Abstract
In the developing retina, the gene encoding the β3 subunit of the neuronal nicotinic receptor, a specific marker of retinal ganglion cells, is under the direct control of the atonal homolog 5 (ATH5) basic helix-loop-helix (bHLH) transcription factor. Although quite short (143 bp in length), the β3 promoter has the remarkable capacity to discriminate between ATH5 and the other neuronal bHLH proteins expressed in the developing nervous system. We have identified three amino acids within the basic domain that confer specificity to the ATH5 protein. These residues do not mediate direct DNA binding but are required for interaction between ATH5 and chromatin-associated proteins during retina development. When misexpressed in neurons, the myogenic bHLH factor MyoD is also able to activate the β3 gene. This, however, is achieved not by binding of the protein to the promoter but by dimerization of MyoD with a partner, a process that depends not on the basic domain but on the HLH domain. By sequestering an E-box-binding protein, MyoD relieves the active repression that blocks the β3 promoter in most neurons. The mechanisms used by bHLH proteins to activate β3 thus highlight how ATH5 is selected by the β3 promoter and coordinates the derepression and transcriptional activation of the β3 gene during the specification of retinal ganglion cells.
The basic helix-loop-helix (bHLH) transcription factors have emerged as a major class of positive and negative regulators in the processes of neural cell fate specification and differentiation (1, 3, 36). bHLH proteins share extensive homology within the basic and helix-loop-helix domains that mediate, respectively, binding to the DNA consensus E-box sequence CANNTG and dimerization (17, 25). Structural and biochemical analyses show that tissue-specific bHLH proteins (class B) form functional, DNA-binding heterodimers with the ubiquitously expressed bHLH proteins (class A) (14, 19, 26). Some HLH transcription factors lack the DNA binding “b” domain (Id proteins) and act as dominant-negative regulators by forming inactive heterodimers with bHLH proteins (2). Id proteins are expressed in proliferating neural progenitors during neurogenesis, and their expression is maintained in some tissues late in development and into adulthood (reviewed in reference 35).
Structure-function studies of bHLH proteins have revealed that important groups of residues are located in the basic domain, as exemplified by the conserved myogenic motif (4, 7), which can be transferred from a myogenic factor into the basic domain of a class A protein such as E12, thereby imparting myogenic potential on the hybrid (4, 8). No such motif has been identified in the neuronal bHLH factors. Domain swapping experiments between Drosophila melanogaster atonal and achaete-scute proteins have shown that the basic domain of atonal is able to promote an atonal-specific neuronal cell fate (6) and that all differing residues are functionally important. A few studies have reported on the contributions of the HLH domain to the cell type determination process. In one such report, the HLH domain of achaete-scute was transferred into MyoD and did not affect the myogenic activity of the hybrid protein (7). The first piece of evidence to show a contribution of the HLH domain came from the demonstration that an achaete-scute/atonal hybrid protein containing both the basic and the HLH domains of atonal is more effective in promoting an atonal-specific neuronal cell fate than a hybrid protein that only contains the atonal basic domain, even though the HLH domain of atonal alone is ineffective (6). Recent studies on the atonal homolog 1 (ATH1) protein have demonstrated that the HLH domain specifies functional information in the developing neural tube (27).
In the developing retina, several bHLH proteins are expressed in a distinctive spatiotemporal order, including neurogenin 2 (Ngn2) (34), atonal homolog 5 (ATH5) (12, 22), NeuroM and NeuroD (11, 18, 30), and others (36). Among those, ATH5 has by far the smallest expression domain and is involved in the specification and differentiation of retinal ganglion cells (RGCs). ATH5 is directly involved in its own regulation and is able to activate genes, such as the β3 subunit of the neuronal nicotinic receptor and Brn3c, which specify neuronal identity traits (16, 22, 33). The relatively simple β3 promoter is sufficient to restrict reporter gene expression to RGCs (20) and is neither bound nor activated by neuronal bHLH transcription factors other than ATH5 present in the developing retina (31, 33).
We decided to determine how the β3 promoter selects ATH5 from among the other members of the bHLH protein family. β3 is coexpressed with ATH5 and NeuroM in newborn RGCs (23), and here we show that, whereas the ATH5 protein binds the β3 promoter in vivo, NeuroM does not. To unravel the mechanism of this selection, we recombined the functional domains of ATH5 into NeuroM and asked which exchange, if any, is sufficient to turn the hybrid protein into an activator of the β3 promoter. Replacing the basic domain of NeuroM with the corresponding domain of ATH5 turned the hybrid protein into a potent activator of the β3 promoter in retinal and telencephalic cells. Only three amino acids differ between the basic domains of NeuroM and ATH5, and we show that all three residues are required for full protein activity. These residues are not involved in DNA binding but mediate interactions between ATH5 and chromatin. Previously, we had shown that ectopic expression of the myogenic factor MyoD stimulates expression of the β3 gene in central neurons that never express it in physiological conditions (31). One of the three amino acids conferring specificity on ATH5 is not conserved in the basic domain of MyoD, suggesting that MyoD activates β3 by a different mechanism. Analyzing how MyoD activates the β3 promoter reveals that a mechanism of active repression is operating in neural cells that do not express β3. Recombining domains of MyoD into NeuroM, we find that the HLH domain of MyoD turns the hybrid protein into a potent activator of the β3 promoter. The ability of the MyoD HLH domain to stimulate the promoter is independent of DNA binding but does require nuclear localization of the protein. We propose a mechanism whereby MyoD, when misexpressed in neurons, dimerizes with a factor that normally represses β3 expression. A comparison of the strategies used by ATH5 and MyoD to activate the β3 promoter suggests that the spatiotemporal control of the β3 gene is affected by the interplay of positive and negative regulators. We suggest as a corollary that, in the developing nervous system, activation of a neuron-type-specific gene by a bHLH protein may have to be accompanied by sequestration or inactivation of a repressor.
MATERIALS AND METHODS
Cell cultures, transfection and chloramphenicol acetyltransferase (CAT) assays, hybrid protein constructions, and mutagenesis.
Chicken embryos were staged according to the method of Hamburger and Hamilton (10). Cells were prepared and transfected as previously described (20, 21). The data were plotted as the means of the results from at least three independent experiments. All constructs (Table 1) were prepared using standard molecular biology techniques and checked by sequencing.
TABLE 1.
