Abstract
Angiotensin II (AII), a potent vasoactive hormone, acts on numerous organs via G-protein-coupled receptors and elicits cell-specific responses. At the level of the heart, AII stimulation alters gene transcription and leads to cardiomyocyte hypertrophy. Numerous intracellular signaling pathways are activated in this process; however, which of these directly link receptor activation to transcriptional regulation remains undefined. We used the atrial natriuretic factor (ANF) gene (NPPA) as a marker to elucidate the signaling cascades involved in AII transcriptional responses. We show that ANF transcription is activated directly by the AII type 1 receptor and precedes the development of myocyte hypertrophy. This response maps to STAT and GATA binding sites, and the two elements transcriptionally cooperate to mediate signaling through the JAK-STAT and protein kinase C (PKC)-GATA-4 pathways. PKC phosphorylation enhances GATA-4 DNA binding activity, and STAT-1 functionally and physically interacts with GATA-4 to synergistically activate AII and other growth factor-inducible promoters. Moreover, GATA factors are able to recruit STAT proteins to target promoters via GATA binding sites, which are sufficient to support synergy. Thus, STAT proteins can act as growth factor-inducible coactivators of tissue-specific transcription factors. Interactions between STAT and GATA proteins may provide a general paradigm for understanding cell specificity of cytokine and growth factor signaling.
Hormones and growth factors acting through cell surface receptors activate multiple signaling cascades, leading to diverse biological responses that depend largely on the cellular context. Considerable understanding has been achieved regarding the mechanisms that couple receptor activation to cytoplasmic effectors. However, the mechanisms by which specific outcomes are generated from common signaling molecules remain incompletely understood. The discovery of complex interconnections between different signaling pathways, combined with the observation that similar cytoplasmic events are associated with or relay distinct biological effects, has led to the suggestion that specificity may be achieved at the level of target genes (4, 69).
G-protein-coupled receptors (GPCR) constitute the largest family of transmembrane receptors in mammals (77). The angiotensin II (AII) type 1 receptor (AT1R), which transduces the biologic effects of AII, is one of the most extensively studied GPCR (18), and drugs that target AT1R are widely used for the treatment of cardiovascular diseases, such as hypertension and cardiac hypertrophy (17). AT1Rs activate a plethora of signaling cascades, including those of mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K), protein kinase C (PKC), Janus kinase (JAK)-STAT, and calcineurin, resulting in apoptosis, proliferation, hypertrophy, or differentiation depending on the cell type and developmental stage (35). At the level of the nucleus, AT1R activation has been shown to alter expression of some ubiquitous as well as tissue-specific transcription factors. They include the immediate-early genes c-fos, c-jun, and egr1 (reviewed in reference 8), and in smooth muscle and adrenal cells, tissue-restricted transcription factors like the homeobox factors MHOX and DAX-1 (27, 52) and the zinc finger proteins KLF5 and SF-1 (52, 65). AII also enhances nuclear accumulation of STAT family members (reviewed in reference 9), NF-κB (59), and nuclear factor of activated T cells 3 (72). However, the exact role of these factors in mediating AII actions remains largely controversial.
At the level of the heart, AT1R activation causes myocyte hypertrophy and apoptosis (55) and is associated with upregulation of c-jun, c-fos, and the cardiac-specific atrial natriuretic factor (ANF) gene, which is induced during hypertrophy (55, 74) AII also activates several STAT family members which have themselves been implicated in cardioprotection, apoptosis, and hypertrophy through mechanisms and effectors that remain to be elucidated (reviewed in reference 9). Similarly, the various signaling pathways activated by AII have all been implicated in cardiac hypertrophy, apoptosis, or both (1, 24, 46). Their importance in mediating specific AII effects as well as their involvement in AII regulation of cardiac genes has not been firmly elucidated.
STAT (signal transducers and activators of transcription) proteins are evolutionary conserved transcription factors that reside in the cytoplasm until they are activated by tyrosine phosphorylation, which leads to their dimerization and nuclear accumulations. Initially identified as the targets of interferon, the STAT family in mammals now comprises seven members that are activated by cytokine and growth factor signaling through receptor coupling to the JAK family of proteins (reviewed in reference 40). STATs can also be activated by receptor tyrosine kinases, like epidermal growth factor receptor, and by some members of the GPCR family, such as the AT1 receptor (9). STAT proteins play important roles in developmental decisions as well as in stress response and host defense (40). Although most STAT proteins are widely expressed, gene targeting in mice revealed nonredundant functions for the different family members. For example, lack of STAT1 or STAT2 leads to an impaired response to interferon and hypersensitivity to infections, while lack of STAT4 and STAT6 is associated with impaired T-cell differentiation. Interestingly, while loss of STAT3 causes early embryonic lethality, tissue-specific inactivation of the STAT3 gene revealed distinct and at times opposing effects in cell survival, proliferation, and acute-phase response, depending on the targeted tissue or organ. Similarly, inactivation of each of the two highly homologous STAT5 genes revealed hematopoietic defects, growth retardation (for STAT5a), and mammary gland development (in STAT5b null).
At the level of the heart, targeted overexpression of constitutively active STAT3 has been shown to enhance vascular endothelial growth factor (VEGF) expression and causes hypervascularization (53), which would enhance cardiac adaptation to stress. Others found that overexpression of STAT3 leads to cardiac hypertrophy but also protects against drug-induced cardiotoxicity (37). More recently, mice with cardiac-specific inactivation of STAT6 were generated and found to have an impaired response to pressure overload (29). In human, STAT3 is constitutively active in several tumors (10) and mutation of STAT1 lead to impaired bacterial immunity (20). The mechanisms by which STAT proteins can transduce such a wide spectrum of cellular responses and biologic effects remain to be determined.
The activities of STAT proteins can be modulated by protein-protein interaction, and a growing number of STAT-interacting proteins is being reported (66). In addition to STAT-interacting kinases and phosphatases, several transcription factors and coactivators/corepressors were shown to associate with and modulate STAT activity. They include CBP/p300 (31) and histone deacetylases (82), as well as inducible regulators such as c-Jun (85) and members of the nuclear receptor family (22, 67, 71). Finally, a role for tissue-specific transcription factors in STAT transcriptional regulation is emerging. In T cells, STAT proteins have been shown to physically and functionally interact with members of the Ets family of transcription factors over composite STAT-Ets DNA elements required for cytokine regulation of target genes (60, 76). In hepatocytes, STAT3 was shown to cooperate with HFN1 in mediating the transcriptional response to interleukin 6 during liver regeneration (38).
GATA proteins are tissue-specific transcription factors that play crucial roles in organogenesis. In mammals, the six members are divided into two subfamilies based on sequence homology and tissue distribution. GATA-1, -2, and -3 play essential roles in hematopoietic cell survival, proliferation, and differentiation, whereas GATA-4, -5, and -6 are key regulators of endodermal and cardiovascular development (56).
In the present study, we show that STAT and GATA proteins cooperatively mediate AII responsiveness of the ANF (NPPA) promoter through direct physical interaction. Moreover, we found that GATA-4 is essential for transcriptional activation by AII, which enhances GATA-4 binding to DNA through PKC-mediated phosphorylation. In turn, GATA-4 is able to recruit activated STAT to target promoters, suggesting that STAT proteins can act as inducible coactivators of the tissue-specific GATA transcription factors. This unravels a novel GATA-dependent mechanism for STAT action which could account for cell specificity of cytokine and growth factor action. Interaction with GATA-4 and activation of ANF may also explain some of the cardioprotective effects of STAT proteins.
MATERIALS AND METHODS
Materials.
The following reagents were purchased from Calbiochem: SB 203580, PD 98059, LY 294002, U-73122, GF 109203X, and AG490. Except for the anti-GATA-4 antibody (14), the antibodies used were purchased from Santa Cruz: Pol II (N20), STAT1 (E23), STAT3 (C20), and STAT5 (C17). The anti-GATA-4, antihemagglutinin (anti-HA), and anti-Flag antibodies have been described previously (14).
Plasmids and adenovirus vectors.
The luciferase reporter constructs used and most GATA expression vectors were described previously (3, 49). The SV-sport1-HA-GATA-4 plasmids were prepared based on the original rat GATA-4 cDNA (25). Heterologous promoters were generated by multimerizing the relevant oligonucleotides flanked by BamHI and BglII sites upstream of the minimal (−57 bp) ANF or BNP (B-type natriuretic peptide) luciferase reporter as described previously (48). Expression vectors for rat AT1aR and its mutants were gifts from Sadashiva S. Karnik (45). Erk1/2 expression vectors were described previously (43). The VEGF-luciferase reporter was a kind gift of Darren Richard. It contains the upstream sequence from the human VEGF gene (44). STAT cDNA vectors expressing wild-type proteins from Svsport1 were provided by Juergen A. Ripperger. HA-tagged wild-type and mutant STAT1α vectors were generated using PCR-mediated amplification and cloning in the cytomegalovirus-driven pCGN plasmid. All constructs were confirmed by sequencing. Ad-LacZ has been described previously (3), and the AII type 1a receptor adenovirus vector (Ad-AT1aR) was a gift of Walter G. Thomas (74). The virus was amplified and quantified as described previously (14).
Cell cultures, adenovirus infection, and transient transfections.
Myoblast C2C12 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 20% fetal bovine serum. Cotransfections were carried out using calcium phosphate 24 h after plating. At 16 h posttransfection, the medium was changed to serum free and supplemented with 0.1 μM AII for overnight stimulation. Cells were harvested, and luciferase activity assays were performed with a Berthold LB953 luminometer. The amount of reporter construct was kept at 1 μg per 20-mm dish and the total amount of DNA at 3 μg. One microgram of rat AT1aR expression vector was used unless otherwise stated. Primary cardiomyocyte cultures were prepared from 4-day-old Sprague-Dawley rats and cotransfected as described previously with minor modifications (14). In brief, cardiomyocytes were cotransfected and treated with 100 nM AII or left untreated in serum-free medium for 24 h. For adenovirus infections, cardiomyocytes were infected with 2 PFU of Ad-AT1aR or the control Ad-LacZ per cardiomyocyte as described previously (14) and treated with 100 nM AII or left untreated in serum-free medium for 20 min or 48 h. When required, pharmacologic inhibitors were added to the cells 30 min prior to AII stimulation. Unless otherwise indicated, the data shown for transfections are the means for at least three independent experiments carried out in duplicate and with different DNA preparations.
Chromatin immunoprecipitation assays and QPCR analysis.
