Abstract
WRSd1 is a Shigella dysenteriae 1 vaccine containing deletions of the virG(icsA) gene required for intercellular spreading and a 20-kb chromosomal region encompassing the Shiga toxin genes (stxAB). WRSd1 was constructed from S. dysenteriae 1 strain 1617 that was originally isolated during the 1968 to 1969 epidemic of Shiga dysentery in Guatemala. The virG(icsA) deletion was constructed from a streptomycin-resistant (Strr) mutant of 1617 by a filter mating procedures using a virG(icsA) deletion derivative, pΔvirG2. A colony that was invasive for HeLa cells and negative for the virG(icsA) gene by Southern blotting was grown anaerobically on plates containing chlorate for selection of resistant colonies that had lost the entire Shiga toxin gene. A virG(icsA) stxAB Strr mutant selected from the chlorate plates was designated WRSd1. This candidate vaccine was evaluated for safety, immunogenicity, and protective efficacy using the guinea pig keratoconjunctivitis model. WRSd1 was Sereny negative, and two applications of this strain to the cornea elicited a significant protective immune response against the S. dysenteriae 1 O antigen. Vaccination with WRSd1 conferred protection against challenge with each of three virulent S. dysenteriae 1 strains. Since a vaccine protecting against multiple Shigella species is required for most areas where Shigella is endemic, protection studies using a combination vaccine of Shigella sonnei vaccine strain WRSS1, Shigella flexneri 2a vaccine strain SC602, and WRSd1 were also performed. Guinea pigs vaccinated with a mixture of equal amounts of the three vaccine strains were protected against challenge with each of the homologous virulent strains. Unlike WRSS1 and SC602, however, the level of protection afforded by WRSd1 in a combination vaccine was lower than the protection elicited by a pure culture. A current Good Manufacturing Practice product of WRSd1 given intragastrically to rhesus monkeys proved safe and immunogenic.
Shigella spp. are the most important cause of acute bloody diarrhea (dysentery), and 600,000 deaths occur globally each year due to shigellosis (14). The emergence of strains resistant to multiple antibiotics and the increasing number of infected persons in some areas of the world emphasize the need for an effective vaccine. The most important Shigella strains to be targeted for vaccine development are S. flexneri 2a, S. dysenteriae 1, and S. sonnei. S. dysenteriae 1 (Shiga bacillus) has been an important cause of epidemic dysentery in Latin America, Asia, and Africa since the 1960s. Infection with S. dysenteriae 1 causes a more severe and prolonged illness with a much higher mortality rate, particularly in young children, infants, the elderly, and the malnourished, than does infection with other Shigella serogroups. Due to the presence of the potent Shiga toxin, S. dysenteriae 1 infection complications include hemolytic-uremic syndrome, seizures, sepsis, rectal prolapse, and toxic megacolon.
The recent success seen in human trials of S. flexneri 2a vaccine strain SC602 (4) and S. sonnei vaccine strain WRSS1 (14a) has indicated that the strategy of using live attenuated Shigella strains with mutations in specific virulence-associated genes produces effective, low-dose oral vaccines for diarrhea and dysentery. SC602 has mutations in the aerobactin gene iuc and a deletion of the virG(icsA) gene that is required for intercellular dissemination. A single oral dose of 104 CFU of SC602 was safe and immunogenic and protected volunteers against the severe symptoms of dysentery when challenged with wild-type S. flexneri 2a strain 2457T (4). WRSS1, which has a 212-bp deletion in the virG(icsA) gene, proved safe and immunogenic when given as a single oral dose ranging from 103 to 106, eliciting a vigorous immune response against S. sonnei lipopolysaccharide (LPS) even at the 103 dose (14a). An S. dysenteriae 1 vaccine will form an important component of a polyvalent Shigella vaccine that will effectively protect volunteers against the major serotypes of Shigella causing disease.
Several vaccine-related products have been described for the purposes of countering S. dysenteriae 1 infection. These include synthetic S. dysenteriae 1 O-antigen-specific saccharides conjugated to human serum albumin (24) and O-specific polysaccharides covalently bound to bacterial toxoids (28). Other approaches that have been tried are an stxB-hlyA fusion integrated into a recombinant bivalent S. flexneri Y vaccine strain expressing S. dysenteriae 1 O antigen (29), expression of S. dysenteriae 1 O-specific polysaccharides in S. flexneri aroD vaccine strains (6), characterization of aroA and aroD genes of S. dysenteriae 1 to be engineered in live vaccines (32), and construction of vaccine candidates based on the loss of virulence genes virG(icsA) and aerobactin production and transport (iuc and iut) which support growth within tissues (7).
Some but not all of these products have undergone limited studies in animals, and only one has undergone clinical testing in humans (28). Here we report the construction of WRSd1, a live, attenuated S. dysenteriae 1 vaccine strain containing a 10-kb deletion on the invasion plasmid encoding the virG(icsA) gene and a 20-kb chromosomal deletion encompassing the stx genes that is being readied for a phase 1 safety and immunogenicity study.
MATERIALS AND METHODS
Bacterial strains.
The parent S. dysenteriae 1 strain 1617 was obtained from the culture collection of Samuel B. Formal, Walter Reed Army Institute of Research (WRAIR) (21) (Table 1). The strain was originally isolated from an outbreak of epidemic Shiga bacillus dysentery in Guatemala, Central America, in 1968 or early 1969 (20). The isolated strain was lyophilized and stored in ampoules in the laboratory of S. Formal. Strain 1617 lacks any antibiotic resistance marker. Shigella strains S. flexneri 2a 2457T and S. sonnei 53G, SC602, and WRSS1 are from the WRAIR culture collection (Table 1). The Shiga strain was obtained from P. Sansonetti, Pasteur Institute, Paris, France, and Ubon378 was obtained from the Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand, culture collection (Table 1).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristicsa | Source or referenceb |
|---|---|---|
| Strains | ||
| 1617 | Virulent S. dysenteriae 1 | WRAIR (20) |
| 1617ΔG | 1617 with virG(icsA) deletion (Str) | WRAIR |
| Ubon 378 | Virulent S. dysenteriae 1 | AFRIMS |
| Shiga | Virulent S. dysenteriae 1 | P. Sansonetti, Pasteur Institute |
| 2457T | Virulent S. flexneri 2a | WRAIR |
| M9OT (M9OT-W) | Virulent S. flexneri 5a | WRAIR |
| 53G | Virulent S. sonnei | WRAIR |
| WRSS1 | S. sonnei vaccine | WRAIR (14a) |
| SC602 | S. flexneri 2a vaccine | WRAIR (4) |
| WRSd1 | S. dysenteriae 1 vaccine (Str) | WRAIR (this study) |
| Plasmids | ||
| pCVD442 | Suicide vector (AmprsacB) | M. Donnenberg, CVD (2, 10) |
| pΔvirG2 | virG deletion construct in pCVD442 | WRAIR (2, 10) |
| pHS3188 | 7.6-kb EcoRI fragment encoding virG(icsA) in pBR322 | P. Sansonetti, Pasteur Institute |
Str, streptomycin resistant (300 μg/ml).
