain of the V protein are responsible for the observed difference in the abilities of CPI− and CPI + to block IFN signaling (8).
An important question that needs to be addressed is when IFN induces an antiviral state within a cell, what effect does this have on SV5 replication? To begin to address this question, we have taken advantage of the fact that Vero cells do not produce, but can respond to, IFN and that the CPI− isolate of SV5 does not block IFN signaling (8). Thus, by adding IFN to Vero cells after they had been infected with CPI−, it was possible to analyze the effects of IFN on virus mRNA and protein synthesis after initiation of the virus replication cycle. Using this approach, we demonstrate that IFN can induce cellular antiviral responses, which significantly alter the pattern and levels of virus transcription and protein synthesis, and the formation of virus inclusion bodies.
MATERIALS AND METHODS
Cells, viruses, and interferon.
Vero cells were grown as monolayers in 25-cm2 or 75-cm2 tissue culture flasks in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum (growth medium) at 37°C. When required, cells were treated with recombinant human IFN-αA/D (rHuIFN-αA/D; PBL Biomedical Labs) (32) at 1,000 units/ml. Strains of SV5, CPI− and CPI+ (5, 6, 15), mumps virus (Jeryl Lynn vaccine strain), and human parainfluenza virus type 2 (hPIV2; Colindale prototype strain) were grown and titrated under appropriate conditions in Vero cells.
CPI− and CPI+ infection and IFN treatment time course.
Vero cell monolayers were infected with CPI− or CPI+ at a multiplicity of infection (MOI) of 10 to 50 PFU/cell (or mock infected). After an adsorption period of 1 to 2 h on a rocking platform at 37°C, the virus inoculum (or growth medium, for mock infections) was removed and replaced with fresh maintenance medium (Dulbecco's modified Eagle's medium containing 2% newborn calf serum). At 12 h postinfection (p.i.), the medium was either supplemented with rHuIFN-αA/D or left untreated as a −IFN control. Cells were either harvested immediately (0 h) or incubated for a further 6, 12, or 24 h (as indicated on the figures) and then harvested for protein or RNA analysis, as described below.
Preparation of radiolabeled antigen extracts, immunoprecipitation, and SDS-PAGE.
At various times after the addition or no addition of IFN, cells were metabolically labeled for 1 h with l-[35S]methionine (500 Ci/mmol; Amersham International Ltd., United Kingdom) in methionine-free tissue culture medium. At the end of the labeling interval, cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in immunoprecipitation buffer (10 mM Tris-HCl, pH 7.8, 5 mM EDTA, 0.3% Nonidet P-40, and 0.65 M NaCl; 4 × 106 to 6 × 106 cells per ml buffer) by sonication with an ultrasonic probe. Fifty microliters of total cell antigen extract sample was kept as a control to examine total cellular protein synthesis. Soluble antigen extract was prepared by subjecting the remainder of the cell lysate to centrifugation at 12,000 × g for 1 h to remove particulate material. Immune complexes were formed by incubating 1-ml samples of the soluble antigen extracts with a pool of monoclonal antibodies (MAbs) to the NP, P, M, and HN proteins of SV5 and a polyclonal antiserum to the P/V proteins of SV5 for 2 h at 4°C. The immune complexes were isolated by incubation with protein G-Sepharose 4B Fast Flow (Sigma) (1 h at 4°C). The proteins in the immune complexes were dissociated by heating in gel electrophoresis sample buffer at 100°C for 5 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, gels were fixed, stained, and dried. The resolved labeled polypeptide bands were visualized by autoradiography and quantitated by phosphorimager analysis.
RNA isolation and Northern blot hybridization.
Cells were washed twice in ice-cold PBS and lysed in Trizol reagent (Invitrogen), and total intracellular RNA was extracted from cell pellets according to the Trizol reagent supplier's protocol, except that the RNAs were extracted with phenol-chloroform and ethanol precipitated after the isopropanol precipitation. The isolated RNA was subjected to denaturing gel electrophoresis and transferred to nitrocellulose membranes as described by Grosfeld and coworkers (18). The nitrocellulose filters were prehybridized for 1 h at 65°C in 6× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) containing 0.1% SDS, 5× Denhardt's solution, and 0.5 mg of sheared DNA per ml. Hybridization was performed overnight under the same conditions with the addition of the [32P]dCTP-labeled negative-sense DNA probes specific for HN, NP, P, M, or actin mRNAs, which were prepared by runoff polymerization, as described below. The blots were washed once in 2× SSC-0.1% SDS at room temperature and three times in 2× SSC-0.1% SDS at 65°C for 30 min. The labeled RNA bands were visualized by autoradiography and quantitated by phosphorimager analysis.
Synthesis of single-stranded DNA hybridization probes.