Hybrid protein sequences
| Protein | Sequence segmenta
|
|||
|---|---|---|---|---|
| N terminus | Basic | Helix 1-loop-helix 2 | C terminus | |
| Ath5 | ...ESAAK | RRLAANARERRR | MQGLNTAFDRLRKVV PQWGQDKKLS KYETLQMALSYIMALTRIL | AEAER... |
| NeuroM | ...ERFRA | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | ETGQT... |
| MyoD | ...TTNAD | RRKAATMRERRR | LSKVNEAFETLKRCTSSNPNQRLPKVEILRNAIRYIEGLQALL | RDQDA... |
| N-ATH5 | ...ESAAK | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | ETGQT... |
| C-ATH5 | ...ERFRA | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | AEAER... |
| bHLH-ATH5 | ...ERFRA | RRLAANARERRR | MQGLNTAFDRLRKVV PQWGQDKKLS KYETLQMALSYIMALTRIL | ETGQT... |
| b-ATH5 | ...ERFRA | RRLAANARERRR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | ETGQT... |
| N-MyoD | ...TTNAD | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | ETGQT... |
| C-MyoD | ...ERFRA | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | RDQDA... |
| bHLH-MyoD | ...ERFRA | RRKAATMRERRR | LSKVNEAFETLKRCTSSNPNQRLPKVEILRNAIRYIEGLQALL | ETGQT... |
| b-MyoD | ...ERFRA | RRKAATMRERRR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | ETGQT... |
| HLH-MyoD | ...ERFRA | RRVKANARERTR | LSKVNEAFETLKRCTSSNPNQRLPKVEILRNAIRYIEGLQALL | ETGQT... |
| bHLH-NeuroM | ...TTNAD | RRVKANARERTR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | RDQDA... |
| b-NeuroM | ...TTNAD | RRVKANARERTR | LSKVNEAFETLKRCTSSNPNQRLPKVEILRNAIRYIEGLQALL | RDQDA... |
| HLH-NeuroM | ...TTNAD | RRKAATMRERRR | MHGLNDALDNLRRVM PCYSKTQKLS KIETLRLARNYIWALSEVL | RDQDA... |
The ATH5, NeuroM, and MyoD protein sequences are, respectively, in boldface, regular, and underlined characters.
Electroporation of genetic material in the retina and mRNA quantification by real-time PCR.
Retina electroporations were as described in reference 22. Briefly, embryonic eyes were collected at stage 22 to 23, and the pigmented epithelium was removed. Whole retinas were immersed at room temperature in about 100 μl of phosphate-buffered saline containing a green fluorescent protein (GFP) reporter plasmid and expression vectors (0.5 μg/μl of each construct). Tissues were set between two electrodes (0.5 cm apart) and submitted to 5 50-V pulses of 50- to 70-ms duration spaced 1 s apart. The electroporated tissues were cultured as floating explants for 24 h at 37°C. Electroporation efficiency was monitored by GFP-positive cells, and tissues were collected in Trizol (Gibco) for RNA extraction. cDNA was synthesized from 1 μg of RNA using random hexamer primers and Superscript II reverse transcriptase (Invitrogen). Real-time PCR using the iCycler iQ real-time PCR detection system (Bio-Rad) and a SYBR green-based kit for quantitative PCR (Bio-Rad) was performed to quantify the amounts of β3 mRNA. Amounts of mRNA were normalized to TBP levels. Primers used are listed in Table 2.
TABLE 2.
Primer sequences
| Primer | Sequence (5′-3′) |
|---|---|
| Beta3 RT fwd | CATCAGGCAGGTTGTCCAAGA |
| Beta3 RT rev | CCACCAGGAATAACCACAGGA |
| TBP RT fwd | GCCGCCCTACGCTCAAG |
| TBP RT rev | CGGCATCATCGGGCTAAA |
Antibodies.
A rabbit antibody against chicken NeuroM (no. Y09597) was raised against the bacterially expressed His-tagged domain of the protein (amino acids 176 to 298). Anti-TATA-binding protein (TBP) antibody (3G3) was kindly provided by Làszlò Tora (IGBMC, Inserm, Strasbourg). Anti-mouse fluorescein isothiocyanate-conjugated antibody and anti-FLAG antibody were purchased from Sigma (no. F5897 and M2, respectively).
ChIP.
Chromatin immunoprecipitation (ChIP) from whole neuroretinas and optic tecta was performed as previously described (33). Embryonic day 9 (E9) telencephalons were dissociated into single cells. Cells (20 × 106) were resuspended in 1 ml of culture medium (Dulbecco's modified Eagle medium, 10% fetal bovine serum) containing a cytomegalovirus promoter-GFP reporter plasmid and expression vectors (0.1 μg/μl and 0.2 μg/μl, respectively). Electroporation consisted of 5 700-V/cm pulses of 20-ms duration spaced 10 s apart. Electroporated cells were plated on 100-mm-diameter polyornithine-coated culture dishes (20 × 106 cells/dish) and cultured for 24 h at 37°C. About 25% of electroporated cells were GFP positive. Cells were collected and processed for the ChIP assay as previously described (33). The data were plotted as means of the results from at least two independent ChIP assays and three independent real-time PCR amplifications. The immunoprecipitation efficiency was calculated as the ratio of precipitated sequence over total sequence amount in input chromatin.
Immunostaining.
Cells were fixed in 100% methanol for 4 min at −20°C. After 15 min of blocking with phosphate-buffered saline containing 0.5% bovine serum albumin and 0.1% Tween 20, cells were incubated with anti-FLAG (1:1,000 dilution) antibody for 1 h at room temperature and revealed with fluorescein isothiocyanate-conjugated anti-mouse antibody (1:1,000 dilution).
Western blotting.
Proteins were analyzed by immunoblotting according to standard protocols. The membranes were immunostained with an anti-FLAG antibody (1:2,500) and anti-TBP antibody (1:2,000).
EMSA.
Protein extractions and electromobility shift assays (EMSAs) were performed essentially as previously described (31). Bacterially expressed glutathione S-transferase (GST) fusion proteins were purified using glutathione beads according to the manufacturer's instructions (Amersham). Promoter DNA fragments (143 bp in length) were end labeled by Klenow filling in the presence of [α-32P]dATP. Usually, 0.5 μg of purified GST fusion protein and 3 μg of total protein extract were used.
Structure modeling.
The structure of the bHLH domain of NeuroM was modeled using the Swiss-Model program (28). The crystal structure of the bHLH domain of MyoD (17) was used as a template. The sequence identity between the corresponding subsequences of NeuroM (residues 88 to 143) and MyoD (residues 110 to 164) is 40%.
RESULTS
The basic domain determines the selectivity of ATH5 for the β3 promoter.
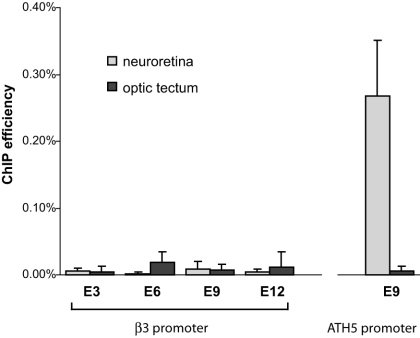
ATH5 is the only neuronal bHLH protein able to activate transcription of the β3 gene (22, 31), and ChIP results demonstrate its in vivo binding to the β3 promoter in the early retina (33). Although it is coexpressed with β3 in newborn RGCs, NeuroM is unable to activate the β3 gene (23, 31), and we wondered if that reflected an inability to bind the promoter during retina development. To address this question, we performed ChIP experiments using an antibody directed against NeuroM and chromatin prepared from E3 to E12 retinas and optic tectum. We did not detect enrichment of the β3 promoter sequences in the NeuroM immunoprecipitates at any stage of retina or optic tectum development (Fig. 1). In contrast, NeuroM specifically binds the ATH5 promoter in the developing retina, in agreement with its role as an activator of ATH5 in newborn RGCs (22; J. Hernandez and M. Ballivet, unpublished).