AFRIMS, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand; CVD, Center for Vaccine Development, Baltimore, Md.
Deletion of virG(icsA) in strain 1617.
A spontaneous streptomycin-resistant derivative of strain 1617 (1617-str) was isolated by growing a log-phase culture of strain 1617 on Luria-Bertani (LB) agar plates containing 300 μg of streptomycin per ml. Colonies that grew were tested for the invasive phenotype in a HeLa cell invasion assay (5). A log-phase culture of 1617-str that was invasive in HeLa cells was the recipient strain in filter mating experiments with donor Escherichia coli strain SM10λpir carrying the suicide plasmid vector construct pΔvirG2 (2, 10). pΔvirG2 has a 212-bp deletion in the virG(icsA) structural gene that was cloned into the suicide vector pCVD442 (2). Dilutions of the mating mixture were plated on LB agar plates containing streptomycin and ampicillin (S/A). Several colonies grew on these plates, representing products of the first recombination event (1617ΔG-S/A). PCR was used to confirm the appropriate integration event as previously described (2, 10). The presence of the sacB gene in pCVD442 inhibits growth of the host cell on 5% sucrose. The sacB gene, encoding sucrase, converts sucrose to levan, which is cytolytic. Therefore, growth on sucrose is used as a positive selection for the loss of the sacB gene and consequently the loss of the vector sequence. Cultures of 1617ΔG-S/A were plated on low-salt LB agar with 5% sucrose and 300 μg of streptomycin per ml as previously described (2, 10). Colonies were tested for loss of ampicillin resistance. Ampicillin-sensitive colonies, 1617ΔG, were further tested with virG(icsA)-specific primers in a PCR to confirm the loss of the vector sequence.
Deletion of the Shiga toxin (stx) gene from 1617ΔG.
1617ΔG was subjected to growth on chlorate plates under anaerobic conditions as described before (7, 21). Suspensions of 1617ΔG were spread on potassium chlorate-containing medium (8, 21) and incubated in an anaerobic jar. After 48 h, isolated colonies were purified twice on the same medium with anaerobic incubation. Several chlorate-resistant strains were isolated and tested for loss of stx gene sequences using colony blot hybridization and radiolabeled Shiga toxin (stx) DNA probe as described (22). The Shiga toxin probe for both colony blot and Southern hybridization procedures was a BamHI 1,142-bp fragment isolated from recombinant plasmid pJN37-19 (22). This fragment encodes 98% of the A subunit and all of the B subunit of SLT1. The fragment was gel purified, and approximately 0.5 μg of the fragment DNA was labeled by nick translation with 5′-[α-32P]dCTP-labeled (New England Nuclear, Boston, Mass.) using reagents supplied by Life Technologies (Gaithersburg, Md.). To confirm the loss of stx structural gene sequence from WRSd1, Southern blots of EcoRI-digested total bacterial DNA from 1617-str and WRSd1 were hybridized to the Shiga toxin probe, the filters were washed under stringent conditions in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C for 2 h and exposed to XAR-5 film (Eastman Kodak, Rochester, N.Y.).
Toxin assay.
Strains WRSd1, 1617, 59 tox (Shiga toxin-positive control), and pMJ100 (stx2 clone) were grown in 3 ml of LB medium overnight at 37°C. Then 1 ml of the culture was removed, sonicated, and clarified by centrifugation. The resulting supernatant was filter sterilized and used as the crude lysate for the cytotoxicity and neutralization assay. This assay was carried out in 96-well microtiter plates seeded with approximately 104 Vero cells in 100 μl of tissue culture medium as previously described (9, 25). WRSd1 lysate was twofold serially diluted, starting with 200 μl of undiluted lysate in the first well, and added to the Vero cells. Each set of dilutions was done in triplicate. In the case of lysates from strains 1617, 59 tox, and pMJ100, 10-fold serial dilutions were made, again in triplicate. One set of dilutions was overlaid with an equal volume of a 1:25 dilution of F45, a polyclonal antibody to αstx1, a second layer was overlaid with a 1:25 dilution of AJ65 polyclonal antibody to αstx2 (AJ65), and the third set of dilutions was overlaid with minimal essential medium (MEM). F45 antitoxin was prepared against purified Shiga toxin from S. dysenteriae 1 strain 60R. AJ65 antitoxin was prepared by immunizing rabbits with crude preparations of sonically disrupted E. coli C600(933W), a Shiga-like toxin II (SLTII)-producing lysogen. After the addition of the lysates, Vero cell monolayers were grown in MEM supplemented with 10% calf serum and gentamicin (50 μg/ml) for 48 h at 37°C in a CO2 incubator. The monolayer was washed, fixed, and stained with crystal violet, and absorbance was read at 620 nm. Fixed and stained toxin-treated tissue culture cells were compared with fixed and stained untreated cells; stain intensity was proportional to the number of viable, attached tissue culture cells present before being fixed to the well. The 50% cytotoxic dose (CD50) is defined as the dilution of the culture supernatant that kills 50% of the Vero cells in a well (9, 25).
PCRs for mapping virG(icsA) and stx deletions.