The single-stranded DNA probes specific for SV5 mRNAs were generated as follows. Total intracellular RNA from CPI−-infected cells was used as a template in a reverse transcription-PCR for synthesis of cDNAs representing specific segments of the HN, NP, P, and M viral genes. For the actin probe, plasmid containing human cytoplasmic β-actin was used as a template for amplification of an actin DNA fragment. Each DNA fragment was approximately 400 to 500 bp in length; the details of primer sequences are available upon request. The DNA fragments were purified by agarose gel electrophoresis and used as templates in a runoff polymerization reaction to produce radioactively labeled single-stranded negative-sense DNA probes. Each runoff reaction mixture contained 100 to 200 ng of DNA template; 2 μM primer; 200 μM (each) unlabeled dATP, dTTP, and dGTP; 6.25 μM unlabeled dCTP; 25 μCi of [α-32P]dCTP (3,000 Ci/mmol); and 2.5 U of Taq DNA polymerase in 1× thermoscript buffer (NEB). The mixture was subjected to 40 cycles under the following conditions: 94°C for 30 s, 55°C for 30 s, 72°C for 30 s. Each probe was extracted with phenol-chloroform and heated to 95°C prior to hybridization. Typically, a quarter of the probe preparation was used in each hybridization reaction; however, to ensure that the probe was saturating the blot, some blots were rehybridized with a twofold-higher concentration of probe. The RNA pattern was similar regardless of the probe concentration in the hybridization mix, indicating that the probe was in excess in these experiments.
Analysis of viral mRNA poly(A) tail length.
The method used to analyze the length of the poly(A) tail of viral mRNA was adapted from that of Rassa et al. (31). Cell monolayers were washed once with PBS, trypsinized, and collected by centrifugation at 300 × g for 5 min. poly(A)+ RNA was isolated directly from the cells using an Oligotex Direct mRNA kit (QIAGEN Ltd., United Kingdom) according to the protocol for direct isolation of mRNA from animal cells, supplied in the manufacturer's instructions. poly(A)+ RNA representing ∼105 cells was heated to 75°C for 5 min and ligated to 50 pmol of phosphorylated oligonucleotide A (5′-P-GGTCACCTTGATCTGAAGC-NH2-3′, the oligonucleotide contains an amino modification to block ligation at its 3′ end) using T4 RNA ligase (New England Biolabs). The ligation was performed in a total volume of 10 μl at 37°C for 60 min. The reaction product was heated to 75°C for 5 min and cooled on ice, and the nucleic acid was extracted with phenol and chloroform, precipitated with ethanol, and resuspended in 12 μl of water. The purified ligation product was then used as a template in a reverse transcription reaction with 50 pmol of oligonucleotide B (5′-GCTTCAGATCAAGGTGACCTTTTT), which is complementary to oligonucleotide A and part of the poly(A) tail, using Sensiscript reverse transcriptase (QIAGEN Ltd., United Kingdom). Reverse transcription was carried out at 37°C for 1 h in a 20-μl reaction volume. The resulting cDNA products were then digested with RNase A/T1 mix and used as a template in a PCR with an mRNA sense SV5 M gene-specific primer (5′-TAACACTACTATTCCAATAACTGG, which anneals 41 to 65 nt from the site of polyadenylation), oligonucleotide B, and 1 μCi of [α-32P]dATP. The PCR consisted of 25 cycles under the following parameters: 95°C for 30 s, 52°C for 60 s, 68°C for 60 s. One-tenth of the resulting PCR products was mixed with loading buffer (5% glycerol, 1 mM Tris, 0.1 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue) and subjected to electrophoresis on a nondenaturing 5% polyacrylamide gel. The gel was dried and analyzed by autoradiography and phosphorimaging.
Immunofluorescence.
For immunofluorescence analysis, cells were grown on 13-mm-diameter coverslips (General Scientific Co. Ltd., Redhill, United Kingdom) in individual wells of 6- or 24-well plates. Cells were infected with CPI−, CPI+, mumps virus, or hPIV2, and the inoculum was adsorbed for 1 h. Treatment with exogenous IFN was as mentioned above. At various times p.i., monolayers were incubated in fixing solution (5% formaldehyde and 2% sucrose in PBS) for 15 min at room temperature, then permeabilized (5% Nonidet-P40 and 10% sucrose in PBS) for 5 min, and washed three times in PBS containing 1% fetal calf serum and 0.1% azide (PBS-1% fetal calf serum-0.1% azide). To detect the proteins of interest, cell monolayers were incubated with 10 to 15 μl of appropriately diluted primary antibody for 1 h. The antibodies used to detect SV5 proteins were the MAbs SV5-NP-a, SV5-P-e, and SV5-HN-4a (29). To detect hPIV2, the MAbs hPIV2-NP-a and hPIV2-NP-b (28) were used, and for mumps virus, a MAb to NP (provided by R. K. Rima, Queens University, Belfast) was used. Cells were subsequently washed (PBS-1% fetal calf serum-0.1% azide) several times, and the antibody-antigen interactions were detected by indirect immunofluorescence (1 h incubation) with a secondary Texas Red-conjugated goat anti-mouse immunoglobulin (Seralab, Oxford, United Kingdom). In addition, cells were stained with the DNA-binding fluorochrome 4′,6′-diamidino-2-phenylindole (DAPI, 0.5 μg/ml; Sigma-Aldrich Co Ltd., United Kingdom) for nuclear staining. Following staining, monolayers were washed with PBS, mounted with coverslips using Citifluor AF-1 mounting solution (Citifluor Ltd., United Kingdom), and examined under a Nikon Microphot-FXA immunofluorescence microscope.