FIG. 1.
In vivo occupancy of the β3 and ATH5 promoters by NeuroM in the course of neuroretina (light gray) and optic tectum (dark gray) development. An antibody directed against NeuroM was used to immunoprecipitate cross-linked chromatin fragments prepared from dissected tissues. Immunoprecipitates were analyzed for the abundance of specific sequences by real-time PCR.
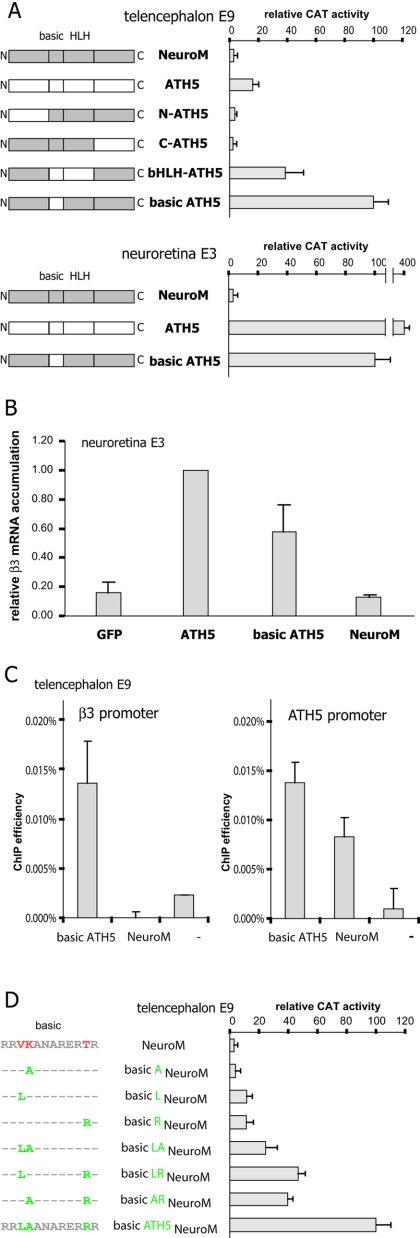
To unravel the mechanism that confers specificity toward the β3 promoter on ATH5, we generated constructs mixing the functional domains of ATH5 and NeuroM. The constructs were transfected along with the β3 promoter (20) fused to the CAT reporter gene into acutely isolated cells from an E3 retina or E9 telencephalon, where ectopic expression of ATH5 is known to activate the β3 promoter (22, 31). Whereas NeuroM is unable to transactivate the β3 promoter, the exchange of its basic domain for that of ATH5 is sufficient to turn the protein into a potent activator of the β3 promoter in both tissues (Fig. 2A). A hybrid protein containing both the basic and HLH domain of ATH5 has a significantly weaker activity in telencephalic cells, suggesting that the hybrid basic-ATH5/NeuroM and wild-type ATH5 dimerize with distinct partners to regulate the β3 promoter. To test whether the different hybrid proteins were synthesized at similar levels in transfected cells, we monitored the activity of the α1 promoter (31), which responds to both ATH5 and NeuroM, and found that it responded similarly to various hybrids (data not shown). The capacity of a hybrid protein containing the basic domain of ATH5 to activate the endogenous β3 gene was independently demonstrated by electroporating a basic-ATH5/NeuroM expression vector into retinas at stage 22 to 23. This led, like ATH5, to a robust accumulation of endogenous β3 transcripts, whereas NeuroM was ineffective (Fig. 2B). We wondered if the ability of the basic-ATH5/NeuroM protein to activate β3 reflects an ability to bind the promoter. To address this question, E9 dissociated telencephalic cells were electroporated with an expression vector encoding the flagged version of the basic-ATH5/NeuroM or NeuroM proteins. Cells were cultured for 24 h and then we performed ChIP experiments using an antibody directed against the FLAG tag. We detected enrichment of the β3 and ATH5 promoter sequences in the basic-ATH5/NeuroM immunoprecipitates (Fig. 2C). In contrast, we found that, whereas NeuroM specifically binds the ATH5 promoter in telencephalic cells, a property consistent with NeuroM's capacity to activate ATH5 expression (22; L. Matter-Sadzinski, unpublished), it does not bind the β3 promoter (Fig. 2C).
FIG. 2.
The specificity of ATH5 for the β3 promoter resides in its basic domain. (A) Expression vectors encoding NeuroM/ATH5 hybrid proteins were constructed and cotransfected with a β3 promoter-CAT reporter plasmid into retinal cells at E3 or into telencephalic cells at E9. CAT activities are normalized relative to that of the basic-ATH5/NeuroM hybrid protein. Exchange of the basic domain of NeuroM for that of ATH5 is sufficient to turn the protein into a potent activator of the β3 promoter. (B) Real-time PCR analysis of the β3 transcript in an electroporated E3 neuroretina. basic-ATH5/NeuroM protein overexpression induces expression of the endogenous gene for the β3 subunit of the nicotinic acetylcholine receptor in the retina. (C) In vivo occupancy of the β3 and ATH5 promoters by FLAG-tagged basic-ATH5/NeuroM or FLAG-tagged NeuroM in E9 telencephalic cells. An antibody directed against the FLAG peptide was used to immunoprecipitate cross-linked chromatin fragments prepared from electroporated telencephalic cells. Immunoprecipitates were analyzed for the abundance of specific sequences by real-time PCR. −, no FLAG-tagged protein. (D) There are three amino acid differences in the basic domains of ATH5 and NeuroM, all combinations of which were constructed and cotransfected into E9 telencephalic cells, as in panel A. The CAT activity of the basic-ATH5/NeuroM protein is set arbitrarily at 100. Note that the three ATH5-specific amino acids are needed to confer full activity.
The amino acids conferring specificity to the ATH5 basic domain are not involved in DNA binding.