For mapping the extent of the virG(icsA) deletion, a 50-μl PCR was prepared with 10 pmol each of primer 6R (5′-GCTTCCGTTGTTCTGACATGAC-3′) and 13F (5′-GTGCTGCCACAGGAAGCGAGTTC-3′) derived from sequences flanking the virG(icsA) region in S. flexneri 5 invasion plasmid pWR501 (31). Single colonies of 1617 and WRSd1 were suspended in 200 μl of water, boiled for 10 min in a water bath, and centrifuged to obtain a lysate. Then 5 μl of the lysate was used in the PCR as the template DNA, and Taq polymerase was used as the enzyme. The reaction cycle consisted of 95°C for 5 min; 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min; followed by an elongation step at 72°C for 5 min. The extent of the stx deletion was mapped using primers JRB196 (5′-CGGGCAATTTGCTGGTAGTGTCGTGCCCATGAC-3′) and JRB199 (5′-CGGCTGGAGGAGCAGCTGGAACATGCTGCTCAC-3′) as described previously (19). In this case, amplification was performed using TaKaRa Ex Taq DNA polymerase (Pan Vera Corporation, Madison, Wis.) with the LA buffer. The reaction cycle consisted of 94°C for 1 min; 30 cycles of 98°C for 20 s and 68°C for 10 min; and a final cycle of 72°C for 10 min. Aliquots of the PCR products in each case were electrophoresed on 1% agarose gels in 1× Tris-borate-EDTA buffer with ethidium bromide (19).
Immunization of guinea pigs.
Guinea pig eyes were inoculated with bacterial cultures grown overnight on LB agar plates and harvested in 1× phosphate-buffered saline (PBS) as previously described (11). The eyes were observed for 5 to 6 days for evaluation of the Sereny reaction. When eyes were immunized for efficacy studies of vaccine candidates, immunization was carried out twice at 2-week intervals. Four weeks after the last immunization, guinea pig eyes were challenged with wild-type strains and scored for development of disease and protection. Development of disease was rated as follows: 0, no disease or mild irritation; 1, mild conjunctivitis or late development and/or rapid clearing of symptoms; 2, keratoconjunctivitis without purulence; 3, fully developed keratoconjunctivitis with purulence. Percent protection is defined as follows: full, percent of eyes rated 0; partial, percent of eyes rated 1; combined, sum of complete and partial (11). Statistical analysis was carried out using the Wilcoxon rank sum test, with P values of ≤0.05 considered significant.
Immunization of rhesus monkeys.
Rhesus monkeys (Macaca mulatta) were immunized according to a WRAIR Institutional Animal Care and Use Committee-approved monkey protocol (WRAIR protocol IO99-99). Monkeys were anesthetized intramuscularly in the caudal thigh with ketamine HCl (10 to 20 mg/kg of body weight) for procedures requiring contact with humans (physical examination, venipuncture, gastric intubation, and colonic biopsies). Then 20 ml of saturated sodium bicarbonate was administered intragastrically through a pediatric stomach tube fitted over a disposable plastic syringe, followed by 20 ml of the bacterial inoculum in water. Complete blood counts were done, and serum and rectal lavage samples were taken by veterinarians at day −7 and at days 2, 7, 14, and 28 as indicated in the protocol. Stool samples were streaked out on Hektoen agar and SS agar plates daily. These plates were scored for lactose-negative colonies. Serum and lavage samples were subjected to enzyme-linked immunosorbent assay (ELISA) using plates coated with serotype-specific LPS. A positive response was ≥2-fold-higher antibody dilution titer over that at day 0.
RESULTS
Construction of 1617ΔG.
The parent strain S. dysenteriae 1617 demonstrated a strong Sereny-positive reaction in all guinea pig test eyes, causing inflammation, keratoconjunctivitis, and purulence within 48 h. A comparison of three virulent S. dysenteriae 1 strains, 1617, Ubon 378, and Shiga (isolated from an S. dysenteriae epidemic in Japan), in a small number of guinea pig eyes indicated that although all three strains were positive in the Sereny test (26), the reaction with 1617 appeared more virulent, with all of the eyes showing the highest rating (Table 2).
TABLE 2.
Sereny reaction of Shigella strains
| Strain | No. of eyesa with rating of:
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 1617 | 0 | 0 | 0 | 6 |
| Ubon 378 | 0 | 0 | 3 | 3 |
| Shiga | 0 | 1 | 2 | 3 |
n = 6.
The 1617ΔG construct, lacking virG(icsA), was made essentially as described previously for the construction of WRSS1 and EcSf2a-3 (2, 10). The construction involved filter mating 1617-str with donor suicide vector strain pΔvirG2. After two sequential recombination events, the wild-type allele in 1617-str is replaced by the deleted version of virG(icsA) in pΔvirG2. Lack of a PCR product with virG(icsA) primers BA114 and BA117 [primers within the structural region of virG(icsA) and cloned in pΔvirG2] indicated that perhaps a larger than expected deletion had occurred in these recombinants. To confirm this, plasmid DNA was isolated from 1617ΔG (ampicillin-sensitive) colonies, digested with EcoRI and SalI, and electrophoresed on 1% agarose gels. The DNA from the gels was transferred onto nitrocellulose filters and probed with the α-32P-radiolabeled 7.6-kb EcoRI fragment from pHS3188 containing the entire structural gene for virG(icsA) and flanking sequences (3). Hybridization analysis confirmed the total loss of the virG(icsA) gene in the 1617ΔG recombinants (data not shown). 1617ΔG was negative in the Sereny reaction in guinea pigs, substantiating the observation that the virG(icsA) gene product was absent.
Complementation of 1617ΔG with virG(icsA) sequence.
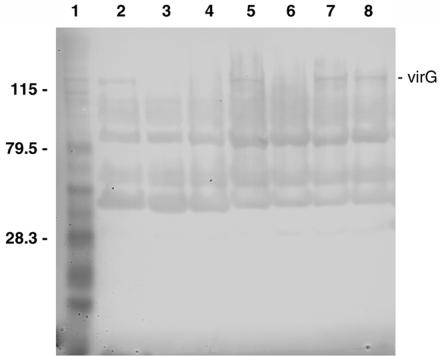
A 1617ΔG isolate that was positive in the HeLa cell invasion assay (5) was electroporated with plasmid pHS3188 (3). Western blot analysis indicated that while there was no detectable expression of the virG(icsA) protein in 1617ΔG, expression of virG(icsA) was restored in 1617ΔG(pHS3188) (Fig.1). An inoculum of 5.5 × 108 CFU of 1617ΔG and 1617ΔG(pHS3188) was placed into the eyes of two animals (four eyes total) and three animals (six eyes total), respectively (Table 3). As a positive control for the Sereny reaction, 4.6 × 108 CFU of the parent strain 1617 was inoculated into one eye of each of six animals. The results here indicated that the loss of the keratoconjunctivitis reaction seen with 1617ΔG was only partially restored by complementation with pHS3188 (Table 3).