Chromatin immunoprecipitation (ChIP) assays were carried out using a modification of a previously described protocol (23). Essentially, 16 million cardiomyocytes per ChIP were plated at 25,500 cells/cm2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 10 μg/ml cytosine β-d-arabino-furanoside hydrochloride (Sigma). On day 2, the medium was changed for a serum-free medium supplemented with 50 mM KCl. Cardiomyocytes were infected with 2 PFU of Ad-AT1aR per cardiomyocyte. The serum-free medium was changed on day 3, and the cells were treated with 100 nM AII for 2 days. Cardiomyocytes were washed with ice-cold phosphate-buffered saline (PBS) and cross-linked with 1% formaldehyde at 4°C for 15 min. Cross-linking was stopped by incubating the cells in a solution of 125 mM glycine for 5 min at 4°C. Cells were harvested in PBS, centrifuged, and then resuspended and lysed in a hypotonic buffer for 5 min in ice. The lysate was centrifuged, and the pellet containing the nuclei was resuspended in a buffer containing 300 mM NaCl and incubated for 30 min at 4°C to extract free nuclear proteins. The pellet containing the cross-linked chromatin was resuspended and sonicated to achieve fragments of about 600 bp. Fragmented chromatin was precleared with protein A/G PLUS-agarose (Santa Cruz Biotechnology) with a 1- to 2-h incubation with agitation at 4°C. A small aliquot was saved as the input, and the remaining was subdivided in equal fractions for ChIP. Immunoprecipitated chromatin was washed several times for 5 min at room temperature. DNA was purified with the QIAquick PCR purification kit (QIAGEN). Samples and input were analyzed by real-time quantitative PCR (QPCR). DNA template and 1 μM oligonucleotides were used at an annealing temperature of 58°C using the Quantitect SYBR green PCR kit (QIAGEN) in an MX4000 real-time PCR machine (Stratagene, La Jolla, CA). The primers used were 5′-AAAGCGGTTTCATCCTCCAGGC-3′ and 5′-ACAGGCTCTAAAGAATTCAGCTACACG-3′ for the distal ANF region and 5′-GCCTTTGTCCGTCACTGTCT-3′ and 5′-GAGCGCCCAGGAAGATAACC-3′ for the proximal ANF promoter. A comparative quantification was used, and the input was set as the calibrator. The results are expressed as the severalfold enrichment of GATA or STAT ChIP over the values obtained with the control immunoglobulin G.
Northern blot and QPCR.
Total RNA was prepared using TRIZOL reagent (Invitrogen Canada, Inc., Burlington, ON, Canada). Northern blots were performed as previously described (55). QPCR was performed on cDNAs obtained by reverse transcription of 2 μg of total RNA using Omniscript reverse transcriptase (QIAGEN, Inc., Mississauga, Canada). Two nanograms of cDNA template and 1 μM oligonucleotides were used. The oligonucleotides for mouse ANF are TGCCGGTAGAAGATGAGGTC (forward) and AGCAGCTGGATCTTCGTAGG (reverse). The oligonucleotides for mouse ribosomal protein S16 are ATCTCAAAGGCCCTGGTAGC (forward) and ACAAAGGTAAACCCCGATCC (reverse).
Electrophoretic mobility shift assays.
Preparation of nuclear extracts from cell culture was as described previously (13, 49). Nuclear proteins were extracted from hearts of human AII type I receptor transgenic and nontransgenic mice essentially as described previously (19). Briefly, the hearts were excised from mice, washed with PBS to remove blood, and then broken into pieces in liquid nitrogen. Afterwards, the heart pieces were homogenized in solution A (0.6% NP-40, 150 mM NaCl, 10 mM HEPES [pH 7.9], 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride) at 4°C, followed by centrifugation for 30 s at 2,000 rpm. The supernatant was then saved and centrifuged for 5 min at 5,000 rpm. The pellets obtained were suspended in solution B (25% glycerol, 20 mM HEPES [pH 7.9], 420 mM NaCl, 1.2 mM MgCl2, 0.2 mM EDTA, 05 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 5 μg/ml aprotinin) at 4°C for 20 min and centrifuged for 15 s at full speed in a microcentrifuge. Extracts were then divided into aliquots and stored at −80°C for future use. Protein concentrations were determined using Bio-Rad assays. Binding assays were performed at room temperature for 30 min in the presence of 500 ng of poly(dI-dC) in 120 mM KCl, 25 mM MgCl2, 20 mM Tris-Cl [pH 7.9], 2 mM DTT, 2 mM EDTA, 8% Ficoll (for GATA-4 and serum response factor (SRF) binding assays), or 5 mM HEPES (pH 7.9), 100 mM NaCl, 0.25 mM EDTA, 0.5 mM DTT, and 5% glycerol (for STAT binding assay).
Western blots and coimmunoprecipitations.
Western blots were performed using whole-cell or nuclear extracts according to standard protocols. Polyclonal antibodies were used at a 1:1,000 dilution and incubated with the membrane for 1 h at room temperature and then with peroxidase-conjugated antibody for another hour at room temperature. The bands were revealed using the ECL Plus standard protocol (Amersham Pharmacia Biotechnology). Coimmunoprecipitation of STAT1 and HA-GATA-4 or Flag-GATA-4 and HA-STAT1 was carried out using nuclear extracts of 293T cells overexpressing the relevant proteins, as described previously (49).
Kinase assays.
The recombinant proteins glutathione S-transferase (GST)-GATA-4 1-207, GST-GATA-4 329-440, and GST-GATA-4 329-440 with a 419-420, SS→AA mutation were produced as described previously (15). Five micrograms of bacterially expressed protein was incubated with 20 ng of the purified catalytic subunit of protein kinase C (Calbiochem) in the reaction buffer (20 mM Tris [pH 7.5], 12.5 mM MgCl2, 0.5 mM EGTA, 50 μM ATP) at 30°C for 1 h. Thereafter, the proteins were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and exposed to X-ray films.
RESULTS
The ANF promoter harbors multiple AII response elements.
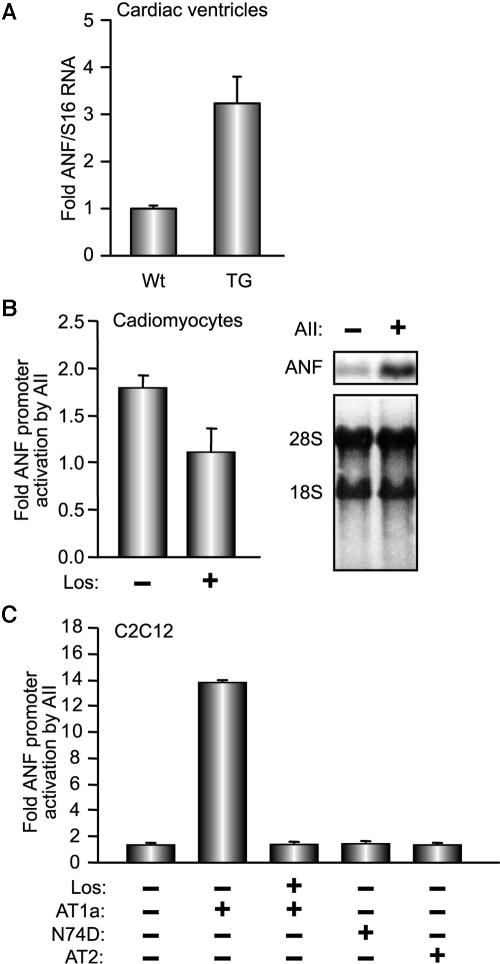
AII-induced cardiac hypertrophy results in upregulation of the ANF gene (55). However, since activation of ANF transcription is a hallmark for the genetic changes that accompany cardiac hypertrophy irrespective of etiology, we first tested whether ANF is a direct target for AII. Previously, we generated transgenic mice (TG) with myocardial specific expression of AT1R; these mice develop progressive cardiac hypertrophy and remodeling (55). We analyzed the levels of ANF transcripts in the ventricles of young TG mice (50 to 60 days old) at a time when cardiac function was intact and when there was no microscopic evidence of cardiomyocyte enlargement or apoptosis. We found that overexpression of AT1R was sufficient to increase ANF transcript levels by threefold (Fig. 1A). This increase could be recapitulated in primary cardiomyocyte cultures at the level of endogenous ANF mRNAs as well as transfected ANF promoter (Fig. 1B). Moreover, coexpression of the ANF-luc reporter with expression vectors encoding rat AT1aR in some noncardiac cells was sufficient to induce promoter activity following treatment with AII (Fig. 1C; also data not shown), indicating that AII-mediated transcriptional activation of ANF is not secondary to cardiomyocyte hypertrophy. The specificity of the response was confirmed by the use of the AT1R antagonist Losartan, which blocked ANF promoter activation (Fig. 1B); additionally, cotransfection of an expression vector encoding the AII type 2 receptor or a dominant-negative AT1 receptor mutant (N74D) failed to activate the ANF promoter in response to AII (Fig. 1C). It should also be noted that expression of AT1R alone or treatment with AII was not sufficient to elicit significant transcriptional effects, and promoter activation required addition of both receptor and ligand. Interestingly, although liganded AT1R could enhance ANF promoter activity in different cell lines, maximal response was achieved in muscle cells, suggesting the involvement of local factors (data not shown). These results indicate that AT1R activation targets ANF transcription directly and that the ANF promoter harbors AII response element(s).
FIG. 1.
ANF is induced by AII in cardiomyocytes. (A) ANF transcript levels were quantified in the ventricles of AT1R transgenic mice (TG) and nontransgenic (Wt) littermates by QPCR. The results shown represent the means ± standard errors of the means for 17 Wt and 8 TG mouse ventricles. (B and C) AII increases the activity of the ANF promoter in cardiomyocytes and in C2C12 cells in an AT1R-dependent fashion. (B) The −695ANF promoter-driven luciferase reporter construct was transfected into cardiomyocytes (left) that were treated for 12 h with 100 nM AII in presence or absence of 100 nM Losartan (Los), an AII antagonist. The right panel is a Northern blot showing changes in endogenous ANF mRNA levels in ventricular cardiomyocyte cultures in response to 24 h of treatment with 100 nM AII. (C) The −695ANF promoter luciferase construct was cotransfected with rat AT1aR, a dominant-negative AT1a mutant (N74D), or an AII type 2 receptor (AT2) expression vector in C2C12 cells. The cells were treated or not with 100 nM AII with or without 100 nM Los for 12 h. The results shown in B and C are the means ± standard errors of at least six independent determinations and represent the ratio of promoter activity plus AII over promoter activity plus vehicle.