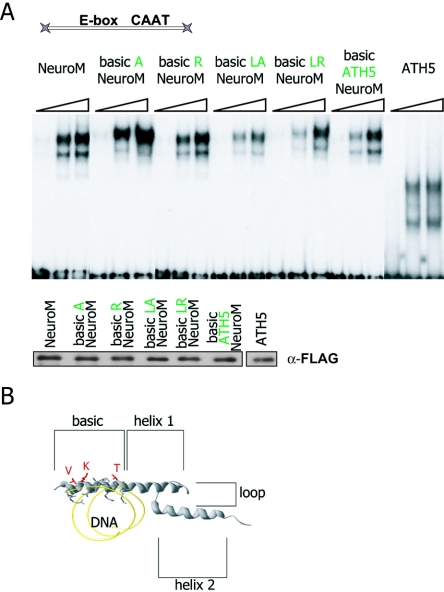
There are only three amino acid differences in the basic domains of ATH5 and NeuroM. To determine how they affect activity, we introduced each of these in the basic domain of NeuroM and tested the mutants in E9 telencephalic cells (Fig. 2D). We observed that the contributions of the three residues are additive, suggesting that they all contribute to the specificity of the ATH5 basic domain. To check if these residues influence binding to the β3 promoter, we performed EMSAs using bacterially expressed, purified GST fusions of ATH5, NeuroM, and basic domain mutants of NeuroM. Even though the hybrid proteins bearing mutations in the basic domain transactivate the β3 promoter poorly (Fig. 2D), they all efficiently bind the promoter sequence in vitro (Fig. 3A). Modeling the secondary structure of the ATH5 basic domain on the structure of MyoD suggests that the three critical amino acids have their side chains facing outward and therefore may not be directly involved in DNA binding (Fig. 3B). The residues in the basic region known to be required for the binding of bHLH proteins to E-box DNA are shared by NeuroM and ATH5, yet ATH5 interacts with the β3 promoter in vivo, whereas NeuroM does not (Fig. 1). Thus, the specificity of ATH5 for the β3 promoter may depend on context-dependent protein-protein interactions between its basic domain and unidentified partners.
FIG. 3.
The three amino acids that confer specificity to the ATH5 basic domain are not involved in DNA binding. (A) Upper panel: schematic structure of the probe (143 bp in length) used in EMSA. Central panel: the various, bacterially expressed bHLH proteins all form stable complexes with the probe. A NeuroM-DNA complex was observed in the same position whether or not the basic domain of NeuroM bore the ATH5-specific mutations. Note that due to the small size of the ATH5 protein (151 residues), its complex with DNA migrates faster than the much larger NeuroM-DNA complex. Lower panel: Western blot showing the bacterially expressed FLAG-tagged recombinant proteins used in the EMSA. α-FLAG, anti-FLAG. (B) Molecular modeling of the bHLH domain of NeuroM suggests that the side chains of the three amino acids face outward and may be available for interactions with cofactors.
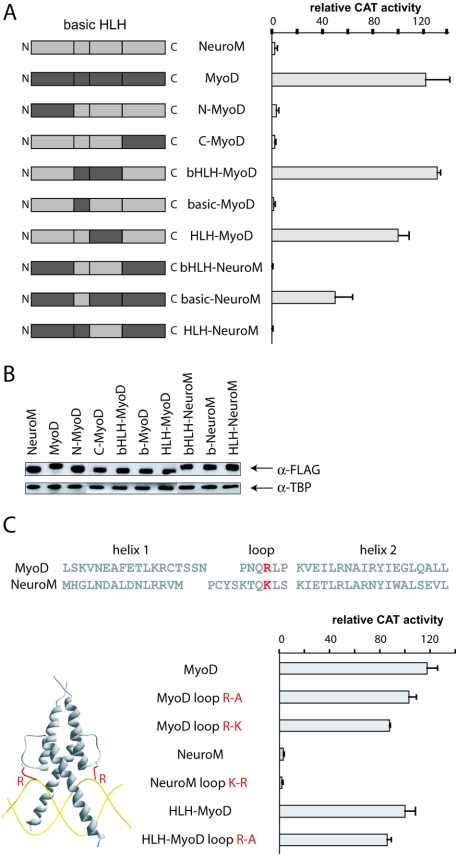
The HLH domain of MyoD is sufficient for activation.
Previous experiments had shown that the β3 promoter is silent in the telencephalon, where it can be transactivated by ectopic MyoD but not by NeuroM (31). There are three amino acid differences between the basic domains of ATH5 and MyoD, one of which contributes to the specificity of ATH5 on the β3 promoter (Table 1; Fig. 2D), suggesting that MyoD might use a different strategy than ATH5 to activate β3. To dissect the minimal domain of MyoD required for activation of the β3 promoter, we generated constructs coding for protein hybrids mixing the N-terminal, C-terminal, and bHLH domains of MyoD and NeuroM (Fig. 4A). The constructs were transfected into acutely isolated cells from E9 chick telencephalon along with the β3 promoter fused to the CAT reporter gene. We found that all hybrid proteins containing the MyoD HLH domain were able to activate the promoter (Fig. 4A). A protein that was NeuroM except for the HLH domain was active and, conversely, a protein that was MyoD except for the HLH domain was inactive, demonstrating that the HLH motif of MyoD is indispensable in this assay. The rest of the protein, including the basic domain, may come indifferently from MyoD or NeuroM. To ensure that the differences in activities between the hybrids were not due to variations in their expression levels, FLAG-tagged proteins were used for transfections and extracts were analyzed by CAT assay and by Western blotting with anti-FLAG antibody. Similar levels of expression were detected for all hybrids (Fig. 4B), and importantly, the tags did not affect the activity of the proteins.
FIG. 4.
MyoD activates the β3 promoter through its HLH domain. (A) Expression vectors encoding NeuroM/MyoD hybrid proteins were constructed and cotransfected with a β3 promoter-CAT reporter plasmid into E9 telencephalic cells. Activities upon the β3 promoter are calculated relative to the HLH-MyoD/NeuroM hybrid protein. Exchanging the HLH domain of NeuroM with the cognate domain of MyoD is sufficient to convert the protein into an activator of the β3 promoter. (B) Expression vectors encoding flagged hybrid and wild-type proteins were transfected in E9 telencephalic cells. The presence of these proteins in cell extracts was revealed by Western blotting and compared to the control protein TBP. The different hybrid and wild-type proteins are expressed at similar levels. α-FLAG, anti-FLAG; α-TBP, anti-TBP. (C) Mutations in the loop do not impair activity of the proteins. Upper panel: alignment of the bHLH domains of MyoD and NeuroM. The highlighted position of the loop is occupied by an arginine (R) in MyoD and a lysine (K) in NeuroM. Left panel: the structure of a MyoD homodimer bound to DNA indicates that the arginine is close to the phosphate backbone of DNA and might contribute to protein-DNA interactions. Right panel: telencephalic cells at E9 were cotransfected with a β3 promoter-CAT reporter plasmid and an expression vector encoding the loop mutant or the wild-type MyoD or NeuroM proteins. Data are calculated relative to the activity of the HLH-MyoD/NeuroM hybrid. The mutation has no detectable effect on the activity of the protein.
Within the HLH domain, the loop region is the most variable in length and amino acid composition among different bHLH proteins. According to the crystallographic data, some of the loop residues make contacts with the DNA backbone, for example, the basic residue at position −3 with respect to helix 2 in MyoD (17). This position is occupied by a lysine (K) in NeuroM and by an arginine (R) in MyoD (Fig. 4C). Such a K for R change has been seen to influence DNA binding in another bHLH protein, deadpan (37). We have altered those residues (R to A, R to K, K to R), and we find that none of the mutations has any significant effect on the activation properties of the proteins (Fig. 4C).
Mutations in the basic domain of MyoD do not affect activity.