TABLE 3.
Complementation of 1617ΔG with pHS3188
| Immunizing strain | No. of eyes | No. of eyes with rating of:
|
|||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| 1617ΔvirG(icsA) | 4 | 4 | 0 | 0 | 0 |
| 1617ΔvirG(icsA)/pHS3188 | 6 | 2 | 4a | 0 | 0 |
| 1617-str | 6 | 2 | 0 | 2 | 2 |
The eyes had enough irritation to rate more than a 0, but total opacity never developed.
PCRs to determine the extent of the virG(icsA) deletion.
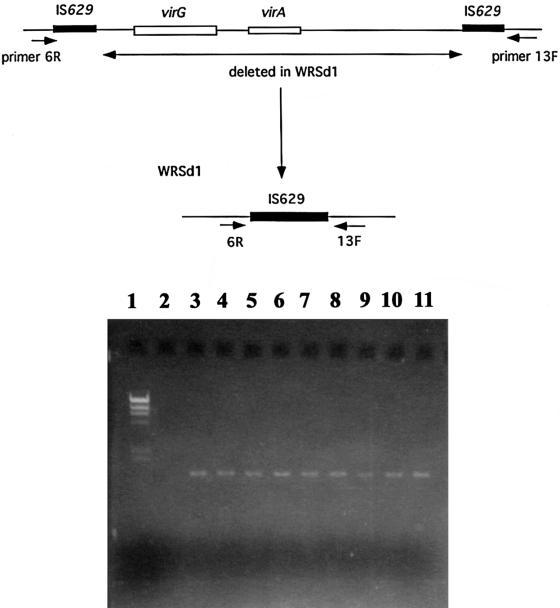
Primers 6R and 13F flank individual IS629 sequences present on either side of a 10-kb fragment on the S. flexneri 5 invasion plasmid. The virG(icsA) gene resides within this 10-kb fragment (Fig. 2) (32). These primers have been used previously in our laboratory to map the large deletion around the virG(icsA) gene in SC602. The deletion in SC602 occurred unexpectedly during its construction. PCR with primers 6R and 13F indicated that the deletion in SC602 had occurred as a result of recombination between the two IS629 sequences, eliminating the virG(icsA) gene, and leaving a single IS629 in place (M. M. Venkatesan, unpublished data). Thus, primers 6R and 13F generate a 1.5-kb PCR product when used with SC602 DNA template but do not amplify a product with the wild-type strain 2457T because of the large size of the product. Since Southern blot analysis of DNA from 1617ΔG indicated complete loss of the virG(icsA) gene, primers 6R and 13F were used to carry out a PCR to determine whether a large deletion, similar to that observed with SC602, had also occurred during the construction of WRSd1. Figure 2 indicates that primers 6R and 13F generated a 1.5-kb PCR product with DNA from WRSd1, while no product was seen with the parent strain 1617. As controls, ipaB-specific primers amplified the ipaB gene in both templates (data not shown). This shows that the extent of deletion in the virG(icsA) region on the invasion plasmid of 1617ΔG spans a 10-kb region. Sequencing of the 1.5-kb PCR-derived product confirmed this and showed that the recombination left a single IS629 sequence in place, as shown for SC602.
FIG. 2.
(Top) Schematic representation of the virG(icsA) and virA regions on the invasion plasmid. The two IS629 sequences are shown on either end of this region, and the positions of primers 6R and 13F are also indicated. The recombination that presumably occurs between the two IS629 sequences resulting in loss of the virA and virG(icsA) genes, as represented in both SC602 and WRSd1, is also shown below the map of the invasion plasmid. (Bottom) The gel picture shows products of a 6R- and 13F-primed PCR with template DNA from (lane 2) 1617-str and (lanes 3 to 11) several colony isolates of WRSd1. Lane 1 contains HindIII DNA molecular size markers.
Loss of stx genes.
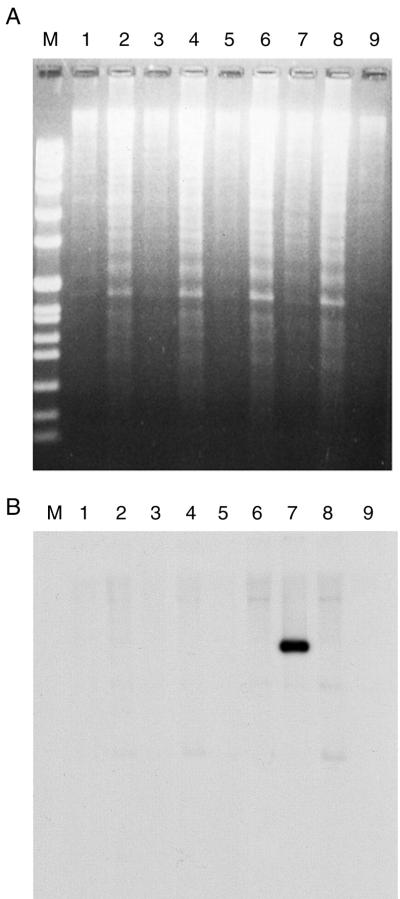
Previous studies had indicated that 10 to 25% of chlorate-resistant mutants of several strains of S. dysenteriae 1 lost the ability to produce toxin (8). Later experiments indicated that this loss was accompanied by loss of the complete structural genes for Shiga toxin (19). 1617ΔG was grown on plates containing chlorate in an anaerobic chamber as previously described (21). Several colonies were tested for loss of the stx genes by colony blot and Southern blot analysis with radiolabeled stx probes (22). The results indicated loss of the entire Shiga toxin gene (Fig. 3). Several of these isolates were then tested in the HeLa cell invasion assay and shown to be as invasive as strain 1617. One such isolate, which was virG(icsA) negative and stx negative, was chosen as vaccine strain WRSd1.
FIG. 3.
Hybridization of labeled Shiga toxin genes to Southern blots of 1617ΔG chlorate-resistant strains. Genomic DNA from several isolates (lanes 1 to 6 and 8 to 9) representing chlorate-resistant derivatives of 1617ΔG were restricted with enzymes, Southern blotted, and hybridized to radioactive Shiga toxin gene probes. Lane 7 represents DNA from strain 1617. Lane M contained DNA molecular size markers. (A) Ethidium-stained gel. (B) Southern blot after exposure to X-ray film.