RESULTS
Interferon induces alteration in CPI− virus protein synthesis.
The aim of this study was to examine the effect of IFN on SV5 replication. The SV5 variant CPI− was used in this study because this virus is unable to block IFN signaling, so any effects of IFN would be expected to be more evident than with a wild-type (wt) virus. CPI− was used to infect Vero cells, which although able to respond to IFN, do not produce IFN due to spontaneous gene deletions (11, 24), allowing IFN to be added to the cells under controlled conditions. So that the effect of IFN could be examined under conditions where virus replication had already been established and that IFN-induced effects on early events, such as virus entry, could be ruled out, IFN was added to cells 12 h following infection. At different times after IFN treatment, cells were metabolically labeled with [35S]methionine and the relative levels of the individual virus protein being synthesized were estimated. Analysis of virus proteins isolated by immunoprecipitation showed that the relative expression levels of NP, P, and V proteins were not significantly different in IFN-treated cells compared to nontreated cells. However, there was an obvious reduction in the relative levels of M, HN, and L (Fig. 1b). To confirm that the difference in protein expression was not an artifact of the immunoprecipitation step, total cell extracts were also examined (Fig. 1a). This analysis confirmed that IFN treatment significantly inhibited expression of the M protein but had little effect on synthesis of the NP, P, or V proteins or cellular proteins (Fig. 1a). These data suggest that IFN causes a specific downregulation in the expression of genes that are downstream of the V/P gene. In contrast, the addition of IFN to cells infected with a strain of SV5 (CPI+) that blocks IFN signaling had no effect on the relative levels of virus protein synthesis (Fig. 1c).
FIG. 1.
IFN treatment has an affect on CPI− viral protein synthesis but not on CPI+ protein synthesis in Vero cells. Vero cells were infected with CPI− (a and b) or CPI+ (c) at an MOI of 50 PFU/cell and either treated with exogenous rHuIFN-αA/D at 12 h p.i. or left untreated as a −IFN control. Cells were metabolically labeled with [35S]methionine for 1 h at various times after the addition of IFN, as indicated on the figure. Virus proteins were precipitated from soluble antigen extracts of these cells with a pool of MAbs to the NP, P, M, and HN proteins and a polyclonal antiserum to the P/V proteins. The precipitated proteins from CPI−- or CPI+-infected cells were subsequently separated on a 4 to 12% gradient polyacrylamide gel (Invitrogen) and visualized by phosphorimager analysis (panels b and c, respectively). Panel a shows the labeled polypeptides present in the total cell extracts from CPI−-infected cells.
Interferon induces alteration in CPI− virus transcription.
Given the striking change in the pattern of CPI− protein synthesis in response to IFN, we next examined the effect of IFN on the pattern of CPI− transcription. Vero cells were mock infected or infected with CPI− and either treated with IFN at 12 h p.i. or left untreated. At 6 and 12 h posttreatment, total intracellular RNA was isolated from the cells and analyzed by Northern blotting with 32P-labeled single-stranded DNA probes specific for viral mRNAs (Fig. 2a to d). A replicate blot was probed for actin mRNA to confirm that similar amounts of RNA were loaded in each lane (Fig. 2e). RNAs corresponding to monocistronic NP, P, M, and HN mRNAs, as well as larger RNAs corresponding to polycistronic readthrough transcription products, were detected. The monocistronic mRNAs were quantitated by phosphorimager analysis (Fig. 2f; note that relative levels of the various viral mRNAs are compared with respect to their levels in the absence of IFN at either 6 or 12 h posttreatment, which were each given a value of 100%). The results from this analysis revealed an alteration in both the levels and pattern of virus transcription in the presence of IFN. Surprisingly, there was an increase in levels of NP and P mRNA in cells treated with IFN compared to untreated cells (Fig. 2a and b, respectively). However, similar levels of M mRNA were detected in untreated and IFN-treated infected cells (Fig. 2c), but there was a significant decrease in HN mRNA in the presence of IFN (Fig. 2d). These data suggest that IFN has different effects on virus gene expression, depending on the position of the gene in the genome, causing an increase in transcription of genes at the 3′ end of the genome and a decrease in transcription of genes the further they are from the 3′ terminus. Comparison of the readthrough multicistronic RNAs on the blot probed for M mRNA support this observation; thus, the multicistronic mRNAs starting at the P gene (P/M and P/M/F) had a pattern similar to that of monocistronic P mRNA, but the multicistronic mRNAs starting at the M gene (M/F and M/F/SH) had a pattern similar to that of monocistronic M mRNA (Fig. 2c). Analysis of the HN blot revealed a surprising effect of IFN expression. In the absence of IFN, the HN probe detected a major band which migrated appropriately to be monocistronic HN (Fig. 2d; data not shown). A smaller faint band, labeled HN (x), could also be detected. Neither of these bands could be detected in the mock-infected control lane, indicating that both were virus specific. Following 12 h of treatment with IFN, the HN (x) band became prominent and was present at a higher level than the HN band. These data suggest that IFN treatment affects the size of the RNA produced from the HN gene.