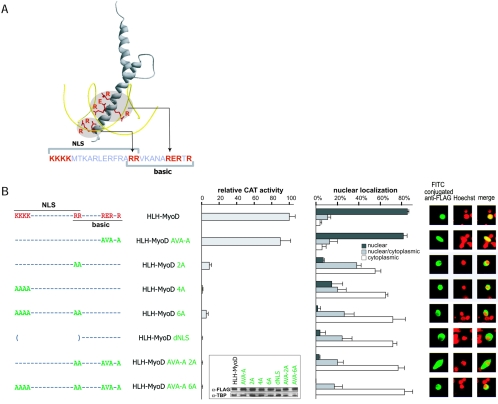
A requirement for the HLH domain of MyoD suggests a mechanism involving dimerization with a specific partner. However, we also wanted to know if the basic domain had to bind E-box DNA for the hybrid to be active. Using the available crystallographic and functional data for MyoD and other bHLH proteins (4, 5, 9, 17, 24, 32), we mutated the basic domain residues most likely involved in DNA binding (Fig. 5). Surprisingly, mutation of the RER motif, which is believed to interact directly with E-box DNA, did not affect the activity of the protein. In contrast, mutation of the RR motif, also mapped as important for interaction with DNA, abolished activity. We noticed, however, that the RR motif in NeuroM is part of a putative bipartite nuclear localization signal (NLS) that overlaps the basic domain (Fig. 5A). We introduced additional mutations in the motif and observed that all mutations affecting the putative NLS abolished activity (Fig. 5B). This raised the possibility that the NLS is indeed functional and that nuclear localization is essential for protein activity. To test this notion, we transfected the FLAG-tagged versions of the proteins and visualized their localization by immunofluorescence. We observed that all mutations in the NLS, including those in the RR motif, affected the subcellular localization of the proteins. The intact hybrid protein was found almost exclusively in the nucleus, while proteins bearing mutated versions of the NLS were excluded from nuclei. Most importantly, proteins that entered the nucleus activated the β3 promoter, even if they contained mutations affecting the residues directly involved in DNA binding (RER motif) (Fig. 5B). The correlation between nuclear localization and promoter activity was not always perfect (e.g., compare HLH-MyoD 2A with HLH-MyoD 4A) (Fig. 5B), perhaps reflecting an inaccurate discrimination between nuclear and cytoplasmic labeling in a fraction of counted cells.
FIG. 5.
Nuclear localization is needed for activity. (A) Model of the bHLH domain of NeuroM. Residues known to interact with DNA are highlighted. The RR motif is part of both the NLS and the basic domain. (B) Telencephalic cells at E9 were cotransfected with a β3 promoter-CAT reporter plasmid and expression vectors encoding proteins bearing mutations in the NLS and/or in the DNA binding domain. Data are normalized relative to the activity of the HLH-MyoD/NeuroM hybrid. In parallel experiments, expression vectors encoding flagged hybrid and wild-type proteins were transfected in E9 telencephalic cells. Inset: Western blot indicating that the mutated proteins are synthesized at similar levels. The subcellular distribution of these proteins was revealed with a fluorescent anti-FLAG antibody. Mutations in the RER motif influence neither the activity of the proteins nor their nuclear localization, whereas all mutations in the NLS domain influence both the activity and the nuclear localization of the proteins. α-FLAG, anti-FLAG; α-TBP, anti-TBP; FITC, fluorescein isothiocyanate.
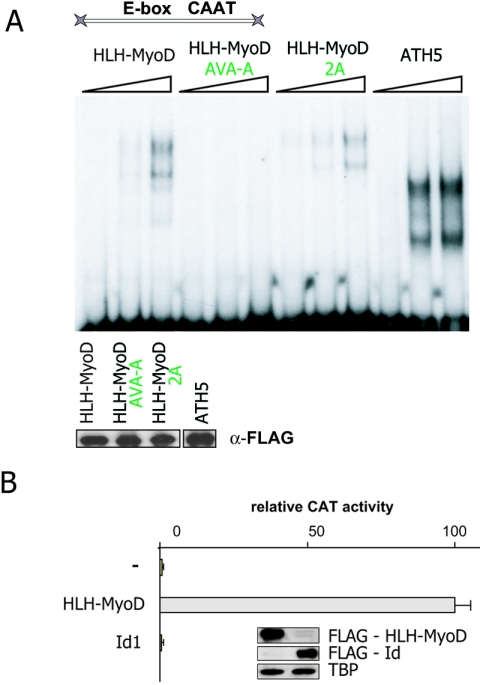
These findings suggested that DNA binding is not necessary for these hybrids to be active. To test this, we checked whether E-box DNA binding is affected in hybrid proteins bearing mutations in the basic region. EMSAs were performed using purified, bacterially expressed GST fusion proteins mixed with the radioactively end-labeled 143-bp β3 promoter (Fig. 6A). We observed that hybrid proteins harboring an intact basic domain could interact with the promoter, whereas the protein bearing mutations within the RER motif was unable to bind. Thus, DNA binding is not necessary for activation of the β3 promoter by the HLH-MyoD hybrid.
FIG. 6.
(A) MyoD activity does not require DNA binding. The bacterially expressed HLH-MyoD/NeuroM and HLH-MyoD 2A/NeuroM proteins bind the E-box in the β3 promoter, whereas the HLH-MyoD AVA-A/NeuroM does not. Mutation in the NLS domain does not prevent the protein from binding DNA but blocks activity. In contrast, mutation of the RER motif blocks DNA binding but not activity (Fig. 5). Hence, activity depends on nuclear localization but not on DNA binding. Lower panel: Western blot showing the bacterially expressed FLAG-tagged recombinant proteins used in the EMSA. α-FLAG, anti-FLAG. (B) Telencephalic cells at E9 were cotransfected with a β3 promoter-CAT reporter plasmid and expression vectors encoding the Id or the HLH-MyoD/NeuroM proteins. Overexpression of Id does not activate the β3 promoter. Inset: Western blot indicating that Id and MyoD are synthesized at similar levels.
Next, we asked if transfection by Id1, a protein naturally lacking a basic domain, would lead to the activation of the β3 promoter. As shown in Fig. 6B, the Id1 protein is not able to activate the β3 promoter, suggesting that the interaction mediated by the MyoD HLH domain is specific.
A factor bound to the E-box prevents other proteins from binding to the β3 promoter.
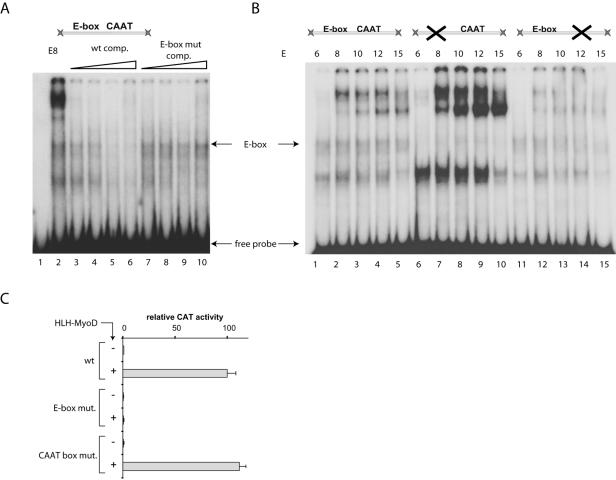
We hypothesize that dimerization of the HLH-MyoD proteins with a specific partner leads to activation of the β3 promoter, which is normally silent in the telencephalon. We envisage a mechanism involving the sequestration of a repressor of the β3 promoter. To detect the repressor, we tested whether protein extracts from various stages of telencephalon development were able to bind the E-box in the β3 promoter. Competition by unlabeled wild-type and mutant E-box DNA allowed us to detect the E-box-specific complex (Fig. 7A).