PCRs to determine extent of stx deletion in WRSd1.
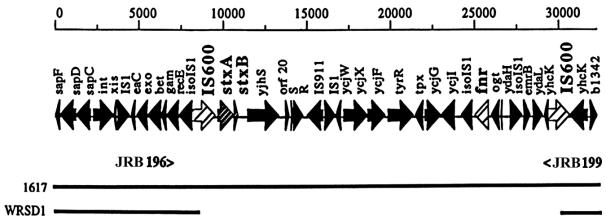
Previously it has been shown that the chlorate-induced loss of stx genes during anaerobic growth of S. dysenteriae 1 strains occurs due to a deletion of approximately 20 kb on the chromosome that includes the loss of the entire stx genes as well as other genes, such as fnr (19). This recombination event is facilitated by two IS600 elements flanking the 20-kb region. Primers JRB196 and JRB199 can be used to differentiate fnr+ stx+ strains (such as strain 1617) from those that have undergone the 20-kb deletion (such as WRSd1, Fig. 4). Using primers JRB196 and JRB199, a 3.1-kbp PCR product was amplified from WRSd1 but not from the parent strain 1617 (17). In wild-type strain 1617 the primers are 19 to 20 kb apart and unable to amplify such a large fragment under the PCR conditions used in this experiment (Fig. 4). Sequence analysis of the 3.1-kb PCR product obtained from WRSd1 confirmed that the deletion in WRSd1 occurred by a recombination between the two IS600 elements as previously described (17), leaving a single IS600 in place.
FIG. 4.
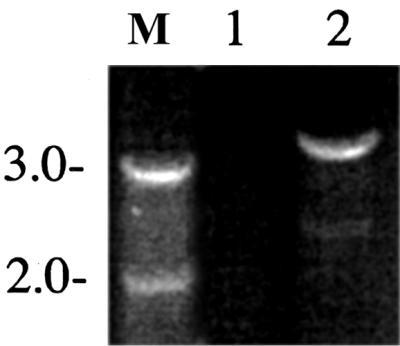
(Top) Schematic representation of a 30-kb region on the S. dysenteriae 1 chromosome, indicating the locations of phage genes, the stx genes, and the fnr gene (19). The locations of the primers JRB196 and JRB199 are shown below, and the extent of the gene map in S. dysenteriae 1 strains 1617 and chlorate-resistant derivative WRSd1 is also indicated below the map. (Bottom) PCR products primed by primers JRB196 and JRB199 using templates (lane 1) 1617 and (lane 2) WRSd1 were electrophoresed on agarose gels and stained with ethidium bromide. The numbers on the left-hand side refer to positions of the DNA molecular size markers run in lane M (in kilobase pairs).
Shiga toxin activity in WRSd1.
Southern blot analysis indicated that WRSd1 contained a complete deletion of the Shiga toxin gene. However, in order to ensure the loss of toxin activity, bacterial supernatants were used in an assay that has been described previously (9, 25). The results of the cytotoxicity and neutralization assay using antibodies to Shiga toxin indicated that the CD50s for the crude lysates were 4.6 × 10−2 per ml for WRSd1, 1 × 10−6 per ml for strain 1617, 2.3 × 10−6 per ml for strain 59 tox, and 1 × 10−7 per ml for pMJ100. The neutralization assay indicated that, as expected, the toxicity of 1617 lysate was neutralized by polyclonal antibody to Shiga toxin F45 but not by polyclonal antibody to Shiga-like toxin II AJ65.
Efficacy test of WRSd1 in guinea pig eyes.
Twelve animals were immunized ocularly on days 0 and 14 with 2 × 108 CFU of WRSd1. Four weeks after the last immunization, animals were challenged at the same dose using one of three virulent S. dysenteriae 1 strains: 1617, the parent strain of WRSd1; Ubon 378; and Shiga. WRSd1 protected fully against challenge by Ubon 378 and Shiga (Table 4). At the single dose used for immunization in this experiment, WRSd1 protected partially against the parent strain 1617 (Table 4). This could be an indication of the virulence of strain 1617.
TABLE 4.
Efficacy of WRSd1 in guinea pig eyesa
| Challenge strain | No. of eyesb with rating of:
|
% Protection
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | Full | Partial | Combined | |
| Ubon 378 | 6 | 2 | 0 | 0 | 75 | 25 | 100 |
| 1617 | 1 | 4 | 0 | 3 | 13 | 50 | 66 |
| Shiga | 5 | 3 | 0 | 0 | 63 | 38 | 100 |
The immunizing strain was WRSd1.
n = 8.
Immunization of guinea pigs with a combination of SC602, WRSS1, and WRSd1.
The immunogenicity and efficacy of SC602 and WRSS1 indicate that a mixture of virG(icsA) mutants of different serotypes of Shigella might constitute an effective polyvalent vaccine. Guinea pigs were immunized twice with 2 × 108 to 3 × 108 CFU/eye of vaccine 2 weeks apart and challenged 4 weeks after the last immunization with 4 × 108 CFU/eye of virulent S. flexneri 2a strain 2457T for SC602, S. sonnei strain 53G for WRSS1, and S. dysenteriae 1 strain Ubon378 for WRSd1 (Table 5). Animals immunized with a mixture of SC602, WRSS1, and WRSd1 were given 2 × 108 to 3 × 108 CFU/eye of each strain by taking equal volumes of each strain and concentrating the vaccine solution. Eyes were monitored for 5 days for development of disease.
TABLE 5.
Efficacy of a polyvalent mixture of Shigella vaccines in guinea pig eyes
| Immunizing strain | na | Challenge strain | No. of eyes with rating of:
|
% Protection
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | Full | Partial | Combined | |||
| SC602 | 8 | 2457T | 6 | 2 | 0 | 0 | 75 | 25 | 100 |
| WRSS1 | 8 | 53G | 8 | 0 | 0 | 0 | 100 | 0 | 100 |
| WRSd1 | 20 | Ubon 378 | 18 | 0 | 0 | 2 | 95 | 0 | 95 |
| Combination of SC602, WRSS1, and WRSd1 | |||||||||
| 8 | 2457T | 6 | 2 | 0 | 0 | 75 | 25 | 100 | |
| 8 | 53G | 7 | 1 | 0 | 0 | 88 | 13 | 100 | |
| 12 | Ubon 378 | 2 | 3 | 4 | 3 | 17 | 25 | 42 | |
| Controlsb | 4 | 2457T | 0 | 0 | 0 | 4 | |||
| 4 | 53G | 0 | 0 | 2 | 2 | ||||
| 8 | Ubon 378 | 0 | 1 | 3 | 4 | ||||
n, number of eyes.