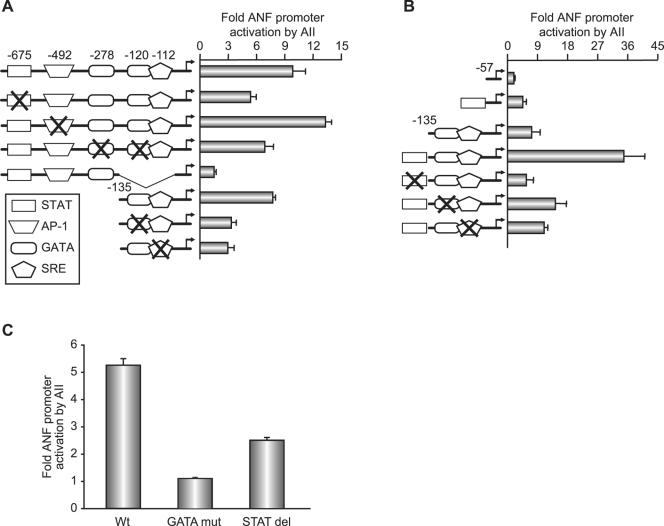
Next, we used mutational analysis to map the response elements. The ANF promoter contains numerous regulatory elements that confer tissue specificity and/or hormonal response (13, 14, 21, 43, 49). They include the composite GATA/serum response element (SRE) site that was shown to be targeted by endothelin 1 (49), a bona fide AP1 site (43) that was reported to be essential for in vivo response to pressure overload (79), a putative binding site for STAT proteins located around −675 bp. and an A/T-rich region that was shown to bind MEF2 proteins (48). Deletion scanning revealed that two regions are required for maximal AII response: the bp −695 to −500 distal region whose deletion reduced AII response by twofold and the proximal region (bp −50 to −135) whose deletion in the context of the bp −695 promoter resulted in the loss of 75% of the response (Fig. 2A; also data not shown). Finer mutagenesis was then used; within the distal region, mutation of the AP1 site as well as the distal MEF-like element had no effect on promoter activation by the liganded AT1R. In contrast, mutation or deletion of the putative STAT element resulted in a 50% decrease in the AT1R response. Mutation of two high-affinity GATA-4 binding sites also reduced promoter activation by 50% (Fig. 2A). Within the proximal promoter, mutation of either the GATA or the low-affinity SRE also reduced the hormone response. Together, these results show that at the level of the ANF promoter, maximal AT1R responsiveness requires the STAT element as well as the GATA sites. Moreover, cooperative interaction between these elements was suggested with the finding that an artificial promoter containing the STAT element upstream of the composite GATA/SRE site mediated a synergistic response to AT1R activation (Fig. 2B). The relative importance of the GATA and STAT elements in ANF promoter activation in response to AII was confirmed with neonatal cardiomyocytes. Consistent with the results obtained in C2C12 cells, deletion of the STAT element resulted in reduced AII responsiveness, but mutation of the GATA elements completely abrogated AII-dependent transcriptional activation (Fig. 2C). Thus, in cardiomyocytes GATA binding sites appear essential, while the putative STAT element contributes quantitatively to the AII response.
FIG. 2.
Mapping AII response elements on the ANF promoter. (A and B) C2C12 cells were cotransfected with various ANF promoter constructs and the AT1aR expression vector and then treated with or without 100 nM AII for 12 h. In panel A, reporter constructs were driven by the wild-type −695 or −135 ANF promoter and deletion or point mutants thereof. In panel B, C2C12 cells were cotransfected with reporter constructs driven by the minimal −57 or −135ANF promoters plus or minus the distal STAT element and point mutants thereof. Note how the distal STAT-containing domain cooperates with the proximal GATA/SRE. The data shown represent the means ± standard errors of the means of at least six independent determinations. (C) ANF promoter elements required for AII response in cardiomyocytes. Cells were treated with 100 nM AII for 24 h. Wt, −695ANF-luc; GATA mut, −695ANF-luc with mutated GATA sites; STAT del, −640ANF-luc. The data are from one representative experiment carried out in duplicate (out of three).
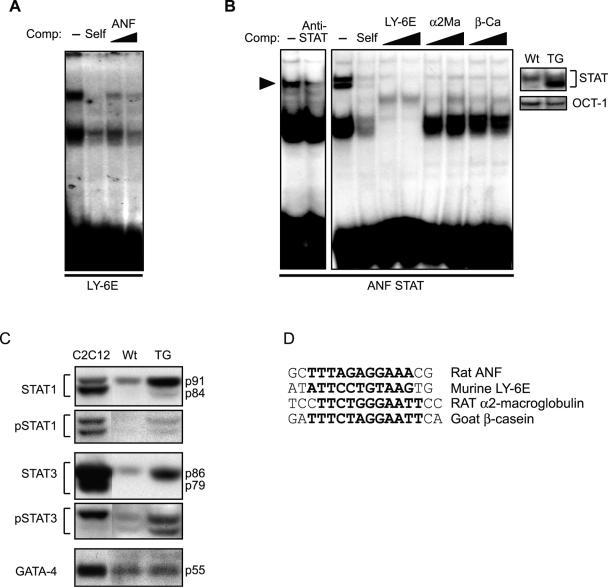
GATA binding over the ANF GATA site reflects GATA-4 interaction (14). To determine the nature of the binding over the putative STAT element, we carried out electrophoretic mobility shift assays using extracts prepared from mouse hearts and myoblast cultures. The sequence of all probes used is shown in Fig. 3D. First, we tested the ability of the putative ANF STAT element to effectively compete binding over a consensus STAT site; these experiments indicated that this region of the ANF promoter is able to displace binding over the well-characterized STAT element of the LY-6E promoter, albeit with lower efficiency than the cold self probe (Fig. 3A). The ANF STAT element also bound transfected STAT1α (Fig. 3B, left panel), as well as endogenous STAT proteins present in nuclear extracts from mouse hearts (Fig. 3B, middle panel); binding over the ANF probe was efficiently displaced by several well-characterized STAT elements (Fig. 3B, middle panel). Interestingly, STAT binding was increased in TG mouse extracts, suggesting that activation of STAT proteins may be an early event in AT1R action in the heart (Fig. 3B, right panel). This was directly confirmed using Western blot analysis, which revealed increased nuclear accumulation of STAT1 and -3 but no detectable change in the level of GATA-4 (Fig. 3C).
FIG. 3.
A) The distal ANF promoter harbors a low-affinity STAT binding site. Electrophoretic mobility shift assays were performed using nuclear extracts of C2C12 cells cotransfected with STAT1α and AT1aR expression vectors and treated with AII. Binding to the 32P-labeled Ly-6E STAT element was competed with 25× cold probe (Self) or with 25× or 50× unlabeled ANF oligonucleotide containing the putative STAT site (ANF). (B) Electrophoretic mobility shift assays were performed using the STAT-containing ANF probe (ANF STAT) and nuclear extracts prepared from STAT1-expressing C2C12 cells (left panel) or from pooled hearts of wild-type (Wt) or AT1R transgenic (TG) mice (middle and right panels). Specific binding was blocked by an anti-STAT1 antibody (left panel). Binding on the ANF STAT probe was competed with a 25× excess of cold probe (Self) or with 25× or 50× labeled probes containing STAT binding sites from the indicated promoters (middle panel). The right panel shows increased STAT binding in AT1R transgenic hearts (TG) compared to wild-type littermates (Wt). Note unchanged OCT-1 binding in the same extracts. (C) Western blots were carried out on whole-cell extracts prepared from the hearts of wild-type (Wt) and AT1R transgenic mice (TG) using the indicated antibodies. In each case, the first lane is a positive control containing extracts from cells overexpressing the relevant transcription factor. (D) Sequences of the various STAT elements used. The core sequences are indicated in bold.
Nuclear action of AII involves the JAK-STAT and PKC-GATA-4 pathways.
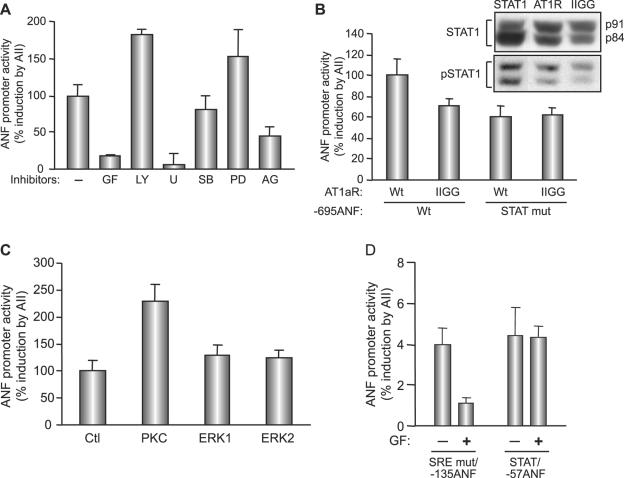
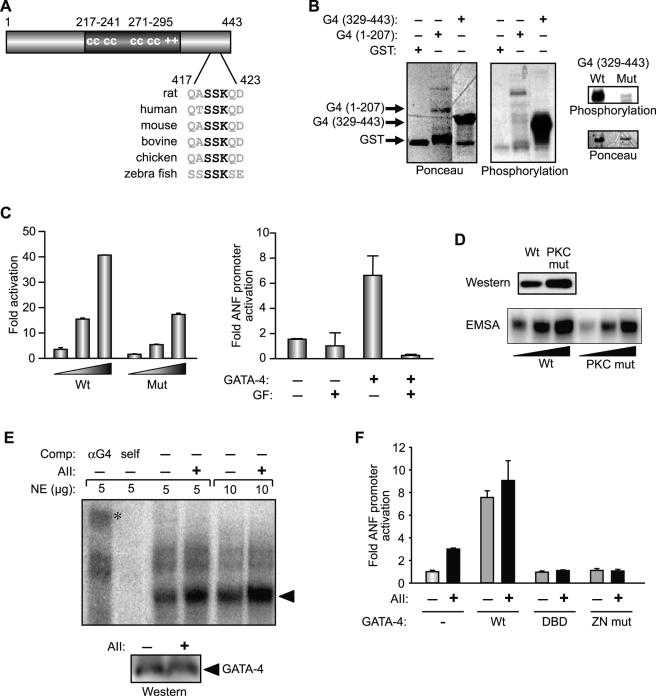
AT1R may activate STAT proteins via JAK2-dependent or -independent pathways (16, 63). On the other hand, contribution of the GATA element in the AII response may be the result of activation of the MAPK cascade, which could lead to phosphorylation and potentiation of GATA-4 activity as reported previously (15, 41). To test the involvement of the various signaling cascades in AII regulation of transcription, we first checked the effects of various pharmacologic inhibitors on the ANF promoter response to AII. As shown in Fig. 4A, inhibition of JAK kinases with AG490 resulted in a 50% loss of promoter activation. Inhibition of p38 MAPK (with SB 203580) or ERK (with PD 98059) did not block the response; in fact, ERK inhibition resulted in a modest but reproducible enhancement of AII responsiveness. Similarly, inhibition of the PI3 kinase pathway (with LY 294002) consistently resulted in a 50 to 60% increase in AII-dependent promoter activation. In contrast, inhibition of phospholipase C (with U-73122) or protein kinase C (GF 109203T) virtually abolished ANF promoter responsiveness to AII. Thus, it appeared that two major signaling pathways, those of JAK-STAT and PKC, were involved in linking AT1R to transcriptional regulation. We further extended these results with the use of a mutant AT1 receptor in which the YIPP motif (amino acids [aa] 319 to 322) required for JAK interaction was mutated (2). Although this mutant receptor (IIGG) could still mediate an AII transcriptional response, the maximal activation achieved was consistently 30 to 40% lower than that of the wild-type receptor when the reporter used was driven by the intact −695ANF promoter; however, when the reporter used contained a mutation in the STAT binding site, the mutant receptor ability to activate transcription was similar to that of the wild-type receptor (Fig. 4B). To ascertain the effect of the mutant AT1 receptor on STAT activation, we analyzed STAT1 levels in whole-cell extracts from AII-treated C2C12 cells transiently transfected with the intact or mutated receptor. STAT1 proteins (p91 and p84) are the predominant STAT proteins in these cells. As shown in the inset of Fig. 4B, the level of endogenous pSTAT1 was lower in extracts prepared from cells transfected with the mutant receptor. Together, these data are consistent with an involvement of JAK2 in linking AT1R to STAT activation and transcriptional effects.