FIG. 7.
Proteins isolated from telencephalon specifically bind the E-box in the β3 promoter. (A) Proteins isolated from the telencephalon at E8 bind the wild-type β3 promoter fragment encompassing the E-box and CAAT box. Competition with increasing amounts of unlabeled wild-type (lanes 3 to 6) and mutated E-box (lanes 7 to 10) probes reveals a complex that is specifically bound to the E-box. Lane 1, no proteins; lane 2, no competitor. wt, wild type; comp., competitor; mut., mutant. (B) Proteins isolated from the telencephalon at E6 to E15 bind both the wild type and the mutated β3 promoter fragment. When the E-box is mutated (lanes 6 to 10) the probe is not bound by the E-box-specific complex, but the assembly of other complexes is strongly enhanced. When the CAAT box is mutated (lanes 11 to 15), binding is much decreased except for the E-box-specific complex. (C) Telencephalic cells at E9 were cotransfected with a mutated or wild-type β3 promoter-CAT reporter plasmid and an expression vector encoding the HLH-MyoD/NeuroM hybrid protein. Data are normalized relative to the activity of the hybrid protein upon the wild-type β3 promoter. The protein does activate the promoter in the absence of a CAAT box but not in the absence of the E-box. +, present; −, absent.
EMSA using protein extracts derived from telencephalon at different stages of development (Fig. 7B) showed that the pattern of bands does not change during development, even though the relative intensity of the signals varies with time. When the same experiment was performed with a DNA probe bearing a mutant E-box, the disappearance of the E-box-specific band was accompanied by a strong enhancement of the bands corresponding to the other complexes. In contrast, a mutation in the CAAT box had no such effect (Fig. 7B). These findings are indicative of an as yet unidentified E-box binding activity whose absence allows for a more pronounced promoter occupation, in agreement with the repressor model.
In telencephalon cotransfections, the CAAT box mutant had full activity, whereas a mutant E-box was completely silent (Fig. 7C). This result is in sharp contrast to what is seen when transfecting retina, where both CAAT and E-box mutants lose all activity (31), demonstrating that the β3 promoter is activated by different means in the two tissues.
DISCUSSION
The mechanism by which neuronal bHLH transcription factors select their targets in the developing nervous system has remained elusive. Our study highlights how the interplay between activators and repressors determines the activity of a neuron-specific promoter during development. That β3 is expressed only in RGCs reflects the high selectivity of the β3 promoter for the bHLH protein ATH5 and also appears to result from a mechanism of active repression operating on the promoter in most other neuronal cell types. We describe a mechanism enabling the β3 promoter to discriminate between ATH5 and other neuronal bHLH proteins expressed in the developing nervous system and provide evidence suggesting that ATH5 coordinates the derepression and the activation of the β3 promoter, a property that involves three amino acids of its basic domain.
Three amino acids confer stringent specificity to ATH5.
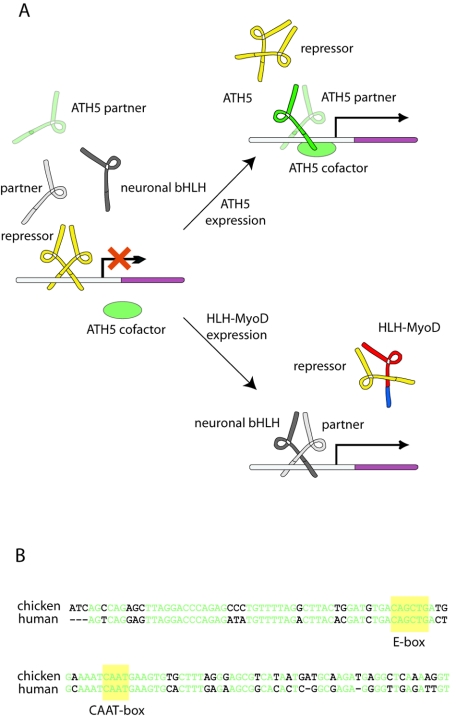
Because of its relatively simple organization, the β3 promoter is a convenient model system in which to elucidate how stringent neuronal type specificity is achieved in the nervous system. A single E-box of sequence CAGCTG mediates the neuronal specificity of the β3 promoter. In the ATH5 promoter, ATH5 interacts with an E-box of sequence CAGATG and mutating the two central bases to GC abolishes promoter activity, yet overexpression of ATH5 compensates for the loss of activity due to the mutation (J. Hernandez and M. Ballivet, unpublished). That E-boxes with different identities mediate the effect of ATH5 on the β3 and ATH5 promoters suggests that sequences in the vicinity of these elements play a crucial role for the recruitment of ATH5 and its partners. Moreover, the β3 promoter and, in particular, the sequences flanking its E-box are conserved in species as distant as chickens and humans (Fig. 8B), suggesting that the ATH5 protein is cooperating with other, conserved transcription factors to regulate the β3 gene. We constructed hybrid proteins to identify the protein domains conferring specificity upon ATH5. NeuroM was selected for this analysis because, even though it is coexpressed with β3 in newborn retinal ganglion cells, it is not involved in the regulation of β3 (23, 31). Congruent with these results, we show here that NeuroM does not bind the β3 promoter in the developing retina (Fig. 1). The hybrid proteins were tested both in the retina at the stage when the ATH5 protein is a strong activator of the β3 promoter and in the telencephalon, a tissue in which β3 is not expressed (Fig. 2A). We found that exchanging the basic domain of NeuroM with the corresponding domain of ATH5 was sufficient to convert the hybrid protein into a potent activator of the β3 promoter in both tissues, indicating that the basic domain of ATH5 is sufficient to confer specificity upon the hybrid protein. In agreement with this idea, overexpression of the hybrid protein in the early retina leads to a robust accumulation of β3 mRNA (Fig. 2B). Only three amino acids differ between the basic domains of ATH5 and NeuroM, and mutagenesis demonstrates that all three are required to impart the full properties of the ATH5 basic domain (Fig. 2D). Molecular modeling (Fig. 3B) indicates that the side chains of these amino acids are oriented outward, and EMSA confirms that they do not affect DNA binding (Fig. 3A). The absence of NeuroM on the β3 promoter in vivo suggests, however, that these amino acids are probably required for ATH5 to interact with chromatin-associated proteins on the β3 promoter.
FIG. 8.