Unimmunized animals.
Animals immunized with SC602 showed transient mild irritation following SC602 administration, which cleared by 24 to 36 h. Animals immunized with WRSS1 and WRSd1 were totally asymptomatic (Table 5). Challenge of these immunized animals with the respective wild-type strains resulted in full protection against disease. When animals were immunized with the mixture of the three vaccine strains and then challenged in separate groups with individual wild-type strains, lower efficacy of protection was seen against S. dysenteriae strain Ubon 378 compared to challenge with S. sonnei 53G and S. flexneri 2a strain 2457T.
Evaluation of the WRSd1 cGMP product in monkeys.
A current good manufacturing practice (cGMP) product of WRSd1 was produced at the Walter Reed Pilot Bioproduction facility. Five rhesus monkeys housed within the animal facility at WRAIR were each immunized intragastrically with approximately 7 × 109 CFU of WRSd1 obtained by hydrating the lyophilized vaccine product. No symptoms were detected for 14 days in any of the five monkeys. Two of five monkeys excreted the vaccine strain in stool cultures for 48 h after the administration of the vaccine, as evidenced by transparent colonies on Hektoen agar plates. These colonies were agglutinated by S. dysenteriae 1 antiserum. Four monkeys showed a 2- to 3-fold rise in serum immunoglobulin G (IgG) response to S. dysenteriae 1 LPS at day 7 or day 14 compared to day 0 or day 2 and one monkey showed a 3-fold rise in serum IgA response to S. dysenteriae LPS. None of the monkeys had a detectable rectal lavage sample immune response.
LPS profile of WRSd1.
LPS was extracted from S. flexneri 5 strains M9OT, 1617, and 1617ΔG and the cGMP product of WRSd1 by previously published methods (33). The LPS was electrophoresed on polyacrylamide gels and silver stained. The results indicated that the LPS profiles of 1617, 1617ΔG, and WRSd1 were identical but different from that of M9OT, as expected (data not shown).
DISCUSSION
The consequence of the recent improved understanding of bacterial virulence-related genes has been to introduce multiple, attenuating and stable mutations into the genome of bacterial pathogens. The generation of modern live oral vaccines involves constructing genetically defined attenuated bacteria capable of inducing a protective immune response without causing disease. WRSd1 is a S. dysenteriae 1 vaccine strain that was derived from clinical isolate 1617. Based on the Sereny reaction and efficacy studies in guinea pigs, the parent strain, 1617, appears to be somewhat more virulent than other S. dysenteriae 1 isolates such as Shiga and Ubon 378. WRSd1 has a 10-kb deletion on the invasion plasmid that encompasses the virG(icsA) gene as well as a 20-kb deletion on the chromosome that deletes not only the entire Shiga toxin gene but several other genes, including fnr (19).
The virG(icsA) deletion on the plasmid occurred spontaneously during the genetic mating experiments and is similar to the one that occurred during the construction of SC602. Based on the complete sequence of the invasion plasmid from S. flexneri 5 (31), this deletion occurs as a consequence of a recombination event between two IS629 sequences that flank the virG(icsA) gene in this region. As a result of mapping the deletion endpoint, two primers, 6R and 13F, which can be used to differentiate the vaccine strain from the parent strain have been designed. These primers will be useful to track the vaccine strain during clinical trials, particularly in regions where dysentery is endemic.
The B subunit of Shiga toxin is potentially immunogenic and may generate protective humoral responses against the action of these toxins. Whether WRSd1 vaccine should have retained the stxB subunit is debatable. Shiga toxin from S. dysenteriae 1 belongs to a family of cytotoxins, also elaborated by Shiga toxin-producing E. coli (STEC) that contribute to the development of enterohemorrhagic colitis and hemolytic-uremic syndrome in humans (24). stx of S. dysenteriae is essentially identical to stx1 of STEC strains. After binding to human glycolipid receptor Gb3, the holotoxin is endocytosed and transported to the Golgi apparatus and then to the endoplasmic reticulum. The A subunit inhibits protein synthesis by acting on the 28S rRNA of the 60S ribosomal subunit, resulting in cell cytotoxicity.
Antibodies to the StxB subunit alone are protective against challenge with whole toxin (17). However, the role of serum antitoxic antibody in protection against dysentery caused by Shigella dysenteriae 1 is unclear. Monkeys fed 1010 virulent organisms after parenteral immunization with a formalin-inactivated Shiga toxoid preparation responded to orally administered Shiga bacilli by development of diarrhea and dysentery that was as severe as the response of unimmunized controls (15). Also, the levels of Shiga toxin antibodies in the sera of patients clinically diagnosed with S. dysenteriae infection are low and variable, and similar observations have been made for Stx1 and Stx2 immune response in people infected with STEC.
To date, most of the studies investigating human antibody responses against Shiga toxins have been based on the detection of anti-Stx1 neutralizing antibody (NAb) in cell culture and anti-Stx1 IgG by ELISA (13). Using the latter, only about one-third of patients with STEC infection have been found to develop NAbs or IgG antibodies against Stx1 (19). The low anti-Stx1 responses by hemolytic-uremic syndrome patients was attributed to an inadequate antigenic stimulus by a toxin with a very high biological activity (16). There is also some evidence that the StxB subunit triggers cell death of epithelial cells but to a lesser degree than the holotoxin (12).
The questionable value of toxin antibody has to be balanced with the observations that the stx genes are physically linked to phage sequences in S. dysenteriae and STEC. The stx operon can exist in multiple copies on the S. dysenteriae chromosome, and bacteriophage-mediated horizontal transmission of stx genes occurs between STEC and Shigella species (19), accounting for the spread of these toxins among diverse E. coli serotypes and into other bacterial species. Recent evidence supports the notion that the host environment may be the common site where new pathogenic strains evolve (1). The presence of the StxB subunit in a vaccine strain might therefore facilitate such recombination events. The complete loss of the phage genes accompanying the loss of the stx genes, as in WRSd1, may be a safer approach to a vaccine against S. dysenteriae 1.
