Abstract
Only a relatively few mutations in its spike protein allow the murine coronavirus to switch from a murine-restricted tropism to an extended host range by being passaged in vitro. One such virus that we studied had acquired two putative heparan sulfate-binding sites while preserving another site in the furin-cleavage motif. The adaptation of the virus through the use of heparan sulfate as an attachment/entry receptor was demonstrated by increased heparin binding as well as by inhibition of infection through treatment of cells and the virus with heparinase and heparin, respectively.
Coronaviruses are enveloped, plus-stranded RNA viruses, which generally cause respiratory and/or intestinal infections, although some may spread systemically. They are pathogens of veterinary importance, often associated with great economic losses. Their relevance has increased considerably since the recent emergence of new human coronaviruses (HCoV), such as the severe acute respiratory syndrome coronavirus (SARS-CoV). In general, coronaviruses have a limited host range and cause disease in a single or a few closely related host species. However, the emergence of SARS-CoV, which resulted from a zoonotic transmission event (19), demonstrates the need for a better understanding of coronavirus interspecies transmission.
The interaction of the coronavirus spike (S) protein, a class I fusion protein (5), with its receptor is the major determinant for virus entry and host range restriction. While nonpermissive cell lines can be rendered susceptible by making them express the receptor (see references below), coronaviruses can also be retargeted to specific cells by exchanging the ectodomain of the S protein with that of another appropriate coronavirus, as was demonstrated for mouse hepatitis virus (MHV) (24) and feline infectious peritonitis virus (20). Receptors have so far been identified for the group 2 coronavirus MHV (murine carcinoembryonic antigen-related cell adhesion molecule 1 [CEACAM1]) (16, 38), for SARS-CoV (ACE2) (26), for the group 1 coronaviruses transmissible gastroenteritis virus and porcine respiratory coronavirus (porcine APN) (12, 13), for feline infectious peritonitis virus (feline APN) (36), for HCoV-229E (human APN) (40), and for HCoV-NL63 (ACE2) (21).
The S protein is synthesized as a heavily glycosylated polypeptide, which oligomerizes in the endoplasmic reticulum to form trimers (14, 27). As a late maturation step during its transport to the cell surface, cleavage of the MHV S protein into an amino-terminal S1 and a carboxy-terminal S2 domain can occur. A basic amino acid sequence resembling the furin consensus sequence motif occurs approximately in the middle of the protein and was shown to be the target of a furin-like enzyme in the case of MHV-A59 (11). While cleavage of the MHV S protein generally correlates strongly with cell-cell fusion (7), virus-cell fusion appears not to be affected by the prevention of S protein cleavage, indicating that these fusion events have different requirements (11).
The amino-terminal S1 domain (or its equivalent part in viruses with uncleaved S proteins) is responsible for receptor binding, and the carboxy-terminal S2 domain is responsible for membrane fusion. For several coronaviruses, the receptor-binding site in the S1 domain has been mapped. For MHV strain JHM (MHV-JHM), for instance, it was mapped to the domain composed of the 330 amino-terminal residues of the S molecule (23). This amino-terminal domain also determines CEACAM receptor specificity of other MHV strains (37). For transmissible gastroenteritis virus (18), HCoV-229E (4, 6), and SARS-CoV (1, 39), the receptor-binding domains have also been mapped to the S1 subunit, though to different regions therein.
Although MHV is critically dependent on murine CEACAM for cell entry and therefore only infects murine cells, MHV variants capable of infecting nonmurine cells were obtained from persistently infected cell cultures (2, 3, 31, 33). The viruses generated by Baric and coworkers (2) still used murine CEACAM as a receptor but were dependent on human CEACAM for entry into human cells. The receptor determinant of the MHV variant (MHV/BHK) generated by Sawicki and Schickli and coworkers (31, 33) has not been determined yet. Strikingly, this variant is no longer dependent on murine CEACAM for entry and appears to exhibit an even more extended host range, being able to infect cells from many different species (33). The MHV/BHK S protein (GenBank accession number AY497331) differs from the S protein of the parental MHV-A59 strain (GenBank accession number AY497328) at 57 residues and, additionally, contains a 7-amino-acid insert. Analysis of several viruses resulting from recombination between MHV-A59 and MHV/BHK demonstrated a correlation between 21 amino acid substitutions and the 7-amino-acid insert, all located in the S1 domain, with the extended host range (32). However, although introduction of these mutations into an isogenic background permitted MHV-A59 to interact with alternative receptors on murine and nonmurine cells, these viruses failed to induce a second round of infection in nonmurine cells under liquid medium, indicating that additional substitutions in S or mutations in other viral genes may be needed for efficient infection of these cells (35).