FIG. 2.
Northern blot analysis of positive-sense RNAs synthesized in Vero cells infected with CPI− in the presence of IFN. Vero cells were infected with CPI− at a high MOI and treated with IFN at 12 h p.i. or left untreated as a −IFN control. Total intracellular RNA was isolated from these cells at 6 and 12 h after addition of IFN and subjected to Northern blot analysis. Hybridization was performed with 32P-labeled negative-sense DNA probes specific for viral mRNA NP (a), P (b), M (c), or HN (d) or the cellular actin mRNA (e). Quantitation was performed on the monocistronic mRNAs, and for each indicated gene, the amount of radioactivity in the RNA band in IFN-treated samples was normalized to the amount of RNA, at the same time point, for the same gene in untreated samples (which was given a value of 100%, indicated by a dotted line in panel f). Vero cells were mock infected or infected with CPI− at a high MOI, treated with IFN as described above, and metabolically labeled with [35S]methionine for 1 h at 0, 6, or 12 h following addition of IFN. Virus proteins were immunoprecipitated with a pool of MAbs to the NP, P, V, M, and HN proteins, which were then separated on a 4 to 12% polyacrylamide gel gradient (Invitrogen) and visualized by phosphorimager analysis (g). Quantitation of the labeled precipitated polypeptides in panel g was carried out, and the protein bands in each lane of IFN-treated samples (+) was normalized so that the respective bands in the lane of untreated samples (−) equaled 100% (indicated by a dotted line in panel h).
To allow a direct comparison of the IFN-induced effects on virus transcription and protein synthesis in this experiment, virus protein synthesis was monitored in cells that were infected and treated with IFN in parallel with those used for the RNA analysis (Fig. 2g). Quantitation of these results is presented in Fig. 2h. From these results, it is clear that although IFN alters the level of transcription and protein expression of each gene, there was not complete concordance between these two processes. Thus, although there was a considerable increase in the relative levels of NP and P mRNAs (3-fold and 8-fold, respectively) in the presence of IFN, the expression levels of NP and P proteins were slightly decreased (approximately 2-fold and 1.5-fold, respectively). Moreover, while there were similar levels of mRNA from the M gene, the expression levels of M protein were drastically decreased in the presence of IFN (approximately 30-fold in infected cells treated with IFN for 12 h). Significantly, while IFN considerably reduced the levels of HN mRNA (5-fold), a much stronger effect was observed in HN protein synthesis (20-fold decrease). Thus, these data suggest that IFN affected SV5 protein expression in at least two ways, (i) by altering the virus transcription gradient, resulting in an increase in transcription of genes at the 3′ end of the genome and a decrease in transcription of genes at the 5′ end of the genome, and (ii) by inhibiting protein synthesis from all virus mRNAs.
Interferon-induced increase in the length of the poly(A) tail of CPI− mRNA.
Careful examination of the Northern blots shown in Fig. 2 and in replicate experiments suggested that the monocistronic mRNAs isolated from cells treated with IFN migrated slightly more slowly than mRNA from untreated cells, especially at 12 h posttreatment. One possible explanation for an increase in the apparent size of the mRNAs was that there was an increase in the length of each of their poly(A) tails. To examine this possibility, we compared the length of the poly(A) tail on M mRNA from IFN-treated or untreated cells using an assay described by Rassa and coworkers (31). This assay involved ligating an oligonucleotide to the 3′ end of the mRNA and amplifying the region from 65 nt upstream of the gene end to the ligated oligonucleotide by reverse transcription and PCR. The PCR step incorporated [32P]dATP into the product, allowing it to be analyzed by polyacrylamide gel electrophoresis and autoradiography or phosphorimager analysis (Fig. 3). In this assay, the DNA band that migrated slightly below the 0.1-kb marker represents M mRNA that was not polyadenylated and the smear of DNA that migrated more slowly represents polyadenylated M mRNA, as described previously (Fig. 3a) (31). To allow quantitative analysis of the different sized products, the gel was analyzed by performing a lane intensity profile analysis in which the phosphorimager software generates a plot of the signal intensity along the length of each lane of the gel (Fig. 3b). Based on the sizes of the PCR products, it was clear that there was a significant increase in the average length of the poly(A) tails on M mRNA in the presence of IFN (by approximately 50 to 100 nucleotides, as estimated by PAGE). Also, whereas the amount of radioactivity incorporated into the product representing nonpolyadenylated RNA was slightly higher for the −IFN sample than for the +IFN sample, the amount of radioactivity contained in the products representing polyadenylated M mRNA was higher in the IFN-treated samples than in the untreated samples, indicating that a larger amount of [32P]dATP was incorporated, which is consistent with a longer A tract in the PCR product. These data demonstrate that IFN treatment caused an increase in the length of the poly(A) tail of the SV5 M mRNA.
FIG. 3.