The mechanism of chlorate-induced deletion of the toxin genes in the S. dysenteriae 1 chromosome has recently been determined (19). The stx genes in the S. dysenteriae chromosome are located close to fnr, which is the fumarate and nitrate reductase regulator gene. fnr is a global transcriptional regulator that mediates anaerobic induction of respiratory gene expression, including expression of fumarate, nitrite, and nitrate reductases. Deletion of fnr would lead to the repression of nitrate reductase expression. Anaerobic growth of S. dysenteriae 1 strains on medium containing chlorate, which is reduced to the toxic metabolite chlorite by nitrate reductases when cells are grown under anaerobic conditions, was found to lead to a high percentage (10 to 25%) of spontaneous chlorate-resistant mutants that coincidentally have lost the ability to produce Stx (8). The linkage of stx to fnr on the chromosome provides a genetic explanation for the observed loss of the stx genes in a high percentage of chlorate-resistant strains of S. dysenteriae 1 (19). Primers have been designed to test this 20-kb deletion that accompanies the loss of the stx-fnr genes. These primers can also serve to distinguish the vaccine strain from the wild-type bacteria by PCR.
A recent study has shown that fnr is one of several genes that is required by an E. coli K1 strain for colonization of the rat gastrointestinal tract (18). WRSd1 lacks the fnr gene as well as several others that are located on the 20-kb chromosomal fragment that is lost during chlorate-induced growth. However, the loss of the 20-kb chromosomal sequence does not impair bacterial invasion in tissue culture cells. Furthermore, previous experiments have shown that administration of chlorate-induced, stx-negative S. dysenteriae 1 strains to human volunteers and monkeys did not compromise the ability of the bacteria to cause dysentery (8, 15, 21), indicating that loss of fnr and associated genes does not prevent the bacteria from colonizing and causing disease. In these previous experiments, however, the deletion in the chlorate-induced stx mutants had not been specifically mapped (8, 15, 21).
Two of five monkeys excreted WRSd1 for 2 days when given 6 × 109 CFU of the cGMP product, indicating that the vaccine could colonize the primate gastrointestinal tract. The immune response generated was lower than that observed in monkeys challenged with 2 × 1010 CFU of SC602 vaccine which had been freshly harvested from overnight growth plates (unpublished data). The higher response seen with SC602 administration could be due to several variables, including higher dose, manner of harvesting the strain, and possibly the presence of the fnr gene. The loss of the fnr and associated genes in WRSd1 may be one explanation for the lower efficacy of WRSd1 in guinea pigs compared to WRSS1 and SC602 when administered together at the same dose and challenged with the individual homologous virulent strains. These results indicate that in a mixture of the three vaccines, WRSd1 may have to be given at a higher dose than SC602 and WRSS1.
Based on the loss of gene sequences, one prediction is that WRSd1 will be safer at doses that proved reactogenic for SC602 and WRSS1. Whether it will be equally immunogenic in human volunteers at doses that were safe and immunogenic for SC602 and WRSS1 remains to be seen. Both SC602 and WRSS1 have undergone successful inpatient and outpatient safety trials in humans in the United States (4, 14a). The strength of these vaccines is in their the ability to evoke a protective immune response at a fairly low oral dose resulting in a safe and efficacious approach to making dysentery vaccines. virG(icsA) mutations can be engineered in several Shigella serotypes, providing for the rapid development of a polyvalent mixture of Shigella vaccine strains effective against all of the major serotypes that are being isolated from clinical samples around the world.
The low dosage of this oral vaccine makes a polyvalent Shigella vaccine approach feasible. Furthermore, if needed, the technology of making these vaccines is easily transferable to lesser-developed countries. SC602 has proven to be safe and immunogenic when administered at a single oral dose of 104 CFU but reactogenic at the 106 CFU dose. WRSS1 is safe and highly immunogenic in doses from 103 to 106 CFU. While 15 to 20% of the volunteers receiving either vaccine have indicated one or more symptoms such as fever, headache, malaise, nausea, and mild diarrhea, none of these symptoms have been incapacitating and neither vaccine has provoked dysentery in volunteers. Immunization with SC602 has afforded protection in human volunteers against dysentery, indicating that it is both safe and efficacious (4). Phase I trials of WRSd1 will determine the safe dosage for WRSd1. It remains to be seen whether, compared to SC602 and WRSS1, a higher dose of WRSd1 will be required to generate an equivalent immune response.
FIG. 1.
Western blot of S. dysenteriae 1 strains complemented with a recombinant virG(icsA)-expressing clone. Whole-cell lysates were run on 10% acrylamide gels (Novex) and hybridized to a VirG(IcsA) peptide-specific antiserum. The lysates were from (lane 2) S. flexneri 2a strain 2457T, (lane 3) S. flexneri 2a vaccine strain SC602, (lane 4) plasmid-cured S. flexneri 2a strain M4243A, (lane 5) strain 1617-str, (lane 6) strain 1617ΔG, and (lanes 7 and 8) two different isolates of 1617ΔG complemented with pHS3188. Lane 1 contained protein molecular size markers, with positions marked on the left-hand side of the figure (in kilodaltons). The position of the VirG(IcsA) protein band on the gel is marked on the right-hand side of the figure.