These studies raised the questions of how these viruses have overcome the apparent dependence on a specific receptor and by what interactions the S protein is triggered to undergo the conformational changes required to initiate the fusion process. In the present study, we determined the attachment/entry receptor of the extended host range variant generated by Sawicki and coworkers (MHV/BHK). In addition, we demonstrated that the S gene of MHV/BHK is sufficient to confer the extended host range phenotype.
To this end, we generated recombinant viruses which differ only in their S gene sequences by targeted recombination, as described previously (9), and compared their growth on murine and nonmurine cells. Thus, while MHV-2aFLS, which has been described previously (8), contains the parental MHV-A59 S gene, MHV-2aFLSrec contains the MHV/BHK S gene (Fig. 1A) in the isogenic MHV-A59 background. The MHV/BHK S gene was assembled from reverse transcription (RT)-PCR products (virus kindly provided by Stan Sawicki) and cloned into transcription vector pMH S21BHK+i (kindly provided by Kay Holmes) (35), in which a firefly luciferase (FL) expression cassette had been introduced at the position of the hemagglutinin esterase pseudogene, as will be described in more detail elsewhere. The MHV/BHK S gene is identical to the published sequence with the exception of two silent mutations. MHV-2aFLSrec was purified twice, using endpoint dilutions on LR7 cells before a passage 1 stock was grown, the S gene of which was checked for integrity before use for RT-PCR and sequence analysis. For both MHV-2aFLS and MHV-2aFLSrec, two independent recombinants were generated to verify that the observed phenotypic characteristics were the result of the intended genomic modifications.
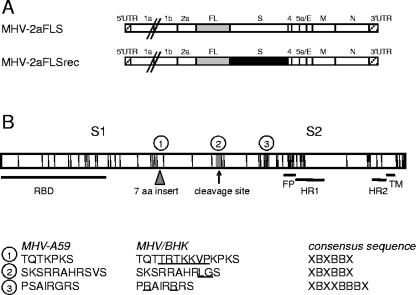
FIG. 1.
Genomic organization of the recombinant viruses. (A) The genome structures of the recombinant MHVs containing either the parental MHV-A59 or the MHV/BHK spike gene and an FL expression cassette are depicted (MHV-2aFLS and MHV-2aFLSrec, respectively). Numbers and lowercase letters designate the genes encoding nonstructural proteins, while genes encoding spike (S) protein, envelope (E) protein, membrane (M) protein, or nucleocapsid (N) protein are marked by the protein abbreviation. The 5′ and 3′ untranslated regions (UTR) are also indicated. (B) The spike protein is depicted as an elongated box. Each vertical line in this box indicates an amino acid substitution in the MHV/BHK S protein compared to the parental MHV-A59 spike protein. The triangle indicates a 7-amino-acid insertion. The MHV-A59 S protein can be cleaved at the position of the arrow into an amino-terminal S1 and a carboxy-terminal S2 domain. Horizontal lines designate the approximate locations of the receptor-binding domain (RBD), putative fusion peptide (FP) (5), heptad repeat region 1 (HR1) and HR2, and the transmembrane domain (TM). The encircled numbers specify the heparin-binding consensus sequences, the locations of which are indicated by gray boxes, while their sequences are given below for the MHV-A59 and the MHV/BHK spike proteins. The amino acid insertions and substitutions in the MHV/BHK spike protein compared to the MHV-A59 spike protein are underlined. The heparin consensus sequences themselves are also shown (X, any amino acid; B, basic amino acid).