FIG. 4.
Involvement of JAK, PKC, and phospholipase C but not PI3K or MAPK in AT1aR-dependent ANF transcriptional activation. (A) C2C12 cells were cotransfected with the −695ANF-luc construct and the AT1aR expression vector and treated or not with AII in the presence of inhibitors for various pathways: p38 MAPK (SB 203580; 10 μM [SB]), ERK (PD 98059; 50 μM [PD]), PI3K (LY 294002; 50 μM [LY]), phospholipase C (U-73122; 5 μM [U]), PKC (GF 109203X; 5 μM [GF]), and JAK2 tyrosine kinase (AG490; 25 μM [AG]). Inhibitors were applied 30 min before the AII treatment. (B) Maximal AII-induced ANF transactivation requires JAK binding to AT1aR. C2C12 cells were cotransfected with wild-type AT1aR (Wt) or YIPP motif mutant (IIGG) and −695ANF-luc constructs containing the wild type (Wt) or a STAT mutant (STAT mut). The inset shows total and phospho-STAT1 (pSTAT1) levels in AII-treated C2C12 cells transiently transfected with intact AT1aR or the IIGG mutant. The first lane is a positive control from AII-treated C2C12 cells overexpressing STAT1α. Note decreased pSTAT1 levels in IIGG-expressing cells relative to those in cells expressing wild-type receptor. (C) PKCβ enhances AII-induced ANF transactivation. C2C12 cells were cotransfected with the −695ANF-luc construct and the AT1aR expression vector plus or minus the PKC (the catalytic subunit), ERK1, or ERK2 expression vector. The expression vectors are described in Material and Methods. (D) AII-induced PKC targets GATA elements. C2C12 cells were cotransfected with AT1aR and the indicated ANF-luc constructs and stimulated with AII in presence or absence of the PKC inhibitor (GF). In all cases, AII treatment was for 12 h. Data shown are means ± standard errors of the means of six independent determinations.
We also used genetic approaches to confirm the involvement of PKC in AII transcriptional effects. Coexpression of the catalytic subunit of PKCβ resulted in a 2.5-fold enhancement of the AII response, whereas expression of constitutively active ERK1/2 had no effect on the level of ANF activation in response to AII (Fig. 4C). Finally, we tested which regulatory element(s) are targeted by PKC. As shown in Fig. 4D, a minimal promoter driven by the ANF STAT element was activated fourfold in response to AII, and this response was not altered in the presence of the PKC inhibitor; in contrast, the ability of the proximal promoter region, and more specifically the GATA element therein, to respond to AII was abrogated by pharmacologic inhibition of PKC. Together, these results suggest that two pathways, those of JAK-STAT and PKC-GATA-4, mediate the AT1R transcriptional response.
We further examined the effect of PKC on GATA-4. Bioinformatics-assisted analysis of the GATA-4 protein revealed a putative PKC phosphorylation site within its C-terminal activation domain (aa 417 to 423); this motif is conserved across all species (Fig. 5A). To test whether this or other domains of GATA-4 are phosphorylatable by PKC, we performed in vitro phosphorylation analysis on bacterially expressed GST fusion proteins containing either the N- or C-terminal transactivation domain of GATA-4. As shown in Fig. 5B, the recombinant protein containing the C-terminal region of GATA-4 (aa 329 to 443) was efficiently phosphorylated by the purified catalytic subunit of PKC. Mutation of the serine residues within the PKC recognition motif (S419A S420A) dramatically decreased PKC phosphorylation, suggesting that this site is the major PKC phosphorylation target on GATA-4. Next, the functional consequences of PKC phosphorylation on GATA-4 were assessed. Pharmacologic inhibition of PKC significantly decreased GATA-4 activation of the ANF promoter (Fig. 5C, right panel), and S419A S420A mutation in GATA-4 reduced its transcriptional activity by more than 50% (Fig. 5C, left panel). To elucidate the mechanisms by which PKC activates GATA-4, we analyzed the effect of the S419A S420A mutation on nuclear protein accumulation and DNA binding properties. As shown in Fig. 5D, although the S419A S420A mutation was consistently expressed at a slightly higher level (top panel), the ability of this mutant to interact with the ANF GATA binding site (bottom panel) was markedly decreased (2.5- to 3-fold, without correcting for protein expression). This result suggested that AII stimulation enhances GATA-4 activity, in part by increasing its DNA-binding capacity. This hypothesis was directly tested by analyzing the level and DNA-binding activity of endogenous GATA-4 in AII-stimulated cardiomyocytes. Treatment of neonate cardiomyocyte cultures with AII did not affect the level of nuclear GATA-4 protein (Fig. 5E, lower panel), a finding consistent with that obtained in extracts from the hearts of AT1R transgenics (Fig. 3C). Nevertheless, gel shift analysis revealed a 2.7-fold increase in GATA binding activity that was confirmed to correspond to GATA-4 with antibody supershift assay (Fig. 5E). That nuclear signaling by AII involved GATA-4 was also confirmed by the ability of two dominant-negative GATA-4 proteins to block the AII response in cardiomyocytes, while addition of the intact GATA-4 protein could substitute for AII stimulation (Fig. 5F). Together, the above results reveal that GATA-4 is an essential mediator of nuclear AII action and that AII activates GATA-4, in part, through PKC-mediated phosphorylation and enhancement of its DNA binding activity.
FIG. 5.
(A) GATA-4 contains a conserved PKC phosphorylation site within the C-terminal activation domain. The putative PKC phosphorylation site is shown in bold. (B) In vitro PKC phosphorylation of GST-GATA-4 fusion proteins containing either the N-(1-207) or C-terminal (329-443) transactivation domain. Ponceau staining was used to show protein loading. The right panel shows the effect of serine-to-alanine mutation within the conserved PKC motif (S419A S420A) on PKC phosphorylation. (C) Effect of mutating the PKC phosphorylation residues on GATA-4 transcriptional activity. In the left panel, C2C12 cells were cotransfected with a 3× GATA-luc reporter and increasing concentrations of wild-type (Wt) HA-GATA-4 (1-425) or mutated (Mut) HA-GATA-4 (S419A S420A). The right panel shows the effect of PKC inhibition with 5 μM GF 109203X on the −135ANF promoter activation with GATA-4. The data shown represent the means ± standard errors of the means of at least four independent determinations. (D) Effect of the same mutation on GATA-4 nuclear levels (top panel) and DNA binding activity (lower panel). Nuclear extracts were prepared from C2C12 cells transfected with wild-type (Wt) or mutant (PKC mut) GATA-4 (described above). The amount of GATA-4 was determined by Western blotting using anti-HA antibody (upper panel). The binding activity was determined by electrophoretic mobility shift assay using increasing concentrations of nuclear extracts and the −120ANF GATA site as a probe. (E) Effect of AII on endogenous GATA-4 levels and DNA-binding activity. Nuclear extracts (NE) were prepared from cardiomyocytes infected with Ad-AT1aR and treated or not with 100 nM AII for 20 min. DNA binding (upper panel) was performed as described above. The specificity of GATA-4 binding was determined by supershift assay with the anti-GATA-4 (αG4) antibody or by competition with the cold ANF GATA oligonucleotide (self). GATA binding is indicated by the arrowhead and the supershift by the asterisk. The amount of GATA-4 in the nuclear extracts was determined by Western blotting using anti-GATA-4 antibody. (F) The AII-GATA-4 pathway in cardiomyocytes. Cardiomyocytes were cotransfected with the −695ANF-luc construct and wild-type (Wt) or two dominant-negative forms of GATA-4, one with the ability to bind DNA but lacking transcriptional activation domains (DBD) and one lacking DNA binding activity but retaining the ability to interact with cofactors (ZN mut). The data are the means ± standard errors of the means of one representative experiment out of four that were qualitatively identical.
Synergistic activation of transcription by GATA-4 and STAT1.