Posttreatment with IFN affects the length of the poly(A) tail on CPI− M mRNA. poly(A)+ mRNA isolated from CPI−-infected Vero cells that were treated with rHuIFN-αA/D at 12 h p.i. for 6 h or left untreated as a −IFN control was used in the poly(A) tail length assay as described in the text, and the labeled products were analyzed by electrophoresis on a 5% polyacrylamide gel (a). The lanes containing samples derived from the −IFN- or +IFN-treated cells were analyzed with a phosphorimager to generate a lane intensity profile, in which the signal intensity (y axis) is plotted against the distance migrated (x axis) for each lane (b). The lane intensity profiles in panel b are aligned with the polyacrylamide gel shown in panel a.
IFN induces virus inclusion body formation in SV5 (CPI−) cells.
Given that IFN significantly alters the pattern of CPI− virus transcription and protein synthesis, we next examined what effect IFN had on the distribution of virus proteins. Vero cells were infected with CPI− for 12 h before the addition of IFN to the culture medium, and the distribution of virus proteins observed at 1, 3, and 6 days p.i. by immunofluorescence (the staining patterns for NP and P are shown in Fig. 4a). In the absence of IFN, NP and P were distributed diffusely throughout the cytoplasm and also in small cytoplasmic inclusion bodies at all times in the infection time course. However, following the addition of exogenous IFN, the distribution of virus proteins was dramatically altered; the NP and P proteins were primarily found in cytoplasmic inclusion bodies, which increased in size with time post-IFN treatment, while the M and HN proteins became undetectable (data not shown). As expected, in parallel experiments, the addition of IFN to CPI+-infected cells did not have a significant effect on the distribution of virus proteins, which had a pattern similar to that observed in cells infected with CPI− in the absence of IFN (Fig. 4b). Upon continued passage of CPI−-infected Vero cells in the continuous presence of IFN, the majority of cells remained infected, but virus replication was repressed and the NP and P proteins were primarily located in cytoplasmic inclusion bodies. However, within a few days following the removal of IFN, the NP and P proteins primarily had a diffuse cytoplasmic distribution, similar to that observed at early times p.i. in the absence of IFN (Fig. 5), and HN could be readily detected on the cell surface (data not shown).
FIG. 4.
IFN induces accumulation of the NP and P proteins into large cytoplasmic inclusion bodies in cells infected with CPI− but not in cells infected with CPI+. Vero cells were infected with either CPI− (a) or CPI+ (b) at an MOI of 50 PFU/cell, and at 12 h p.i., rHuIFN-αA/D was added to the culture medium or cells were left untreated as a −IFN control. Monolayers were fixed at 1, 3, and 6 days p.i. (CPI−) or 3 days p.i. (CPI+), and the distribution of NP and P proteins was analyzed by immunofluorescence using a pool of specific MAbs.
FIG. 5.
Distribution of the NP and P proteins in cells persistently infected with CPI− following removal of IFN from the culture medium. Vero cells were infected with CPI− at an MOI of 50 PFU/cell, and at 12 h p.i., rHuIFN-αA/D was added to the culture medium. The cells were cultured for 14 days (which required them to be passaged twice) in the continuous presence of IFN. At 14 days p.i., the cells were passaged in the presence or absence of IFN, and the distribution of the NP and P proteins was visualized at 15 and 17 days p.i., i.e., 1 or 3 days following the removal of IFN.
Restriction in the replication of strains of SV5 (CPI+) that block IFN signaling following infection of cells in an IFN-induced antiviral state.
Although most wt strains of SV5, including CPI+, block IFN signaling and limit the production of IFN by infected cells, their ability to circumvent the IFN response is not absolute. This has been demonstrated by the fact that such strains form larger plaques in monolayers of MRC5 and HEp2 cells, which have been engineered to be nonresponsive to IFN (37). To observe how IFN may limit the spread of SV5 strains that block IFN signaling, Vero cells were treated with IFN for 14 h or left untreated, as a control, and subsequently infected with either CPI− or CPI+ in the continued presence or absence of IFN as appropriate. At 12 h, 18 h, and 24 h p.i., the cells were metabolically labeled with [35S]methionine and the virus proteins were isolated by immunoprecipitation to estimate the relative levels of virus protein synthesis (Fig. 6). As expected, in the absence of IFN, both CPI− and CPI+ produced high levels of virus proteins at each time point. However, in IFN-pretreated cells, the pattern of both CPI− and CPI+ protein synthesis was dramatically altered. At 12 h p.i., there was an overall reduction in NP and P protein synthesis, and very little M and no HN or L could be detected. This pattern remained the same throughout the time course in CPI−-infected cells. However, in CPI+-infected cells, the levels of virus protein synthesis increased as the infection progressed, with the pattern becoming closer to that observed in untreated cells by 24 h p.i. We have previously shown that wt SV5 can degrade STAT1 in cells which have entered an antiviral state (13). Therefore, presumably, STAT1 was degraded in CPI+ (but not CPI−)-infected cells, and as a consequence, these cells could not maintain an antiviral state indefinitely in the absence of IFN signaling. We next examined the effect of IFN pretreatment on the distribution of virus proteins in infected cells. Vero cells were or were not pretreated with IFN for 14 h and then infected at a high MOI with either CPI− or CPI+ virus. Cell monolayers were fixed at 1 and 3 days p.i. and examined by immunofluorescence. In IFN-pretreated cells infected with CPI−, the NP and P were distributed in cytoplasmic inclusion bodies at both 1 and 3 days p.i. (Fig. 7a). However, the size of the inclusion bodies got slightly larger with time, consistent with the finding that although CPI− protein synthesis was inhibited in IFN-pretreated cells, low levels of NP and P synthesis occurred. In IFN-pretreated cells infected with CPI+, at 1 day p.i., the majority of NP and P were located in small cytoplasmic inclusion bodies, although in a few cells, NP and P were distributed throughout the cytoplasm. (Fig. 7b). By 3 days p.i., the proportion of cells strongly positive for virus antigen was much higher, although in a significant proportion of cells, NP and P were still primarily located in cytoplasmic inclusion bodies. Also, an increasing proportion of CPI+ (but not CPI−)-infected cells became positive for HN as the time course progressed (data not shown).