As a first step in characterizing the recombinant viruses, comparisons of their growth characteristics on murine cells were made. Cultures of LR7 cells infected with the different recombinants were lysed at 6.5 and 8.5 h postinfection, after which luciferase expression levels were determined (Fig. 2A). The recombinant viruses expressed similar levels of luciferase at both time points, indicating that the changes in the spike protein did not affect the ability of the viruses to enter LR7 cells. However, the recombinant viruses clearly differed in their ability to form plaques (Fig. 2B). While infection with MHV-2aFLS resulted in the formation of large syncytia, no cell-cell fusion was observed for MHV-2aFLSrec; only single cells appeared to be infected, as visualized by immunocytochemistry using anti-MHV antibodies (K134) 30).
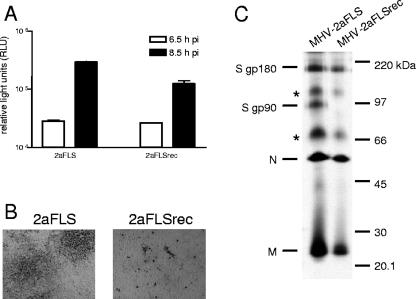
FIG. 2.
Infection of murine cells. (A) LR7 cells were inoculated with MHV-2aFLS or MHV-2aFLSrec at a multiplicity of infection of 1. At the indicated time points postinfection (pi), the cells were lysed and intracellular luciferase expression was determined by using a luminometer (values are expressed in relative light units, RLU). Standard deviations are indicated. (B) Plaque phenotypes of recombinant viruses on LR7 cells. At 18 h postinfection, cells were fixed with a 3% formaldehyde solution, after which the agar overlay was removed. After being permeabilized with 1% Triton X-100 in phosphate-buffered saline, viral antigen was detected with the anti-MHV serum k134 at a 1:400 dilution. Peroxidase-conjugated swine immunoglobulins to rabbit immunoglobulins (Dakopatts) were used as secondary antibodies at a 1:100 dilution. Peroxidase was visualized, using an AEC substrate kit from Vector laboratories. Pictures were taken using bright-field microscopy and a Nikon DS-L1 digital camera. (C) LR7 cells were infected with the recombinant viruses indicated on the top of the gel at a multiplicity of infection of 10. Cells were labeled for 3 h with 35S-labeled amino acids (Amersham), starting 5 h postinfection. At the end of the labeling period, culture media were collected and prepared for immunoprecipitation with the anti-MHV serum k134, followed by polyacrylamide gel electrophoresis, followed by fluorography, as previously described (10). The positions of the different viral proteins are indicated on the left, while the molecular mass marker is indicated on the right. Asterisks indicate higher-order structures of the M and/or N proteins.
Previously, we have demonstrated that, while virus-cell fusion was not affected by cleavage inhibition of the MHV-A59 spike protein, cell-cell fusion was significantly reduced. Because the mutations in the S protein did not appear to interfere with the infectivity of the recombinant viruses on LR7 cells but had a severe effect on cell-cell fusion, the cleavage of the spike proteins was analyzed (Fig. 2C). The S protein of MHV-2aFLS appeared in its two well-known forms, the mature 180-kDa form (gp180) and its 90-kDa cleavage products (gp90). However, cleavage could not be detected for MHV-2aFLSrec, not even after prolonged exposure of the film to the gel. Although the basic amino acid sequence, resembling the furin consensus sequence, is not changed in the S protein of this virus, the substitution of the serine and valine residues immediately downstream thereof by leucine and glycine residues, respectively (RRAHR↓SV → RRAHRLG), may prevent cleavage. Consistently, the prediction program ProP (15) readily identified the cleavage site in the S gene of MHV-2aFLS, but not after substitution of the serine and valine residues.