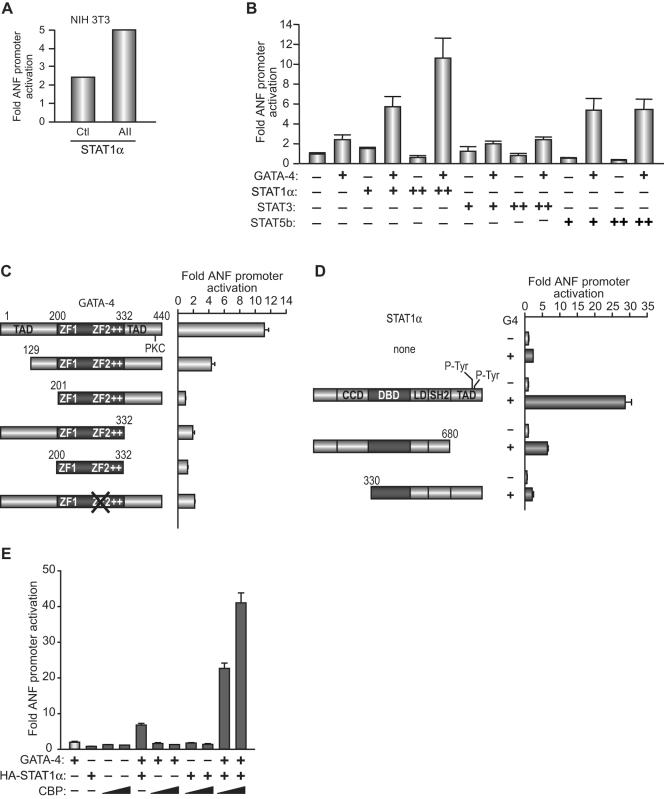
The data presented in Fig. 2B and Fig. 4 also suggested that transcriptional regulation of ANF by AII may involve cooperative interaction between STAT proteins and GATA factors. We tested this hypothesis using cotransfections and immunoprecipitation assays. First, we analyzed the effects of different STAT proteins on ANF promoter activity. STAT1α but not STAT3 or STAT5b consistently activated the ANF promoter in cardiac and noncardiac cells, and this effect was further potentiated in the presence of AII (Fig. 6A). Next, we tested whether STAT1α and GATA-4 cooperate in transcriptional activation. As shown in Fig. 6B, STAT1α and GATA-4 synergistically activated the ANF promoter. Addition of STAT1α consistently enhanced GATA-4 transcriptional activation of this promoter by five- to sixfold (n > 20). In contrast, STAT3, in various amounts, had no effect on GATA-4 activity. Interestingly, although STAT5b by itself did not activate the ANF promoter, it was able to cooperate with GATA-4 in transcriptional activation, though to a lesser extent than STAT1α (Fig. 6B).
FIG. 6.
(A) AII potentiates STAT1α-induced transactivation of ANF. NIH 3T3 cells were cotransfected with the −695ANF-luc construct and the STAT1α expression vector and treated with 100 nM AII (AII) or vehicle (Ctl) for 12 h. (B) Synergistic activation of ANF transcription by GATA-4 and STAT1α. Cells were cotransfected with the −695ANF-luc and GATA-4 vectors in the presence or absence of various STAT expression vectors. The results shown are from C2C12 cells, but similar results were also obtained in NIH 3T3 cells. (C and D) Structure-function analysis of GATA-4 and STAT1α using the −695ANF-luc reporter. The various GATA-4 or STAT1α expression vectors are shown schematically. (E) Effect of the CBP coactivator on GATA-4/STAT1 synergy. The experiments were done as described in panels C and D above. The data shown represent the means ± standard errors of the means of several experiments (n = 6 for panels A and B; n = 4 for panels C, D, and E).
To better understand the mechanisms involved in STAT/GATA synergy, we carried out structure-function analysis of GATA-4 and STAT1α. The GATA-4 protein contains two transcriptional activation domains flanking its two-zinc-finger DNA-binding domain. As shown in Fig. 6C, removal of the first 129 aa, which decreased GATA-4 transcriptional activity, reduced but did not abrogate synergy; deletion of the C-terminal activation domain significantly reduced synergy, indicating that intact GATA-4 transcriptional activity is required for functional interaction with STAT1. Consistent with this, the DNA binding domain (aa 200 to 332) was unable to support synergy. Mutations in the second zinc finger, which abolish DNA binding, also interfered with STAT1 synergy (Fig. 6C), suggesting that GATA-4 binding to its site is necessary for functional interaction with STAT over the ANF promoter. Next, we tested whether the transactivation domains of STAT1 were required for synergy with GATA-4. Various STAT1 functional domains have been identified (12) and are schematically depicted in Fig. 6D. Removal of the entire C-terminal transactivation domain significantly reduced but did not abolish synergy; however, removal of the N-terminal region, which harbors a domain required for STAT dimer-dimer formation as well as a coiled-coiled domain involved in CBP/p300 interaction, abrogated synergy (Fig. 6D). From this analysis, we conclude that the C-terminal activation domain of STAT1 is required for maximal synergy but that the N-terminal region is essential for functional interaction with GATA-4. The finding that the activation domains of both GATA-4 and STAT1 are needed for maximal synergy, given that both proteins interact with the CBP/p300 coactivators (6, 31), led us to investigate whether GATA-4/STAT1 cooperativity involves corecruitment of CBP. As shown in Fig. 6E, addition of increasing amounts of CBP greatly potentiated the synergistic action of GATA-4 and STAT1 on the ANF promoter.
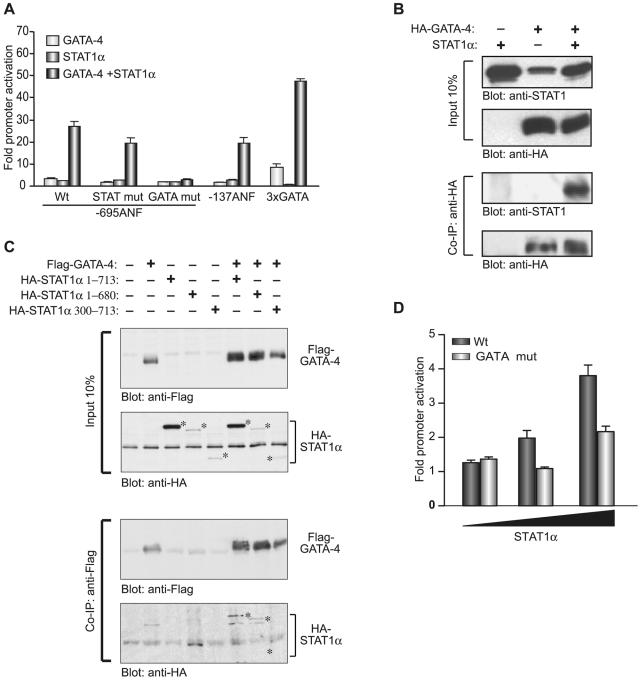
Next, we determined which promoter elements were required for GATA-4 and STAT1 cooperativity. As shown in Fig. 7A, deletion of sequences between bp −695 and −135 decreased maximal activation levels but did not impair GATA-4/STAT1 synergy, suggesting that binding of STAT1 to DNA was dispensable. Consistent with this, mutation of the STAT element in the context of the full-length promoter reduced but did not abolish synergy. In contrast, mutation of the GATA sites abrogated both GATA responsiveness and GATA/STAT synergy. A minimal promoter driven by multimerized GATA binding sites was cooperatively activated by GATA-4 and STAT1. Together, these results indicate that GATA elements are necessary and sufficient for GATA/STAT synergy and suggest that GATA factors may be able to recruit STAT proteins to target promoters. To test this, we performed coimmunoprecipitation assays which revealed that GATA-4 was indeed able to physically interact with STAT1 in vivo (Fig. 7B). Interestingly, removal of the N-terminal but not the C-terminal domain of STAT1 abolished physical association with GATA-4 (Fig. 7C), a finding that would explain the inability of N-terminally deleted STAT1 (330 to 713) to synergize with GATA-4 (Fig. 6D). Finally, to confirm the relevance of a GATA-dependent pathway for STAT1 action, ANF-luc constructs containing the −695 wild-type or GATA element-mutated ANF promoter were tested for their ability to respond to STAT1 in cardiomyocytes. As shown in Fig. 7A, whereas the wild-type promoter was dose-dependently activated by cotransfected STAT1, mutation of the GATA sites abrogated STAT1 activation. These results—which are consistent with those of Fig. 2C—indicate that in vivo, interaction with GATA factors bound to the promoter is required for STAT1 activation of the ANF promoter.
FIG. 7.
(A) Mapping of the DNA elements required for GATA-4/STAT1α synergy. NIH 3T3 cells were cotransfected with the indicated ANF-luc constructs or a minimal BNP promoter driven by multimerized GATA binding sites (3xGATA) and with GATA-4, STAT1α, or both expression vectors. The data shown represent the means ± standard errors of the means of at least six independent determinations. (B and C) GATA-4 interacts physically with STAT1α in vivo. In panel B, nuclear extracts from 293T cells transfected with STAT1α and/or HA-GATA-4 were immunoprecipitated using an anti-HA antibody, separated on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and subjected to immunoblotting using either anti-STAT1 or anti-HA antibodies (lower two panels). The top panels are Western blots carried out on the nuclear extracts used for the immunoprecipitation. In panel C, similar experiments were done using Flag-GATA-4 and HA-tagged full-length or N- or C-terminally deleted STAT1α. Immunoprecipitation was carried out with the anti-Flag antibody. The positions of the different STAT1 proteins as revealed with the anti-HA antibody are indicated by asterisks. (D) GATA elements are essential for ANF transactivation by STAT1α in cardiomyocytes. Increasing amounts of the STAT1α expression vector were cotransfected in cardiomyocytes with the −695ANF-luc construct (Wt) or a similar construct containing point mutations in the two high-affinity GATA sites (GATA mut). The data shown represent the means ± standard errors of the means of four independent determinations.
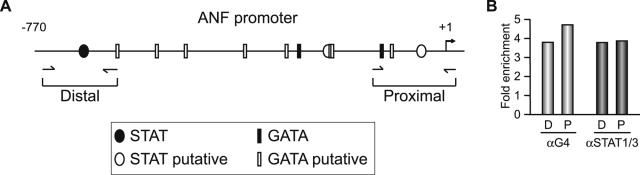
Next, we probed the in vivo association of the GATA-4 and STAT proteins with the endogenous ANF gene in cardiomyocytes. For this, we performed chromatin immunoprecipitation assays using AT1aR-activated neonate cardiomyocytes in primary cultures. As shown in Fig. 8, we found specific enrichment of GATA-4 and STAT1 and -3 over both the distal andproximal domains of the ANF promoter. These results indicate that the ANF gene is a direct transcriptional target for GATA-4 and STAT1 and -3.
FIG. 8.
In vivo association of the GATA-4 and STAT proteins with the ANF promoter. (A) Schematic representation of the various STAT and GATA elements on the ANF promoter. Putative elements correspond to in silico-identified consensus sequences. The location of the sets of primers used in the QPCR is depicted below. (B) Enrichment of GATA-4 and STAT1 and -3 proteins on the endogenous ANF promoter as revealed from ChIP experiments. The results shown are from one representative experiment out of two carried out in duplicate on primary cardiomyocytes stimulated for 48 h with 100 nM AII. The data are expressed as the ratio of ChIP with the respective antibodies over that obtained with immunoglobulin G. For details, see Materials and Methods.