FIG. 6.
Effect of IFN pretreatment on CPI− and CPI+ protein synthesis in Vero cells. Cells were (pretreat) or were not (untreat) pretreated with rHuIFN-αA/D for 14 h prior to infection with CPI− or CPI+ at an MOI of 10 PFU/cell. Cells pretreated with IFN were infected and subsequently cultured in the continuous presence of IFN. At 12, 18, and 24 h p.i., cells were metabolically labeled with [35S]methionine for 1 h. Virus proteins were immunoprecipitated using a mixture of MAbs to the NP, P, M, and HN proteins, which were separated on a 4 to 12% gradient polyacrylamide gel and visualized by phosphorimager analysis.
FIG. 7.
Pretreatment with IFN affects the distribution of NP and P proteins of both strains of SV5, CPI− and CPI+. Vero cells were or were not pretreated with rHuIFN-αA/D for 14 h and then infected with CPI− (a) or CPI+ (b) at an MOI of 10 PFU/cell. Cells pretreated with IFN were both infected and subsequently cultured in the continuous presence of IFN. At 1 and 3 days p.i., the cells were fixed, and the distribution of viral proteins was analyzed by immunofluorescence using a pool of MAbs to the NP and P proteins.
IFN can induce inclusion body formation in cells infected with mumps virus and hPIV2.
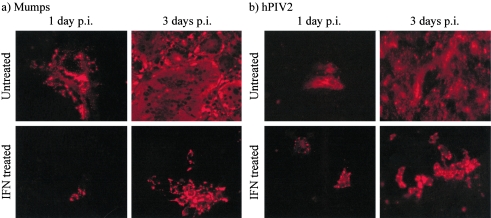
In light of the observed effects of IFN on SV5 protein synthesis and spread, we investigated whether IFN similarly affected the replication of other rubulaviruses. Vero cells were infected with mumps virus or hPIV2 at a low MOI (0.01 PFU per cell) and treated with IFN at 12 h p.i. or left untreated as a control. The spread of these viruses from the initial focus of infection to neighboring cells was monitored by immunofluorescence using antibodies specific for their respective proteins. From these experiments, it was clear that IFN delayed the spread of mumps virus and hPIV2 (Fig. 8). In addition, there was evidence that inclusion body formation was induced, similar to the situation observed with CPI+.
FIG. 8.
Treatment with IFN has an effect on the spread of mumps virus and hPIV2. Vero cells were infected at an MOI of 0.01 PFU/cell with mumps virus (a) or hPIV2 (b), and 12 h later, rHuIFN-αA/D was added to the culture medium or the cells were left untreated. Monolayers were fixed at 1 and 3 days p.i. and stained with appropriate antibodies to detect the respective viruses. (DAPI staining of the monolayers revealed that there were between 150 and 300 cells present in each panel [data not shown]).
DISCUSSION
Although SV5 specifically reduces the production of IFN by infected cells through its interaction with mda-5, it does not completely prevent cells from secreting IFN. In vivo, immune cells, such as plasmacytoid dendritic cells and activated subsets of lymphocytes, will also secrete large amounts of IFN in response to virus infection, which could exert effects in virus-infected cells (9). What is clear from the results presented here and elsewhere (3, 8, 12, 34) is that cells in an IFN-induced antiviral state severely restrict SV5 replication. However, if cells in an IFN-induced antiviral are infected with strains of SV5, such as CPI+, which block IFN signaling by targeting STAT1 for degradation, they cannot maintain the antiviral state once STAT1 has been degraded, thereby allowing the virus to replicate normally and to spread from an initial focus of infection. To further define how IFN inhibits the spread of SV5, we have taken advantage of the facts that CPI− cannot block IFN signaling and that Vero cells do not produce IFN. Thus, by infecting Vero cells with CPI− and adding exogenous IFN to infected cells once virus replication was well established, the effects of IFN on virus macromolecular synthesis could be monitored. These experiments demonstrated that IFN induces an antiviral response in cells, which changes the pattern of SV5 transcription and protein synthesis and results in an altered distribution of virus proteins.