(A) Two possible strategies for the regulation of the β3 promoter. In cells that do not express the β3 gene, a repressor bound to the E-box inhibits promoter activity. If expressed, ATH5 and its dimerization partner displace the repressor and activate the promoter. The specific interaction between ATH5 and the β3 promoter is mediated by a cofactor binding to the basic domain of ATH5. When misexpressed in neuronal cells, the HLH-MyoD hybrid protein specifically dimerizes with the repressor and sequesters it from the promoter. In the absence of repressor, other neuronal bHLH proteins can bind and activate the promoter. (B) Alignment of chicken and human β3 promoter sequences. The E-box and CAAT box are highlighted, as are conserved residues.
Experiments have shown that it is not the basic region but the HLH domain that specifies the necessary information for the functions of ATH1 in neurogenesis (27). Thus, although ATH1 and ATH5 are highly related members of the atonal group, they may use different strategies to effect their functions. Congruent with our findings, the atonal and neurogenin proteins select neural progenitors by distinct mechanisms determined by three amino acids of their basic domains (29), two of which occupy the same positions as those shown here to confer differential activity on ATH5 and NeuroM.
MyoD regulates the β3 promoter by a mechanism independent of DNA binding.
ATH5 and MyoD are the only two bHLH proteins found to activate β3 expression in neural cells. Even though two of three amino acids conferring specificity on ATH5 occupy the same position in the basic domain of MyoD (Table 1), we have excluded a direct activation of the β3 promoter by the basic domain of MyoD (Fig. 4A). This reinforces the notion that all three amino acids found in the basic domain of ATH5 are required for the direct activation of the β3 promoter. It also reinforced the idea that specific interactions between the basic domain and chromatin-associated proteins are required for activation of β3 transcription. Moreover, a comparison of the functional properties of MyoD and ATH5 has unexpectedly revealed that the capacity of a bHLH protein to activate a neural gene might result not only from interactions with a promoter but also from an ability to sequester agents that would otherwise prevent expression of the gene. To dissect the mechanism of MyoD-mediated activation in neurons, we have prepared hybrid proteins containing domains of MyoD and NeuroM in different combinations. We demonstrate that exchange of the HLH domain of NeuroM with the corresponding domain of MyoD is sufficient to convert the hybrid protein into a potent activator of the β3 promoter (Fig. 4A). A hybrid additionally containing the basic domain of MyoD has higher activity in our assay, but the basic domain alone is unable to convert the hybrid into an active protein. Residues known to be crucial for E-box binding by bHLH proteins have been identified (5, 9, 17, 32), and mutation of the very well conserved RER motif renders the basic domain incapable of DNA binding (4, 7, 24). Surprisingly, we found that mutation of this motif in the HLH-MyoD hybrid did not alter activity (Fig. 5B), although it did abolish DNA binding (Fig. 6A). The well-conserved RR motif is also known to influence DNA binding of bHLH protein. When this motif was mutated, the hybrid protein lost its capacity to transactivate the β3 promoter (Fig. 5B), although its ability to bind DNA in vitro was maintained (Fig. 6A). We noticed, however, that the RR motif in NeuroM is part of a bipartite NLS that overlaps the basic domain (Fig. 5A) and that mutation in any part of this signal profoundly affects the nuclear localization of the hybrid proteins (Fig. 5B). Taken together, our results show that activation of the β3 promoter by the HLH-MyoD hybrid requires nuclear localization but not DNA binding. Id proteins do not contain a basic domain and dimerize with E proteins (class A), thus sequestering them and preventing them from forming functional heterodimers with tissue-specific bHLH proteins (2). Given the higher affinity of Id for E proteins than for tissue-specific factors (13), overexpression of E proteins might perhaps be sufficient to circumvent the inhibitory effect of Id. Overexpression of E12 in neural cells does not lead, however, to the activation of the β3 promoter (31), indicating that Id proteins do not dimerize with the putative proteins that inhibit the β3 promoter (Fig. 6B). Our experiments suggest an activation mechanism that involves dimerization of the HLH-MyoD hybrid with a specific nuclear protein. Such a dimerization might lead to the sequestration of a repressor protein acting either through DNA binding or through protein-protein interaction with the activator (Fig. 8A). To distinguish between those two possibilities, we looked for E-box binding activity in neural tissues that do not express β3. EMSAs with nuclear extracts from telencephalon and optic tectum revealed numerous interactions between protein complexes and the β3 promoter (Fig. 7A and B and data not shown). When the E-box is mutated, changes in the EMSA pattern suggest that the protein complex associated with the E-box disappears, whereas binding with sequences located in the vicinity of the mutated E-box is enhanced (Fig. 7B), as if complexes bound to the E-box did diminish the binding of other proteins to the remainder of the promoter. Finally, we have shown that the E-box is needed for activation of the β3 promoter when the HLH-MyoD protein relieves active repression, whereas the CAAT box is dispensable.
ATH5 coordinates derepression with the activation of β3.
Active gene repression has been well documented in other parts of the nervous system, such as the spinal cord. During the specification of motor neuron subtypes, cascades of transcription factors ensure proper cell type development. Most of the transcription factors implicated in this process inhibit transcription, suggesting that repression mechanisms are essential for cell fate specification in the developing spinal cord (15). We propose that in tissues where the β3 gene is not expressed, active repression is imposed on the promoter as a result of competition between putative activators and excess repressors (Fig. 8A). The overexpressed HLH-MyoD protein would displace the equilibrium by dimerization with the repressor protein, thus enabling putative activators to bind the promoter. This model suggests that strict specificity of the promoter is achieved not only by the presence of the appropriate activator but also by the balance between repressors and activators. In the early retina, ATH5 is expressed at a low rate in about one-third of retinal cells, yet β3 is not expressed in these cells. Whereas ATH5 already interacts with its own promoter, it does not yet bind the β3 promoter in this subpopulation of progenitors (33). Overexpression of ATH5 at early stages of development leads to the precocious activation of the β3 gene. That ATH5 should reach a threshold level suggests that it has to sequester an inhibitor before it can bind the promoter and stimulate transcription. Congruent with this idea, the in vivo binding of ATH5 to the β3 promoter correlates very well with the short period of development when ATH5 is upregulated and able to stimulate β3 transcription (33).
ATH5 may overcome a repression and activate transcription through its basic domain (Fig. 8A). We suppose that this domain interacts with specific cofactors that mediate chromatin binding and β3 activation. Moreover, the requirement for a CAAT box properly spaced from the E-box (31) suggests that the activation complex includes factors interacting with the CAAT box. The role of the complex appears to be both to displace a repressor and to trigger transcription. In contrast, the sequestration of a repressor by MyoD may lead to activation of β3 by a different pathway. This view is supported by our data showing that factors interacting with the CAAT box appear not to be required when β3 is activated by MyoD (Fig. 7C).
In sum, we highlight a mechanism by which the β3 promoter selects ATH5 among the other neuronal bHLH proteins expressed in the developing retina. This selection is mediated by highly specific interactions between the basic domain of ATH5 and the β3 promoter during the narrow time window when ATH5 is upregulated and the majority of retinal ganglion cells are specified. The requirement for coordinated basic domain-mediated binding of ATH5 to DNA and to chromatin-associated proteins is likely to be prototypic of bHLH target selection in development.