Acknowledgments
We thank Ross Turbyfill for LPS analysis, Joe Harre and the members of the WRAIR Veterinary Medicine department for help with the monkey experiments, Alison O'Brien's lab at USUHS, Bethesda, Md., for carrying out the Shiga toxin assay, and Lee Tseng with the monkey ELISAs.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, the U.S. Department of the Army, or the U.S. Department of Defense, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, et al. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, W. A., E. V. Oaks, A. B. Hartman, and M. M. Venkatesan. 1996. Construction and characterization of virG(icsA)-deleted Escherichia coli-Shigella flexneri hybrid vaccine strains. Vaccine 14:1053-1061. [DOI] [PubMed] [Google Scholar]
- 3.Bernardini, M. L., J. Mounier, et al. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coster, T. S., C. W. Hoge, L. L. Van DeVerg, A. B. Hartman, E. V. Oaks, M. M. Venkatesan, D. Cohen, G. Robin, A. Fontaine-Thompson, P. J. Sansonetti, and T. L. Hale. 1999. Vaccination against shigellosis with attenuated Shigella flexneri 2a strain SC602. Infect. Immun. 67:3437-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 6.Falt, I. C., E. K. Schweda, S. Klee, et al. 1995. Expression of Shigella dysenteriae serotype 1 O-antigenic polysaccharide by Shigella flexneri aroD vaccine candidates and different S. flexneri serotypes. J. Bacteriol. 177:5310-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontaine, A., J. Arondel, and P. J. Sansonetti. 1990. Construction and evaluation of live attenuated vaccine strains of Shigella flexneri and Shigella dysenteriae 1. Res. Microbiol. 141:907-912. [DOI] [PubMed] [Google Scholar]
- 8.Gemski, P., A. Takeuichi, and O. Washington. 1972. Shigellosis due to Shigella dysenteriae 1: relative importance of mucosal invasion versus toxin production in pathogenesis. J. Infect. Dis. 126:523-530. [DOI] [PubMed] [Google Scholar]
- 9.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman, A. B., and M. M. Venkatesan. 1998. Construction of stable attenuated virG(icsA) Shigella sonnei vaccine strain WRSS1 and protective efficacy and immunogenicity studies in the guinea pig keratoconjunctivitis model. Infect. Immun. 66:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman, A. B., C. P. Powell, et al. 1991. Small animal model to measure efficacy and immunogenicity of Shigella vaccine strains. Infect. Immun. 59:4075-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, N. L., A. Islur, R. Haq, M. Mascarenhas, M. A. Karmali, M. H. Perdue, B. W. Zanke, and P. M. Sherman. 2000. Escherichia coli Shiga toxins induce apoptosis in epithelial cells that is regulated by the Bcl-2 family. AJP Gastrointestinal Liver Physiol. 278:G811-G819. [DOI] [PubMed] [Google Scholar]
- 13.Karmali, M. A., M. Petric, M. Winkler, M. Bielaszewska, J. Brunton, N. van-de-Kar, T. Morooka, G. B. Nair, S. E. Richardson, and G. S. Arbus. 1994. Enzyme-linked immunosorbent assay for detection of immunoglobulin G antibodies to Escherichia coli Vero cytotoxin 1. J. Clin. Microbiol. 32:1457-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, et al. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. WHO 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 14a.Kotloff, K. L., D. N. Taylor, M. B. Sztein, S. S. Wasserman, G. A. Losonsky, J. P. Nataro, M. Venkatesan, A. Hartman, W. D. Picking, D. E. Katz, J. D. Campbell, M. M. Levine, and T. L. Hale. 2002. Phase I evaluation of ΔvirG Shigella sonnei live, attenuated, oral vaccine strain WRSS1 in healthy adults. Infect. Immun. 70:2016-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine, M. M., H. I. DuPont, S. B. Formal, R. B. Hornick, A. Takeuichi, E. J. Gangarosa, M. J. Snyder, and J. P. Libonati. 1971. Pathogenesis of Shigella dysenteriae 1 (Shiga) dysentery. J. Infect. Dis. 127:261-270. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig, K., M. A. Karmali, V. Sarkim, et al. 2001. Antibody response to Shiga toxins Stx2 and Stx1 in children with enteropathic hemolytic-uremic syndrome. J. Clin. Microbiol. 39:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcato, P., G. Mulvey, R. J. Read, et al. 2001. Immunoprophylactic potential of cloned Shiga toxin 2 B subunit. J. Infect. Dis. 183:435-443. [DOI] [PubMed] [Google Scholar]
- 18.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 19.McDonough, M. A., and J. R. Butterton. 1999. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 20.Mendizabal-Morris, C. A., J. Leonardo, J. Mata, et al. 1971. Epidemic Shiga-bacillus dysentery in Central America. Derivation of the epidemic and its progression in Guatemala, 1968-69. Am. J. Trop. Med. Hyg. 10:927-933. [DOI] [PubMed] [Google Scholar]
- 21.Neill, R. J., P. Gemski, S. B. Formal, and J. W. Newland. 1988. Deletion of the Shiga toxin gene in a chlorate-resistant derivative of Shigella dysenteriae type 1 that retains virulence. J. Infect. Dis. 158:737-741. [DOI] [PubMed] [Google Scholar]
- 22.Newland, J. W., and R. J. Neill. 1988. DNA probes for Shiga-like toxins I and II and for toxin-converting bacteriophages. J. Clin. Microbiol. 26:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obrig, T. G. 1997. Shiga toxin mode of action in E. coli O157:H7 disease. Front. Biosci. 2:635-642. [DOI] [PubMed] [Google Scholar]
- 24.Pozsgay, V., C. Chu, L. Pannell, et al. 1999. Protein conjugates of synthetic saccharides elicit higher levels of serum IgG lipopolysaccharide antibodies in mice than do those of the O-specific polysaccharide from Shigella dysenteriae type 1. Proc. Natl. Acad. Sci. USA 96:5194-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in EHEC strains are responsible for the antigenic heterogeneity of the 0157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sereny, B. 1957. Experimental keratoconjunctivitis shigellosa. Acta Microbiol. Acad. Sci. Hung. 4:367-376. [PubMed] [Google Scholar]
- 27.Sturm, S., and K. N. Timmis. 1986. Cloning of the rfb gene region of Shigella dysenteriae 1 and construction of an rfb-rfp gene cassette for the development of lipopolysaccharide-based live anti-dysentery vaccines. Microb. Pathog. 1:289-297. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, D. N., A. C. Trofa, J. Sadoff, C. Chu, D. Bryla, J. Shiloach, D. Cohen, S. Ashkenazi, Y. Lerman, W. Egan, et al. 1993. Synthesis, characterization, and clinical evaluation of conjugate vaccines composed of the O-specific polysaccharides of Shigella dysenteriae type 1, Shigella flexneri type 2a, and Shigella sonnei (Plesiomonas shigelloides) bound to bacterial toxoids. Infect. Immun. 61:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzschaschel, B. D., S. R. Klee, V. de Lorenzo, et al. 1996. Towards a vaccine candidate against Shigella dysenteriae 1: expression of the Shiga toxin B-subunit in an attenuated Shigella flexneri aroD carrier strain. Microb. Pathog. 21:277-288 [DOI] [PubMed] [Google Scholar]
- 30.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker, J. C., and N. K. Verma. 1997. Cloning and characterisation of the aroA and aroD genes of Shigella dysenteriae type 1. Microbiol. Immunol. 41:809-813. [DOI] [PubMed] [Google Scholar]
- 33.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharide extraction with phenol:water and further application of the procedure. Methods Carbohydr. Chem. 2:83-91. [Google Scholar]