These studies showed that IFN treatment had at least three effects on CPI− virus mRNA expression. First, it caused a change in the virus transcription gradient, resulting in augmented levels of mRNA from genes at the 3′ end of the genome, and a decrease in the levels of mRNA from genes at the 5′ end of the genome (Fig. 2). Second, it caused an alteration in the length of the poly(A) tail of virus-specific mRNA (Fig. 2 and 3). Third, it caused a change in the expression of HN mRNA, with a significant increase in the level of a truncated mRNA, HN (x) (Fig. 2d). The nature of HN (x) is not known, but presumably, it is either an HN gene transcription product that has been initiated or terminated inappropriately or an HN mRNA degradation product, perhaps specifically generated through the action of an IFN-induced enzyme such as RNase L. One plausible explanation for each of these three effects on mRNA expression is that IFN treatment results in a change in the processivity of the virus polymerase. If polymerase processivity were reduced, the polymerase/transcript complex might be relatively unstable and prone to disengaging, even during mRNA synthesis within gene coding regions, thus accounting for the truncated HN (x) mRNA species. In addition, a reduction in polymerase processivity could result in excessive stuttering during polyadenylation, thus accounting for the increased length of the poly(A) tails of viral mRNA. Both of these effects could increase the chance of the polymerase disengaging from the template, within genes and at the gene junctions, respectively, thereby explaining the observed general decrease in mRNA levels encoded by genes farther from the single 3′-proximal polymerase entry site. Furthermore, if the polymerase that dissociated from the template prematurely could reinitiate mRNA synthesis at the 3′ end, this might explain why higher levels of NP and P mRNA were transcribed in cells in the presence of exogenous IFN. If this model is correct, it is unclear why there seems to be less chance of the polymerase disengaging the template between the NP and P genes than at the other gene junctions, but this could reflect effects of the different intergenic regions. In this respect, it is of note that it has been reported that sequence diversity at SV5 gene junctions may differentially affect SV5 gene expression and hence provide additional control of virus transcription (19, 30, 31). However, further studies will be necessary to fully ascertain how IFN induces the observed effects on CPI− virus transcription and whether the effects are primarily on termination-reinitiation or chain elongation. Furthermore, it is possible that IFN induces alterations in the stability of viral mRNAs, although it is unlikely that any gross changes in mRNA stability could explain the steeper gradient of mRNA transcription from the 3′ to the 5′ end of the genome observed upon IFN treatment. A direct comparison between mRNA accumulation and protein synthesis showed that there was not a complete correlation between these processes (Fig. 2). For example, in cells infected with CPI−, higher levels of NP and P mRNAs were synthesized in IFN-treated cells than in untreated cells, but the overall level of NP and P protein synthesis was slightly decreased in IFN-treated cells compared to untreated cells. More strikingly, while the levels of M mRNA were similar in IFN-treated and untreated cells, the levels of M protein were dramatically reduced in IFN-treated cells. These results suggest that in addition to IFN-induced effects on virus transcription, virus protein synthesis was also independently repressed, perhaps through the induction of PKR and oligo(A) synthetase. However, since there was little discernible effect of IFN on the overall level of cellular protein synthesis in IFN-treated cells infected with CPI− (Fig. 1a), if IFN does induce specific inhibition of virus protein synthesis, the effector mechanisms must be acting locally within cells in areas where virus replication is occurring, e.g., through double-stranded RNA activation of PKR and oligo(A) synthetase.
In addition to demonstrating that IFN can induce changes to the patterns of virus transcription and protein synthesis, we also show that IFN induces changes in the distribution of the virus proteins synthesized. Thus, following IFN treatment of CPI−-infected Vero cells, the NP and P proteins rapidly became localized in cytoplasmic inclusion bodies, whereas in the absence of IFN, the majority of NP and P in untreated cells was more evenly distributed throughout the cytoplasm. Interestingly, the inclusion bodies in IFN-treated cells increased in size as the infection progressed (Fig. 4), consistent with the observation that the synthesis of NP and P continued over this time period in the presence of IFN (data not shown). Whether the residual virus transcription that occurs in cells in an IFN-induced antiviral state occurs within these inclusion bodies is not known. IFN treatment also induced the formation of inclusion bodies in cells infected with other viruses, CPI+ (Fig. 7), mumps virus (Fig. 8), and hPIV2 (Fig. 8), indicating that this phenomenon is not specific to the CPI− virus. The driving forces for the formation of these inclusion bodies, the role of IFN-induced cellular proteins (e.g., MxA) in their formation, their composition, and their role in the virus life cycle are currently under investigation, as is the molecular basis for the effects that IFN has on SV5 transcription and protein synthesis.
An enormous amount of effort has gone into understanding the molecular basis by which cellular proteins with antiviral activity work and how specific viruses circumvent the IFN response. However, the results presented here highlight that virus pathogenesis may also be influenced by the specific way viruses replicate in cells that have entered an IFN-induced antiviral state. For example, we have suggested that viruses that do not block IFN signaling, such as CPI−, may be selected in vivo because they are better able to establish persistent infections. Our model is that as the virus becomes repressed in response to IFN, virus glycoproteins are lost from the surface of infected cells and virus nucleocapsid proteins accumulate in cytoplasmic inclusion bodies (8). The results presented here suggest that if cytoplasmic inclusion bodies do have a role to play in virus pathogenesis, then they may also be formed when viruses that can block IFN signaling infect cells already in an IFN-induced antiviral state (Fig. 7). Indeed, cytoplasmic inclusion bodies may be a virus defense mechanism in which the virus can hide both from intracellular antiviral responses and adaptive immune responses (8, 16). If this is the case, then it opens up the question as to whether the way in which viruses such as SV5 change their pattern of virus transcription and protein synthesis in response to the programmed IFN response may have been selected for during virus evolution.