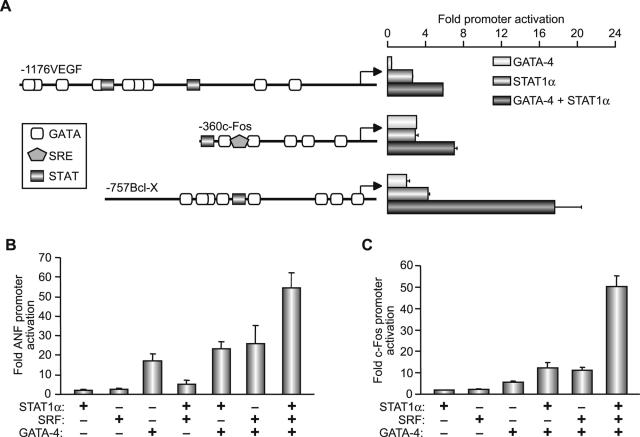
Finally, we tested the effects of GATA-4 and STAT1 on other growth factor-inducible promoters. For this, we used the c-fos, Bcl-xL, and VEGF promoters. The c-fos and Bcl-xL promoters are inducible by various cytokines and/or growth factors that act through the JAK-STAT pathway, and they contain well-characterized STAT binding sites (68, 84). These promoters also contain GATA binding sites and are activated by GATA transcription factors (3, 49). The VEGF promoter was recently shown to be activated by STAT3 (51, 80), and our own results indicate that it is also a GATA target. As shown in Fig. 9A, all three promoters were synergistically activated by GATA-4 and STAT1. Given that in the case of c-fos and ANF we previously showed that GATA-4 cooperates with SRF (49), and since mutation of the proximal ANF SRE also affects cooperative interaction between the STAT element and the proximal promoter (Fig. 2B), we tested for possible functional cooperation between STAT, GATA-4, and SRF. We were unable to detect any cooperativity between STAT proteins and SRF on the ANF or c-fos promoters using several DNA concentrations of STAT and SRF expression vectors (data not shown). However, when GATA-4, STAT1, and SRF were simultaneously added, superactivation of both ANF-driven (Fig. 9B) and c-fos-driven (Fig. 9C) reporters was observed, suggesting that GATA-4 is able to recruit STAT1 to a GATA/SRF ternary complex and that STAT1 may act as an inducible coactivator of this transcriptional complex.
FIG. 9.
(A) Functional cooperation between GATA-4 and STAT1α is not restricted to the ANF promoter. NIH 3T3 cells were cotransfected with the −1176 VEGF, −360 c-fos, or −757 Bcl-x promoter-driven luciferase reporters and GATA-4, STAT1α, or both expression vectors. (B and C) Functional cooperation between STAT1α, GATA-4, and SRF. Cells were cotransfected with −137ANF (B) or −360 c-fos (C) promoter-driven luciferase reporter constructs with various combinations of SRF, GATA-4, and STAT1α. The data shown represent the means ± standard errors of the means for four to six experiments with C2C12 cells.
DISCUSSION
Extracellular stimulus-induced regulation of cell fate requires the execution of a complex program of transcriptional events. How specificity of the transcriptional response is achieved remains largely undefined, although it is now widely accepted that combinatorial interactions of signaling pathways and transcription factors play a major role in this process (reviewed in reference 4). In the present work, we examined the mechanisms that transduce the transcriptional response to a biologically important peptide hormone acting through a G-protein-coupled receptor. Our results indicate that nuclear integration of AII signaling involves cooperative interaction between the signal-regulated STAT transcription factors and the tissue-specific family of GATA proteins whose activity is also directly modulated by various intracellular signaling cascades. These data provide new insight into the mechanisms underlying signaling specificity of AT1R. Moreover, the finding of combinatorial interaction between the STAT and GATA proteins has broad implication for understanding cytokine and growth factor signaling in health and disease.
AII regulation of ANF transcription.
AII is a major cardioregulatory hormone, and drugs that inhibit its biosynthesis or its receptor are widely used for the treatment of human cardiovascular conditions. Surprisingly, the mechanisms by which AII regulates transcription in the heart remain undefined. Within the heart, AII targets both myocytes and nonmyocytes (64, 74). Except for the c-fos gene (61), few direct transcription targets of AII in cardiomyocytes have been identified. The data presented show that ANF is a direct transcription target of AII and that upregulation of ANF does not require AII-induced hypertrophy. Thus, studies of AII regulation of the ANF promoter offered the opportunity to identify AII-mediated transcriptional mechanisms without the confounding input of hypertrophy-induced pathways. The results show that AII, via its type 1 receptor (AT1R), activates the ANF promoter through two major signaling cascades, JAK-STAT and PKC-GATA-4. Interestingly, MAPK signaling did not appear to play a role in relaying AII signaling at the level of the ANF promoter. This may reflect a more specific role for the MAPK pathway in the growth (hypertrophic) effects of AII that are dependent on the EGF receptor (74). In contrast, ANF is a cardioprotective hormone that antagonizes cardiac hypertrophy (30). ANF is also a well-known antagonist of AII (39). Thus, ANF activation by AII may be part of a negative feedback loop that serves to attenuate AII action. In this case, the signaling pathways linking AT1R to ANF transcription, i.e., PKC and JAK-STAT, would be predicted to be cardioprotective. Several studies support this conclusion. First, STAT proteins have been reported to transduce cardioprotective signals (29, 37, 83). Although the mediators of this beneficial effect are not defined, interaction of STAT with GATA-4, a cardioprotective transcription factor (3), and activation of ANF and/or VEGF may explain at least part of STAT cardioprotective effects. Second, although numerous hypertrophic signals were shown to activate different PKC isoforms, the bulk of the evidence argues against an involvement of PKC in initiating and/or maintaining hypertrophy (11, 28). In fact, upregulation of PKCβ in the adult heart has been shown to have beneficial effects (75). Importantly, pharmacologic inhibition of PKC did not block AII-induced hypertrophy (74) but did abrogate AII activation of c-fos transcription (62) and ANF biosynthesis (34). In the future, it will be interesting to map the specific domains of the AT1R that are required for ANF transcription and determine whether they can be dissociated from those involved in mediating the AII hypertrophic response. Such studies could ultimately lead to the development of novel therapeutic drugs that selectively target specific AII-activated signaling pathways.
GATA-4, an integrator of cell signaling in the heart.
Cooperative interactions between cell-specific transcription factors and signal-activated regulators represent one of the most effective means to achieve cell and target gene specificity by signaling pathways. The experiments presented in this paper reveal that GATA-4 plays an essential role for ANF promoter activation in response to AII. In this context, GATA-4 acts as an integrator of two signaling pathways: PKC, which targets its C-terminal domain, resulting in enhanced DNA binding activity, and the JAK-STAT pathway, which potentiates its activity through protein-protein interaction with STATs. GATA-4, a member of the GATA family of tissue-specific zinc finger proteins, is a critical regulator of cardiogenesis (26, 58). In postnatal cardiomyocytes, GATA-4 is essential for maintaining the cardiac genetic program (14) and for the adaptive response of the heart to numerous stimuli, including hormones and work overload (15, 42, 57). GATA-4 actions involve combinatorial interactions with other cell-restricted or inducible transcription factors, including Nkx2.5, Mef2, SRF, nuclear factor of activated T cells, SMAD, and c-fos (reviewed in reference 73). GATA-4 activity is also directly modulated by signaling cascades; ERK and p38 MAPK phosphorylate and enhance GATA-4 transcriptional activation domains (15, 41), while glycogen synthase kinase (GSK3β) phosphorylates the GATA-4 DNA binding domain and inhibits nuclear GATA-4 accumulation (50). Through its cooperative interaction with other transcriptional regulators, GATA-4 serves as a nuclear integrator of several signaling pathways, most notably calcineurin, MAPK, PI3 kinase, and receptor serine-threonine kinases. Our findings that PKC and JAK-STAT also converge on GATA-4 expand the role of GATA-4 as a transcriptional mediator of epigenetic signals and will be important in understanding the mechanisms of action of numerous other cardioregulators. For example, opioid receptor agonists were found to enhance embryonic stem cell differentiation into cardiomyocytes via PKC-dependent activation of GATA-4 (78). On the other hand, some cytokines acting through a gp130-coupled receptor have been shown to require GATA elements for transcriptional regulation of target promoters (47).
Our findings that GATA binding sites were sufficient to support GATA/STAT synergy and that GATA-4 was able to physically associate with STAT1 in living cells are especially noteworthy, since they raise the possibility that STAT proteins can activate target promoters via GATA binding sites. Synergistic action of the GATA/STAT complex does not appear to be due to an effect of STAT on GATA-4 binding to its site (data not shown). The structure-function analysis indicates that at least one of the transcriptional activation domains of GATA-4 is required; in the case of STAT1, the presence of the N-terminal domain appears to contact GATA-4, and the C-terminal activation domain, which is known to interact with CBP/p300 (31), is required for maximal synergy (Fig. 6). GATA-4 and several other GATA factors also associate with CBP/p300 through the DNA binding domain, leading to enhanced transcriptional activity (6, 7, 33). Together with ability of CBP to further enhance synergy, these data suggest that the GATA/STAT functional synergy likely involves corecruitment of CBP/p300.
GATA/STAT interaction and cell specificity of cytokine action.
The involvement of the GATA and STAT proteins in cytokine signaling is well documented in the hematopoietic system. For example, both STAT5 and GATA-1 are essential for the survival effects of erythropoietin on erythroid progenitors (68, 81), and dominant-negative GATA proteins block interleukin 13 action; these proteins also mediate the effects of thrombopoietic cytokines in erythroid and megakaryotic differentiation (32, 36). Similarly, in the lymphoid system, both GATA-3 and STAT5 and -6 mediate cytokine-induced development of CD4+ cells and the differentiation of T-helper type 2 (cells (54). Interestingly, cooperation between GATA and STAT has been suggested by the finding that the T-helper type 2 locus control region requires both GATA-3 and STAT6 for generating/maintaining an open chromatin configuration (70). More recently, GATA-1 was found to mediate interferon 8 induction of the human major histocompatibility complex class 1b gene; in this case, GATA-1 binds to a weak site adjacent to the interferon response element, and the presence of both sites results in superactivation (5). Thus, the GATA/STAT synergy described in this work is likely to represent a general mechanism that could account for cell-specific effects of cytokine/growth factors acting through tyrosine kinase-coupled receptors. This may be particularly relevant to understanding cytokine action in hematopoietic cells. In this respect, we demonstrated that at least two targets of GATA and STAT action in hematopoietic cells, namely the Bcl-x and c-fos promoters, are synergistically activated by GATA and STAT proteins (Fig. 9). Whether this paradigm extends to other target genes in hematopoietic and other cell types deserves to be tested.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (MGP-13056 and MOP36382) and Génome-Québec. H.W. is the recipient of an AstraZeneca Canada/HSFC Research Fellowship, and M.N. holds a Canada Research Chair in Molecular Biology.