Acknowledgments
We thank Michèle Geindre and Christine Alliod for technical assistance. We are indebted to Làszlò Tora for a generous gift of anti-TBP antibody.
The Swiss National Science Foundation, the Eye Hospital Jules Gonin, the Marguerite Vuilleumier Foundation, the Pro Visu Foundation and the State of Geneva support our laboratories.
REFERENCES
- 1.Anderson, D. J., A. Groves, L. Lo, Q. Ma, M. Rao, N. M. Shah, and L. Sommer. 1997. Cell lineage determination and the control of neuronal identity in the neural crest. Cold Spring Harbor Symp. Quant. Biol. 62:493-504. [PubMed] [Google Scholar]
- 2.Benezra, R., R. L. Davis, D. Lockshon, D. L. Turner, and H. Weintraub. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49-59. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3:517-530. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, T. J., T. Chakraborty, and E. N. Olson. 1991. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl. Acad. Sci. USA 88:5675-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownlie, P., T. Ceska, M. Lamers, C. Romier, G. Stier, H. Teo, and D. Suck. 1997. The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure 5:509-520. [DOI] [PubMed] [Google Scholar]
- 6.Chien, C. T., C. D. Hsiao, L. Y. Jan, and Y. N. Jan. 1996. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl. Acad. Sci. USA 93:13239-13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, R. L., P. F. Cheng, A. B. Lassar, and H. Weintraub. 1990. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60:733-746. [DOI] [PubMed] [Google Scholar]
- 8.Davis, R. L., and H. Weintraub. 1992. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science 256:1027-1030. [DOI] [PubMed] [Google Scholar]
- 9.Ellenberger, T., D. Fass, M. Arnaud, and S. C. Harrison. 1994. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8:970-980. [DOI] [PubMed] [Google Scholar]
- 10.Hamburger, V., and H. L. Hamilton. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88:49-92. [PubMed] [Google Scholar]
- 11.Inoue, T., M. Hojo, Y. Bessho, Y. Tano, J. E. Lee, and R. Kageyama. 2002. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development 129:831-842. [DOI] [PubMed] [Google Scholar]
- 12.Kanekar, S., M. Perron, R. Dorsky, W. A. Harris, L. Y. Jan, Y. N. Jan, and M. L. Vetter. 1997. Xath5 participates in a network of bHLH genes in the developing Xenopus retina. Neuron 19:981-994. [DOI] [PubMed] [Google Scholar]
- 13.Langlands, K., X. Yin, G. Anand, and E. V. Prochownik. 1997. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J. Biol. Chem. 272:19785-19793. [DOI] [PubMed] [Google Scholar]
- 14.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. K., and S. L. Pfaff. 2001. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 4(Suppl.):1183-1191. [DOI] [PubMed] [Google Scholar]
- 16.Liu, W., Z. Mo, and M. Xiang. 2001. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl. Acad. Sci. USA 98:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, P. C., M. A. Rould, H. Weintraub, and C. O. Pabo. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77:451-459. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt, T., R. Ashery-Padan, N. Andrejewski, R. Scardigli, F. Guillemot, and P. Gruss. 2001. Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105:43-55. [DOI] [PubMed] [Google Scholar]
- 19.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matter, J. M., L. Matter-Sadzinski, and M. Ballivet. 1995. Activity of the beta 3 nicotinic receptor promoter is a marker of neuron fate determination during retina development. J. Neurosci. 15:5919-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matter-Sadzinski, L., M. C. Hernandez, T. Roztocil, M. Ballivet, and J. M. Matter. 1992. Neuronal specificity of the alpha 7 nicotinic acetylcholine receptor promoter develops during morphogenesis of the central nervous system. EMBO J. 11:4529-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matter-Sadzinski, L., J. M. Matter, M. T. Ong, J. Hernandez, and M. Ballivet. 2001. Specification of neurotransmitter receptor identity in developing retina: the chick ATH5 promoter integrates the positive and negative effects of several bHLH proteins. Development 128:217-231. [DOI] [PubMed] [Google Scholar]
- 23.Matter-Sadzinski, L., M. Puzianowska-Kuznicka, J. Hernandez, M. Ballivet, and J. M. Matter. 2005. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development 132:3907-3921. [DOI] [PubMed] [Google Scholar]
- 24.McFadden, D. G., J. McAnally, J. A. Richardson, J. Charite, and E. N. Olson. 2002. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129:3077-3088. [DOI] [PubMed] [Google Scholar]
- 25.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 26.Nair, S. K., and S. K. Burley. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112:193-205. [DOI] [PubMed] [Google Scholar]
- 27.Nakada, Y., T. L. Hunsaker, R. M. Henke, and J. E. Johnson. 2004. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131:1319-1330. [DOI] [PubMed] [Google Scholar]
- 28.Peitsch, M. C. 1996. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24: 274-279. [DOI] [PubMed] [Google Scholar]
- 29.Quan, X. J., T. Denayer, J. Yan, H. Jafar-Nejad, A. Philippi, O. Lichtarge, K. Vleminckx, and B. A. Hassan. 2004. Evolution of neural precursor selection: functional divergence of proneural proteins. Development 131: 1679-1689. [DOI] [PubMed] [Google Scholar]
- 30.Roztocil, T., L. Matter-Sadzinski, C. Alliod, M. Ballivet, and J. M. Matter. 1997. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development 124:3263-3272. [DOI] [PubMed] [Google Scholar]
- 31.Roztocil, T., L. Matter-Sadzinski, M. Gomez, M. Ballivet, and J. M. Matter. 1998. Functional properties of the neuronal nicotinic acetylcholine receptor beta3 promoter in the developing central nervous system. J. Biol. Chem. 273:15131-15137. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu, T., A. Toumoto, K. Ihara, M. Shimizu, Y. Kyogoku, N. Ogawa, Y. Oshima, and T. Hakoshima. 1997. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 16:4689-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skowronska-Krawczyk, D., M. Ballivet, B. D. Dynlacht, and J. M. Matter. 2004. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development 131:4447-4454. [DOI] [PubMed] [Google Scholar]
- 34.Sommer, L., Q. Ma, and D. J. Anderson. 1996. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8:221-241. [DOI] [PubMed] [Google Scholar]
- 35.Tzeng, S. F. 2003. Inhibitors of DNA binding in neural cell proliferation and differentiation. Neurochem. Res. 28:45-52. [DOI] [PubMed] [Google Scholar]
- 36.Vetter, M. L., and N. L. Brown. 2001. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin. Cell Dev. Biol. 12:491-498. [DOI] [PubMed] [Google Scholar]
- 37.Winston, R. L., and J. M. Gottesfeld. 2000. Rapid identification of key amino-acid-DNA contacts through combinatorial peptide synthesis. Chem. Biol. 7:245-251. [DOI] [PubMed] [Google Scholar]