Acknowledgments
T.S.C. is extremely grateful to Fundacao Ciencia e Tecnologia, Portugal, for a Ph.D. studentship. The work was also supported by grants from the Wellcome Trust and BBSRC.
REFERENCES
- 1.Abraham, G., and A. K. Banerjee. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartner, W., S. Krakowka, and J. R. Blakeslee. 1987. Persistent infection of Vero cells by paramyxoviruses. A morphological and immunoelectron microscopic investigation. Intervirology 27:218-223. [DOI] [PubMed] [Google Scholar]
- 6.Baumgartner, W. K., S. Krakowka, A. Koestner, and J. Evermann. 1982. Acute encephalitis and hydrocephalus in dogs caused by canine parainfluenza virus. Vet. Pathol. 19:79-92. [DOI] [PubMed] [Google Scholar]
- 7.Chatziandreou, N., N. Stock, D. Young, J. Andrejeva, K. Hagmaier, D. J. McGeoch, and R. E. Randall. 2004. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 85:3007-3016. [DOI] [PubMed] [Google Scholar]
- 8.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 9.Colonna, M., G. Trinchieri, and Y. J. Liu. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5:1219-1226. [DOI] [PubMed] [Google Scholar]
- 10.Conzelmann, K. K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson, S. U. 1982. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31:635-642. [DOI] [PubMed] [Google Scholar]
- 15.Evermann, J. F., S. Krakowka, A. J. McKeirnan, and W. Baumgartner. 1981. Properties of an encephalitogenic canine parainfluenza virus. Arch. Virol. 68:165-172. [DOI] [PubMed] [Google Scholar]
- 16.Fearns, R., D. F. Young, and R. E. Randall. 1994. Evidence that the paramyxovirus simian virus 5 can establish quiescent infections by remaining inactive in cytoplasmic inclusion bodies. J. Gen. Virol. 75:3525-3539. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 18.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., and R. A. Lamb. 1999. Effect of inserting paramyxovirus simian virus 5 gene junctions at the HN/L gene junction: analysis of accumulation of mRNAs transcribed from rescued viable viruses. J. Virol. 73:6228-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 21.Horvath, C. M. 2004. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15:117-127. [DOI] [PubMed] [Google Scholar]
- 22.Iverson, L. E., and J. K. Rose. 1982. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J. Virol. 44:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 24.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai, Y., and A. Kato. 2004. Accessory genes of the Paramyxoviridae, a large family of nonsegmented negative-strand RNA viruses, as a focus of active investigation by reverse genetics, p. 197-248. In Y. Kawaoka (ed.), Biology of negative strand RNA viruses, vol. 283. Springer-Verlag, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 26.Parks, G. D., K. R. Ward, and J. C. Rassa. 2001. Increased readthrough transcription across the simian virus 5 M-F gene junction leads to growth defects and a global inhibition of viral mRNA synthesis. J. Virol. 75:2213-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 28.Randall, R. E., and D. F. Young. 1988. Comparison between parainfluenza virus type 2 and simian virus 5: monoclonal antibodies reveal major antigenic differences. J. Gen. Virol. 69:2051-2060. [DOI] [PubMed] [Google Scholar]
- 29.Randall, R. E., D. F. Young, K. K. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 30.Rassa, J. C., and G. D. Parks. 1999. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J. Virol. 73:3904-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rassa, J. C., G. M. Wilson, G. A. Brewer, and G. D. Parks. 2000. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 274:438-449. [DOI] [PubMed] [Google Scholar]
- 32.Rehberg, E., B. Kelder, E. G. Hoal, and S. Pestka. 1982. Specific molecular activities of recombinant and hybrid leukocyte interferons. J. Biol. Chem. 257:11497-11502. [PubMed] [Google Scholar]
- 33.Stock, N., S. Goodbourn, and R. E. Randall. 2004. The anti-interferon mechanisms of paramyxoviruses. Kluwer Plenum, New York, N.Y.
- 34.Wansley, E. K., P. J. Dillon, M. D. Gainey, J. Tam, S. D. Cramer, and G. D. Parks. 2005. Growth sensitivity of a recombinant simian virus 5 P/V mutant to type I interferon differs between tumor cell lines and normal primary cells. Virology 335:131-144. [DOI] [PubMed] [Google Scholar]
- 35.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61-119. [DOI] [PubMed] [Google Scholar]
- 37.Young, D. F., L. Andrejeva, A. Livingstone, S. Goodbourn, R. A. Lamb, P. L. Collins, R. M. Elliott, and R. E. Randall. 2003. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 77:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]