We thank S. Karnik, D. Richard, and T. Hoang for sharing various DNA vectors, Walter G. Thomas for generously providing the adenovirus AT1aR vector, Lise Laroche for secretarial assistance, and the Nemer Lab for helpful discussions.
REFERENCES
- 1.Adams, J. W., and J. H. Brown. 2001. G-proteins in growth and apoptosis: lessons from the heart. Oncogene 20:1626-1634. [DOI] [PubMed] [Google Scholar]
- 2.Ali, M. S., P. P. Sayeski, L. B. Dirksen, D. J. Hayzer, M. B. Marrero, and K. E. Bernstein. 1997. Dependence on the motif YIPP for the physical association of Jak2 kinase with the intracellular carboxyl tail of the angiotensin II AT1 receptor. J. Biol. Chem. 272:23382-23388. [DOI] [PubMed] [Google Scholar]
- 3.Aries, A., P. Paradis, C. Lefebvre, R. J. Schwartz, and M. Nemer. 2004. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc. Natl. Acad. Sci. USA 101:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, D. M., K. S. Gustafson, J. Wang, S. Z. Wang, and G. D. Ginder. 2004. A GATA factor mediates cell type-restricted induction of HLA-E gene transcription by gamma interferon. Mol. Cell. Biol. 24:6194-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhalla, S. S., L. Robitaille, and M. Nemer. 2001. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J. Biol. Chem. 276:11439-11445. [DOI] [PubMed] [Google Scholar]
- 7.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blume, A., T. Herdegen, and T. Unger. 1999. Angiotensin peptides and inducible transcription factors. J. Mol. Med. 77:339-357. [DOI] [PubMed] [Google Scholar]
- 9.Booz, G. W., J. N. Day, and K. M. Baker. 2002. Interplay between the cardiac renin angiotensin system and JAK-STAT signaling: role in cardiac hypertrophy, ischemia/reperfusion dysfunction, and heart failure. J. Mol. Cell Cardiol. 34:1443-1453. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 11.Braz, J. C., K. Gregory, A. Pathak, W. Zhao, B. Sahin, R. Klevitsky, T. F. Kimball, J. N. Lorenz, A. C. Nairn, S. B. Liggett, I. Bodi, S. Wang, A. Schwartz, E. G. Lakatta, A. A. DePaoli-Roach, J. Robbins, T. E. Hewett, J. A. Bibb, M. V. Westfall, E. G. Kranias, and J. D. Molkentin. 2004. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat. Med. 10:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Bromberg, J., and J. E. Darnell, Jr. 2000. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19:2468-2473. [DOI] [PubMed] [Google Scholar]
- 13.Bruneau, B. G., G. Nemer, Schmitt, J. P., F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709-721. [DOI] [PubMed] [Google Scholar]
- 14.Charron, F., P. Paradis, O. Bronchain, G. Nemer, and M. Nemer. 1999. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol. Cell. Biol. 19:4355-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charron, F., G. Tsimiklis, M. Arcand, L. Robitaille, Q. Liang, J. D. Molkentin, S. Meloche, and M. Nemer. 2001. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 15:2702-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaturvedi, P., M. V. Reddy, and E. P. Reddy. 1998. Src kinases and not JAKs activate STATs during IL-3 induced myeloid cell proliferation. Oncogene 16:1749-1758. [DOI] [PubMed] [Google Scholar]
- 17.Dahlof, B., R. B. Devereux, S. E. Kjeldsen, S. Julius, G. Beevers, U. de Faire, F. Fyhrquist, H. Ibsen, K. Kristiansson, O. Lederballe-Pedersen, L. H. Lindholm, M. S. Nieminen, P. Omvik, S. Oparil, and H. Wedel. 2002. Cardiovascular morbidity and mortality in the Losartan Intervention For Endpoint reduction in hypertension study (LIFE): a randomised trial against atenolol. Lancet 359:995-1003. [DOI] [PubMed] [Google Scholar]
- 18.de Gasparo, M., K. J. Catt, T. Inagami, J. W. Wright, and T. Unger. 2000. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol. Rev. 52:415-472. [PubMed] [Google Scholar]
- 19.Deryckere, F., and F. Gannon. 1994. A one-hour minipreparation technique for extraction of DNA-binding proteins from animal tissues. BioTechniques 16:405. [PubMed] [Google Scholar]
- 20.Dupuis, S., C. Dargemont, C. Fieschi, N. Thomassin, S. Rosenzweig, J. Harris, S. M. Holland, R. D. Schreiber, and J. L. Casanova. 2001. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 293:300-303. [DOI] [PubMed] [Google Scholar]
- 21.Durocher, D., F. Charron, R. Warren, R. J. Schwartz, and M. Nemer. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faulds, M. H., K. Pettersson, J. A. Gustafsson, and L. A. Haldosen. 2001. Cross-talk between ERs and signal transducer and activator of transcription 5 is E2 dependent and involves two functionally separate mechanisms. Mol. Endocrinol. 15:1929-1940. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira, R., I. Naguibneva, M. Mathieu, S. Ait-Si-Ali, P. Robin, L. L. Pritchard, and A. Harel-Bellan. 2001. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey, N., and E. N. Olson. 2003. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 65:45-79. [DOI] [PubMed] [Google Scholar]
- 25.Grépin, C., L. Dagnino, L. Robitaille, L. Haberstroh, T. Antakly, and M. Nemer. 1994. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol. Cell. Biol. 14:3115-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grépin, C., G. Nemer, and M. Nemer. 1997. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development 124:2387-2395. [DOI] [PubMed] [Google Scholar]
- 27.Hautmann, M. B., M. M. Thompson, E. A. Swartz, E. N. Olson, and G. K. Owens. 1997. Angiotensin II-induced stimulation of smooth muscle alpha-actin expression by serum response factor and the homeodomain transcription factor MHox. Circ. Res. 81:600-610. [DOI] [PubMed] [Google Scholar]
- 28.Heidkamp, M. C., A. L. Bayer, J. L. Martin, and A. M. Samarel. 2001. Differential activation of mitogen-activated protein kinase cascades and apoptosis by protein kinase C epsilon and delta in neonatal rat ventricular myocytes. Circ. Res. 89:882-890. [DOI] [PubMed] [Google Scholar]
- 29.Hikoso, S., O. Yamaguchi, Y. Higuchi, S. Hirotani, T. Takeda, K. Kashiwase, T. Watanabe, M. Taniike, I. Tsujimoto, M. Asahi, Y. Matsumura, K. Nishida, H. Nakajima, S. Akira, M. Hori, and K. Otsu. 2004. Pressure overload induces cardiac dysfunction and dilation in signal transducer and activator of transcription 6-deficient mice. Circulation 110:2631-2637. [DOI] [PubMed] [Google Scholar]
- 30.Holtwick, R., M. van Eickels, B. V. Skryabin, H. A. Baba, A. Bubikat, F. Begrow, M. D. Schneider, D. L. Garbers, and M. Kuhn. 2003. Pressure-independent cardiac hypertrophy in mice with cardiomyocyte-restricted inactivation of the atrial natriuretic peptide receptor guanylyl cyclase-A. J. Clin. Investig. 111:1399-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvai, A. E., L. Xu, E. Korzus, G. Brard, D. Kalafus, T. M. Mullen, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 1997. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. USA 94:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwasaki, H., S. Mizuno, R. A. Wells, A. B. Cantor, S. Watanabe, and K. Akashi. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19:451-462. [DOI] [PubMed] [Google Scholar]
- 33.Kakita, T., K. Hasegawa, T. Morimoto, S. Kaburagi, H. Wada, and S. Sasayama. 1999. p300 protein as a coactivator of GATA-5 in the transcription of cardiac-restricted atrial natriuretic factor gene. J. Biol. Chem. 274:34096-34102. [DOI] [PubMed] [Google Scholar]
- 34.Kalra, D., N. Sivasubramanian, and D. L. Mann. 2002. Angiotensin II induces tumor necrosis factor biosynthesis in the adult mammalian heart through a protein kinase C-dependent pathway. Circulation 105:2198-2205. [DOI] [PubMed] [Google Scholar]
- 35.Kim, S., and H. Iwao. 2000. Molecular and cellular mechanisms of angiotensin II-mediated cardiovascular and renal diseases. Pharmacol. Rev. 52:11-34. [PubMed] [Google Scholar]
- 36.Kirito, K., M. Osawa, H. Morita, R. Shimizu, M. Yamamoto, A. Oda, H. Fujita, M. Tanaka, K. Nakajima, Y. Miura, K. Ozawa, and N. Komatsu. 2002. A functional role of Stat3 in in vivo megakaryopoiesis. Blood 99:3220-3227. [DOI] [PubMed] [Google Scholar]
- 37.Kunisada, K., S. Negoro, E. Tone, M. Funamoto, T. Osugi, S. Yamada, M. Okabe, T. Kishimoto, and K. Yamauchi-Takihara. 2000. Signal transducer and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin-induced cardiomyopathy. Proc. Natl. Acad. Sci. USA 97:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leu, J. I., M. A. Crissey, J. P. Leu, G. Ciliberto, and R. Taub. 2001. Interleukin-6-induced STAT3 and AP-1 amplify hepatocyte nuclear factor 1-mediated transactivation of hepatic genes, an adaptive response to liver injury. Mol. Cell. Biol. 21:414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin, E. R., D. G. Gardner, and W. K. Samson. 1998. Natriuretic peptides. N. Engl. J. Med. 339:321-328. [DOI] [PubMed] [Google Scholar]
- 40.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 41.Liang, Q., R. J. Wiese, O. F. Bueno, Y. S. Dai, B. E. Markham, and J. D. Molkentin. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marttila, M., N. Hautala, P. Paradis, M. Toth, O. Vuolteenaho, M. Nemer, and H. Ruskoaho. 2001. GATA4 mediates transcriptional activation of the B-type natriuretic peptide gene expression in response to hemodynamic stress. Endocrinology 142:4693-4700. [DOI] [PubMed] [Google Scholar]
- 43.McBride, K., and M. Nemer. 1998. The C-terminal domain of c-fos is required for activation of an AP-1 site specific for jun-fos heterodimers. Mol. Cell. Biol. 18:5073-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milanini, J., F. Vinals, J. Pouyssegur, and G. Pages. 1998. p42/p44 MAP kinase module plays a key role in the transcriptional regulation of the vascular endothelial growth factor gene in fibroblasts. J. Biol. Chem. 273:18165-18172. [DOI] [PubMed] [Google Scholar]