Next, the extended host range phenotypes of the recombinant viruses were analyzed. As a first step, viral infectivity in the passage 2 stocks was determined by a quantal assay on LR7 and HeLa cells and their 50% tissue culture infective dose values were calculated. When inoculated into cultures of murine LR7 cells, MHV-2aFLS and MHV-2aFLSrec appeared not to differ substantially in infectivity. However, a clear difference was observed after inoculation of the HeLa cells. No titer could be determined for MHV-2aFLS containing the parental S gene, while viruses containing the MHV/BHK S gene reached titers that were only 10-fold lower than those obtained on LR7 cells (data not shown). Subsequently, the growth characteristics of the recombinant viruses on HeLa cells were analyzed in more detail. Cultures of HeLa cells infected with the different recombinants were lysed at 8.5 and 18 h postinfection, after which the luciferase expression levels were determined (Fig. 3B). MHV-2aFLSrec was clearly able to infect HeLa cells, while MHV-2aFLS was not. Luciferase levels increased from 8.5 to 18 h postinfection, corresponding with the spread of the virus through the cell culture. These results were confirmed by immunocytochemistry, using the MHV antiserum on HeLa cells at 18 h postinfection (Fig. 3C). These results indicate that the changes in the spike gene are sufficient to confer an extended host range phenotype to MHV-A59.
FIG. 3.
Infection of human cells. (A) HeLa cells were inoculated with MHV-2aFLS and MHV-2aFLSrec, and intracellular luciferase expression was determined as described in the legend to Fig. 2A. (B) HeLa cells were infected with the recombinant viruses as described above. At 18 h postinfection, the cells were fixed and viral antigen was detected as described in the legend to Fig. 2B.
In an attempt to understand the extended host range caused by the mutations in the MHV/BHK S protein, we made a careful study of its features. This revealed the presence of two putative heparan sulfate (HS)-binding sites, which were not present in the parental sequence (Fig. 1B). Furthermore, the predicted furin-cleavage site (SKSRRAHR↓) also corresponds to a HS-binding consensus sequence (XBBXBX) (28). Strikingly, the mutations that prevented the cleavage of the S protein did not affect the furin consensus sequence itself and hence also not the HS-binding consensus motif. These findings prompted us to analyze HS as an attachment/entry receptor for MHV-2aFLSrec.
In order to test the involvement of HS in the entry process of the host range mutant, LR7 and HeLa cells were treated with heparinase I (Sigma), which degrades HS (28), for 1.5 h prior to inoculation with the recombinant MHVs. Strikingly, luciferase expression by MHV-2aFLS, which carries the parental spikes, in LR7 cells was increased approximately 2.5-fold after heparinase I treatment (Fig. 4A). However, treatment of both LR7 and HeLa cells with heparinase I decreased the entry of MHV-2aFLSrec by 50 to 60%. The incomplete inhibition may be explained by the presence of heparinase-resistant oligosaccharides (28). Next, the ability of heparin (ICN Biochemicals) to inhibit viral infectivity was tested. Heparin, a product of mast cells, is commonly used as an analog of cellular HS in receptor-ligand interaction assays, since ligand interactions with heparin and heparan sulfate have little quantitative difference (22). Incubation of the recombinant viruses with heparin prior to infection dramatically decreased the infectivity of MHV-2aFLSrec (>99%) on both LR7 and HeLa cells but not the infectivity of MHV-2aFLS (Fig. 4B). Finally, the ability of virions to bind heparin was tested. Therefore, similar quantities of virions were incubated for 1 h at 4°C with type I heparin-agarose beads in the presence or absence of heparin (500 ng/ml; added 1 h before addition of the beads). After three washes, the amounts of viral genomic RNA adsorbed to the beads were compared by Taqman reverse transcription-PCR (11). The results of a representative experiment are shown in Fig. 4C. As expected, many more MHV-2aFLSrec than MHV-2aFLS virions were found attached to the heparin beads. In both cases, adsorption to the beads could be efficiently blocked by the addition of heparin, indicating the specificity of the assay.
FIG. 4.