- 45.Miura, S., J. Zhang, and S. S. Karnik. 2000. Angiotensin II type 1 receptor-function affected by mutations in cytoplasmic loop CD. FEBS Lett. 470:331-335. [DOI] [PubMed] [Google Scholar]
- 46.Molkentin, J. D., and G. W. Dorn II. 2001. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 63:391-426. [DOI] [PubMed] [Google Scholar]
- 47.Morimoto, T., K. Hasegawa, S. Kaburagi, T. Kakita, H. Masutani, R. N. Kitsis, A. Matsumori, and S. Sasayama. 1999. GATA-5 is involved in leukemia inhibitory factor-responsive transcription of the B-myosin heavy chain gene in cardiac myocytes. J. Biol. Chem. 274:12811-12818. [DOI] [PubMed] [Google Scholar]
- 48.Morin, S., F. Charron, L. Robitaille, and M. Nemer. 2000. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 19:2046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin, S., P. Paradis, A. Aries, and M. Nemer. 2001. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol. Cell. Biol. 21:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morisco, C., K. Seta, S. E. Hardt, Y. Lee, S. F. Vatner, and J. Sadoshima. 2001. Glycogen synthase kinase 3beta regulates GATA4 in cardiac myocytes. J. Biol. Chem. 276:28586-28597. [DOI] [PubMed] [Google Scholar]
- 51.Niu, G., T. Bowman, M. Huang, S. Shivers, D. Reintgen, A. Daud, A. Chang, A. Kraker, R. Jove, and H. Yu. 2002. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene 21:7001-7010. [DOI] [PubMed] [Google Scholar]
- 52.Osman, H., C. Murigande, A. Nadakal, and A. M. Capponi. 2002. Repression of DAX-1 and induction of SF-1 expression. Two mechanisms contributing to the activation of aldosterone biosynthesis in adrenal glomerulosa cells. J. Biol. Chem. 277:41259-41267. [DOI] [PubMed] [Google Scholar]
- 53.Osugi, T., Y. Oshima, Y. Fujio, M. Funamoto, A. Yamashita, S. Negoro, K. Kunisada, M. Izumi, Y. Nakaoka, H. Hirota, M. Okabe, K. Yamauchi-Takihara, I. Kawase, and T. Kishimoto. 2002. Cardiac-specific activation of signal transducer and activator of transcription 3 promotes vascular formation in the heart. J. Biol. Chem. 277:6676-6681. [DOI] [PubMed] [Google Scholar]
- 54.Pai, S. Y., M. L. Truitt, and I. C. Ho. 2004. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc. Natl. Acad. Sci. USA 101:1993-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paradis, P., N. Dali-Youcef, F. W. Paradis, G. Thibault, and M. Nemer. 2000. Overexpression of angiotensin II type 1 receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc. Natl. Acad. Sci. USA 97:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patient, R. K., and J. D. McGhee. 2002. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12:416-422. [DOI] [PubMed] [Google Scholar]
- 57.Pikkarainen, S., H. Tokola, T. Majalahti-Palviainen, R. Kerkela, N. Hautala, S. S. Bhalla, F. Charron, M. Nemer, O. Vuolteenaho, and H. Ruskoaho. 2003. GATA-4 is a nuclear mediator of mechanical stretch-activated hypertrophic program. J. Biol. Chem. 278:23807-23816. [DOI] [PubMed] [Google Scholar]
- 58.Pu, W. T., T. Ishiwata, A. L. Juraszek, Q. Ma, and S. Izumo. 2004. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 275:235-244. [DOI] [PubMed] [Google Scholar]
- 59.Purcell, N. H., G. Tang, C. Yu, F. Mercurio, J. A. DiDonato, and A. Lin. 2001. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc. Natl. Acad. Sci. USA 98:6668-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rameil, P., P. Lecine, J. Ghysdael, F. Gouilleux, B. Kahn-Perles, and J. Imbert. 2000. IL-2 and long-term T cell activation induce physical and functional interaction between STAT5 and ETS transcription factors in human T cells. Oncogene 19:2086-2097. [DOI] [PubMed] [Google Scholar]
- 61.Sadoshima, J., and S. Izumo. 1993. Molecular characterization of angiotensin II-induced hypertrophy of cardiac myocytes and hyperplasia of cardiac fibroblasts. Critical role of the AT1 receptor subtype. Circ. Res. 73:413-423. [DOI] [PubMed] [Google Scholar]
- 62.Sadoshima, J. I., and S. Izumo. 1993. Signal transduction pathways of angiotensin II-induced c-fos gene expression in cardiac myocytes in vitro. Circ. Res. 73:424-438. [DOI] [PubMed] [Google Scholar]
- 63.Sayeski, P. P., M. S. Ali, S. J. Frank, and K. E. Bernstein. 2001. The angiotensin II-dependent nuclear translocation of Stat1 is mediated by the Jak2 protein motif 231YRFRR. J. Biol. Chem. 276:10556-10563. [DOI] [PubMed] [Google Scholar]
- 64.Schnee, J. M., and W. A. Hsueh. 2000. Angiotensin II, adhesion, and cardiac fibrosis. Cardiovasc. Res. 46:264-268. [DOI] [PubMed] [Google Scholar]
- 65.Shindo, T., I. Manabe, Y. Fukushima, K. Tobe, K. Aizawa, S. Miyamoto, K. Kawai-Kowase, N. Moriyama, Y. Imai, H. Kawakami, H. Nishimatsu, T. Ishikawa, T. Suzuki, H. Morita, K. Maemura, M. Sata, Y. Hirata, M. Komukai, H. Kagechika, T. Kadowaki, M. Kurabayashi, and R. Nagai. 2002. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 8:856-863. [DOI] [PubMed] [Google Scholar]
- 66.Shuai, K. 2000. Modulation of STAT signaling by STAT-interacting proteins. Oncogene 19:2638-2644. [DOI] [PubMed] [Google Scholar]
- 67.Si, J., and S. J. Collins. 2002. IL-3-induced enhancement of retinoic acid receptor activity is mediated through Stat5, which physically associates with retinoic acid receptors in an IL-3-dependent manner. Blood 100:4401-4409. [DOI] [PubMed] [Google Scholar]
- 68.Silva, M., A. Benito, C. Sanz, F. Prosper, D. Ekhterae, G. Nunez, and J. L. Fernandez-Luna. 1999. Erythropoietin can induce the expression of bcl-x(L) through Stat5 in erythropoietin-dependent progenitor cell lines. J. Biol. Chem. 274:22165-22169. [DOI] [PubMed] [Google Scholar]
- 69.Simon, M. A. 2000. Receptor tyrosine kinases: specific outcomes from general signals. Cell 103:13-15. [DOI] [PubMed] [Google Scholar]
- 70.Spilianakis, C. G., and R. A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 71.Stocklin, E., M. Wissler, F. Gouilleux, and B. Groner. 1996. Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726-728. [DOI] [PubMed] [Google Scholar]
- 72.Suzuki, E., H. Nishimatsu, H. Satonaka, K. Walsh, A. Goto, M. Omata, T. Fujita, R. Nagai, and Y. Hirata. 2002. Angiotensin II induces myocyte enhancer factor 2- and calcineurin/nuclear factor of activated T cell-dependent transcriptional activation in vascular myocytes. Circ. Res. 90:1004-1011. [DOI] [PubMed] [Google Scholar]
- 73.Temsah, R., and M. Nemer. 2005. GATA factors and transcriptional regulation of cardiac natriuretic peptide genes. Regul. Pept. 128:177-185. [DOI] [PubMed] [Google Scholar]
- 74.Thomas, W. G., Y. Brandenburger, D. J. Autelitano, T. Pham, H. Qian, and R. D. Hannan. 2002. Adenoviral-directed expression of the type 1A angiotensin receptor promotes cardiomyocyte hypertrophy via transactivation of the epidermal growth factor receptor. Circ. Res. 90:135-142. [DOI] [PubMed] [Google Scholar]
- 75.Tian, R., W. Miao, M. Spindler, M. M. Javadpour, R. McKinney, J. C. Bowman, P. M. Buttrick, and J. S. Ingwall. 1999. Long-term expression of protein kinase C in adult mouse hearts improves postischemic recovery. Proc. Natl. Acad. Sci. USA 96:13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Travagli, J., M. Letourneur, J. Bertoglio, and J. Pierre. 2004. STAT6 and Ets-1 form a stable complex that modulates Socs-1 expression by interleukin-4 in keratinocytes. J. Biol. Chem. 279:35183-35192. [DOI] [PubMed] [Google Scholar]
- 77.Vassilatis, D. K., J. G. Hohmann, H. Zeng, F. Li, J. E. Ranchalis, M. T. Mortrud, A. Brown, S. S. Rodriguez, J. R. Weller, A. C. Wright, J. E. Bergmann, and G. A. Gaitanaris. 2003. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 100:4903-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ventura, C., E. Zinellu, E. Maninchedda, M. Fadda, and M. Maioli. 2003. Protein kinase C signaling transduces endorphin-primed cardiogenesis in GTR1 embryonic stem cells. Circ. Res. 92:617-622. [DOI] [PubMed] [Google Scholar]
- 79.von Harsdorf, R., J. G. Edwards, Y. T. Shen, R. K. Kudej, R. Dietz, L. A. Leinwand, B. Nadal Ginard, and S. F. Vatner. 1997. Identification of a cis-acting regulatory element conferring inducibility of the atrial natriuretic factor gene in acute pressure overload. J. Clin. Investig. 100:1294-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei, D., X. Le, L. Zheng, L. Wang, J. A. Frey, A. C. Gao, Z. Peng, S. Huang, H. Q. Xiong, J. L. Abbruzzese, and K. Xie. 2003. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene 22:319-329. [DOI] [PubMed] [Google Scholar]
- 81.Weiss, M. J., and S. H. Orkin. 1995. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. USA 92:9623-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu, M., L. Nie, S. H. Kim, and X. H. Sun. 2003. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 22:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xuan, Y. T., Y. Guo, H. Han, Y. Zhu, and R. Bolli. 2001. An essential role of the JAK-STAT pathway in ischemic preconditioning. Proc. Natl. Acad. Sci. USA 98:9050-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, E., L. Lerner, D. Besser, and J. E. Darnell, Jr. 2003. Independent and cooperative activation of chromosomal c-fos promoter by STAT3. J. Biol. Chem. 278:15794-15799. [DOI] [PubMed] [Google Scholar]
- 85.Zhang, X., M. H. Wrzeszczynska, C. M. Horvath, and J. E. Darnell, Jr. 1999. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol 19:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]