Interaction with heparan sulfate/heparin. (A and B) LR7 and HeLa cells were inoculated with the recombinant viruses as described in the legend to Fig. 2A, except that the cells had been pretreated with heparinase I for 1.5 h before the inoculation (A) or the recombinant viruses had been incubated with different concentrations of heparin for 1 h at 4°C (B). At 5 h post infection, the FL activity in the cultures was determined. Standard deviations are indicated. (C) The percentage of MHV virions adsorbed to heparin-agarose beads was determined by a Taqman reverse transcriptase PCR specific for viral genomic RNA, as previously described (11). The black bars (+ heparin) represent the results when virions were incubated with heparin prior to incubation with the heparin-agarose beads, and the white bars indicate the results when the virions were not incubated with heparin prior to incubation (− heparin). Standard deviations are indicated.
Previously, it was hypothesized that persistent infection of murine cells, which reduces murine CEACAM1a expression (31), selects for virus variants with mutations in the S gene that optimize binding to and entry into murine cells expressing little murine CEACAM1a (32). Our results demonstrate that the selected MHV mutants, which were no longer dependent upon murine CEACAM1a for entry (33, 35), had gained the ability to enter cells in a HS-dependent manner. Clearly, HS is an attractive target for viral binding because of its physiological location on the surface of most animal cells, where the initial interactions with viruses occur. Indeed, viruses from different families, including alpha-, herpes-, flavi-, picorna-, and retroviruses (for a review, see reference 28), have been shown to interact with glycosaminoglycans, in most cases heparan sulfate, often, but not always, as a cell culture adaptation.
Binding of the MHV S1 domain to the (soluble) CEACAM receptor has been shown to trigger conformational changes that are supposed to enable virus entry by activation of the fusion function of the S2 domain (17, 25, 29, 41). Such conformational changes are also expected to be required for mCEACAM1a-independent and HS-dependent infection of MHV. Indeed, a peptide corresponding to the MHV-A59 HR2 region (5) (Fig. 1) was able to efficiently inhibit the infection (data not shown). Although the requirement of a secondary protein receptor cannot be excluded, the acquisition of two putative HS-binding sites and the preservation of another such site in the furin-cleavage motif suggest that multiple interactions of the MHV/BHK S protein are required to induce the necessary conformational changes. In support of this, recombinant MHV carrying only the MHV/BHK S1 domain was unable to spread in nonmurine cells under liquid medium (35), a process for which additional mutations in the S gene are required (results from this study and unpublished results). Furthermore, MHV/pi23, a virus obtained after 23 of the 600 passages that resulted in MHV/BHK, also contains a putative HS-binding site in the S1 domain at the same position as in MHV/BHK, albeit as a smaller insertion, while it lacks the putative HS-binding site immediately upstream of the fusion peptide. MHV/pi23 does infect nonmurine cells to some extent but much less efficiently than MHV/BHK (32). In addition to the multiple HS-binding sites, however, mutations found in other parts of the S protein, such as the HR1 domain and the putative fusion peptide (Fig. 1), might also contribute to the efficient entry into nonmurine cells. We are currently in the process of determining the S protein mutations that are required for the extended host range phenotype.
Our results show that only a relatively few mutations in the S protein can convert MHV from a virus that depends for its cell entry on a highly specific receptor to one than can utilize a relatively nonspecific moiety, heparan sulfate. Since these changes were rapidly acquired in persistently infected cell cultures, S gene mutations might also occur in persistently infected animals in tissues where low levels of the receptor are expressed. Such changes might contribute to interspecies transmission; hence, an increased understanding of this process is desirable. It is noteworthy that, for SARS-CoV as well, genetic variations in the S gene appear to be essential for the transition from a virus capable of animal-to-human transmission to a virus spreading from human to human (34), a transition that eventually caused the severe acute respiratory syndrome outbreak.
Acknowledgments
We thank Stan Sawicki and Kay Holmes for providing MHVrec1 and pMH S21BHK+I, respectively.
This work was supported by grants from The Netherlands Organization for Scientific Research (NWO-VIDI-700.54.421) and the China Scholarship Council to C.A.M.d.H. and Z.L., respectively.
REFERENCES
- 1.Babcock, G. J., D. J. Esshaki, W. D. Thomas, and D. M. Ambrosino. 2004. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 78:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., E. Sullivan, L. Hensley, B. Yount, and W. Chen. 1999. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73:638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., B. Yount, L. Hensley, S. A. Peel, and W. Chen. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 71:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslin, J. J., I. Mork, M. K. Smith, L. K. Vogel, E. M. Hemmila, A. Bonavia, P. J. Talbot, H. Sjostrom, O. Noren, and K. V. Holmes. 2003. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37°C. J. Virol. 77:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-113. In S. G. Siddell (ed.), The coronaviridae. Plenum Press, New York, N.Y.
- 8.de Haan, C. A., B. J. Haijema, D. Boss, F. W. Heuts, and P. J. Rottier. 2005. Coronaviruses as vectors: stability of foreign gene expression. J. Virol. 79:12742-12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., H. Vennema, and P. J. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A. M., K. Stadler, G.-J. Godeke, B. J. Bosch, and P. J. M. Rottier. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78:6048-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas, B., J. Gelfi, R. L'Haridon, L. K. Vogel, H. Sjostrom, O. Noren, and H. Laude. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmas, B., J. Gelfi, H. Sjostrom, O. Noren, and H. Laude. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. [DOI] [PubMed] [Google Scholar]
- 14.Delmas, B., and H. Laude. 1990. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64:5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duckert, P., S. Brunak, and N. Blom. 2004. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17:107-112. [DOI] [PubMed] [Google Scholar]
- 16.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G. S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, T. M. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J. Virol. 71:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 20.Haijema, B. J., H. Volders, and P. J. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjellen, L., and U. Lindahl. 1991. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60:443-475. [DOI] [PubMed] [Google Scholar]
- 23.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, Y., X. Yan, W. Cao, C. Wang, J. Feng, J. Duan, and S. Xie. 2004. Probing the structure of the SARS coronavirus using scanning electron microscopy. Antivir. Ther. 9:287-289. [PubMed] [Google Scholar]
- 28.Liu, J., and S. C. Thorp. 2002. Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 22:1-25. [DOI] [PubMed] [Google Scholar]
- 29.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rottier, P., J. Armstrong, and D. I. Meyer. 1985. Signal recognition particle-dependent insertion of coronavirus E1, an intracellular membrane glycoprotein. J. Biol. Chem. 260:4648-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicki, S. G., J. H. Lu, and K. V. Holmes. 1995. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 69:5535-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schickli, J. H., L. B. Thackray, S. G. Sawicki, and K. V. Holmes. 2004. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 78:9073-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schickli, J. H., B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes. 1997. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 71:9499-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, H. D., C. C. Tu, G. W. Zhang, S. Y. Wang, K. Zheng, L. C. Lei, Q. X. Chen, Y. W. Gao, H. Q. Zhou, H. Xiang, H. J. Zheng, S. W. Chern, F. Cheng, C. M. Pan, H. Xuan, S. J. Chen, H. M. Luo, D. H. Zhou, Y. F. Liu, J. F. He, P. Z. Qin, L. H. Li, Y. Q. Ren, W. J. Liang, Y. D. Yu, L. Anderson, M. Wang, R. H. Xu, X. W. Wu, H. Y. Zheng, J. D. Chen, G. Liang, Y. Gao, M. Liao, L. Fang, L. Y. Jiang, H. Li, F. Chen, B. Di, L. J. He, J. Y. Lin, S. Tong, X. Kong, L. Du, P. Hao, H. Tang, A. Bernini, X. J. Yu, O. Spiga, Z. M. Guo, H. Y. Pan, W. Z. He, J. C. Manuguerra, A. Fontanet, A. Danchin, N. Niccolai, Y. X. Li, C. I. Wu, and G. P. Zhao. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 102:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thackray, L. B., and K. V. Holmes. 2004. Amino acid substitutions and an insertion in the spike glycoprotein extend the host range of the murine coronavirus MHV-A59. Virology 324:510-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tresnan, D. B., R. Levis, and K. V. Holmes. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 70:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams, R. K., G. S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]