Abstract
Interleukin-21 (IL-21) plays important roles in regulating the immune response. IL-21 receptor (IL-21R) mRNA is expressed at a low level in human resting T cells but is rapidly induced by mitogenic stimulation. We now investigate the basis for IL21R gene regulation in T cells. We found that the −80 to −20 region critically regulates IL-21R promoter activity and corresponds to a major DNase I-hypersensitive site. Electrophoretic mobility shift assays, DNA affinity chromatography followed by mass spectrometry, and chromatin immunoprecipitation assays revealed that Sp1 binds to this region in vitro and in vivo. Moreover, mutation of the Sp1 motif markedly reduced IL-21R promoter activity, and Sp1 small interfering RNAs effectively diminished IL-21R expression in activated T cells. Interestingly, upon T-cell receptor (TCR) stimulation, T cells increased IL-21R expression and Sp1 protein levels while decreasing Sp1 phosphorylation. Moreover, phosphatase inhibitors that increased phosphorylation of Sp1 diminished IL-21R transcription. These data indicate that TCR-induced IL-21R expression is driven by TCR-mediated augmentation of Sp1 protein levels and may partly depend on the dephosphorylation of Sp1.
The interleukin-21 receptor (IL-21R) is a type I cytokine receptor that is selectively expressed in lymphoid tissues, particularly by T, B, and NK cells (35, 38). IL-21R is most similar to the IL-2 receptor β chain and the IL-4 receptor α chain (35, 38, 39), and correspondingly, IL-21 is most similar to IL-2, IL-4, and IL-15 (38). Like IL-2, IL-4, IL-7, IL-9, and IL-15, the receptor for IL-21 also contains the common cytokine receptor γ chain (γc), and IL-21 signals in part through the activation of Jak1 and Jak3 (2, 11, 24, 35).
IL-21 is produced by activated CD4+ T cells (38, 39), and corresponding to the expression of its receptor, IL-21 has actions on T, B, and NK cells. It enhances the proliferation of both anti-CD3 activated thymocytes and peripheral T cells (16, 38), and it also acts synergistically with IL-7 or IL-15 to enhance CD8+ T-cell proliferation (38, 57). IL-21 can promote NK cell maturation from bone marrow progenitors and activate the cytolytic activity of peripheral NK cells, and it can reduce IL-15-induced expansion of resting NK cells (16, 38), although IL-21R−/− mice have normal NK cell development (37). IL-21 can augment B-cell death in vitro (31, 36) and in vivo (36), but it also promotes the differentiation of B cells into postswitch and plasma cells and is critical for antigen-specific immunoglobulin (Ig) production in vivo (36, 37). IL-21R−/− mice exhibited normal lymphocyte development but abnormal Ig production, with reduced serum levels of IgG1 and IgG2b but elevated IgE in response to antigen (37). Correspondingly, IL-21 can inhibit antigen-induced IgE production (48). IL-21R−/− IL-4−/− double knockout mice exhibit a severely impaired IgG response as well as diminished IgE levels, indicating that these two cytokines cooperatively regulate Ig production (37). In addition to its physiological roles in lymphoid biology, IL-21 has antitumor actions as well that correlate with its ability to activate NK and cytotoxic CD8+ T cells and to enhance gamma interferon production by these cells (29, 47, 52, 57). Given the range of actions of IL-21 and its importance in regulating the immune system, we investigated the molecular mechanism involved in IL21R gene regulation.
MATERIALS AND METHODS
Cell culture.
Human peripheral blood (PB) lymphocytes were isolated from normal donors by Ficoll density gradient centrifugation. T cells were purified by negative selection (Pan T-cell isolation kit, Miltenyi Biotec, Auburn, CA) and cultured at 37°C in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin G, and 100 μg/ml streptomycin. Jurkat E6.1 cells (American Type Culture Collection, Manassas, VA) were cultured in the same medium. Molt-3 cells (American Type Culture Collection) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM GlutaMAX-1, 1 mM sodium pyruvate, 100 U/ml penicillin G, and 100 μg/ml streptomycin.
Real-time PCR analysis.
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA). First-strand cDNA was made from 2 μg of total RNA using random hexamers and Omniscript reverse transcriptase (QIAGEN, Valencia, CA), following the manufacturer's suggested protocol. Quantitation of specific mRNAs and 18S rRNA (as a control) was performed by real-time PCR using the 7900H sequence detection system (Applied Biosystems, Foster City, CA). cDNAs were amplified using the TaqMan universal PCR master mix (Applied Biosystems). The primers and probes used to detect human IL-21R, Sp1, and 18S rRNA are as follows: IL-21R forward primer (5′-TGTGGAGGCTATGGA AGAAGATATG-3′), reverse primer (5′-GTGCACCCACCCATTTCTTG-3′), and probe (5′-6-carboxyfluorescein [FAM]-CGGTTCTTCATGCCCCTGTAA AGGG-6-carboxytetramethylrhodamine [TAMRA]-3′); Sp1 forward primer (5′-CAGCTTCAGGCTGTTCCAAACT-3′), reverse primer (5′-CTGCCAACT GACCTGTCCATT-3′), and probe (5′-FAM-TGGTCCCATCATCATCCG GACACC-TAMRA-3′); and 18S rRNA forward primer (5′-TTCGGAACT GAGGCATGAT-3′), reverse primer (5′-TTTCGCTCTGGTCCGTCTTG-3′), and probe (5′-FAM-CGCCGCTAGAGGTGAAATTCTTGGACC-TAMRA-3′).
5′ RACE.
Total RNA was isolated from activated human PB T cells, and rapid amplification of 5′ cDNA ends (5′ RACE) was performed with a GeneRACER kit (Invitrogen). A “RACER 5′ primer” complementary to the RNA oligonucleotide sequence and a gene-specific primer (GSP) complementary to IL-21R cDNA (either GSP1, 5′-GAGGAGGGAGACACTTCTTGAGT-3′, or GSP2, 5′-ACTGTCCTGAGCAGGTCACAGTC-3′) were used in the PCR. The “RACER 5′ nested primer” and either IL-21R GSP2 or GSP3 (5′-CGGCTT GATGCTCTCAGCCAGGA-3′) were then used in a second, nested PCR. The PCR product was subcloned into pCR4-TOPO vector with a TOPO TA cloning kit (Invitrogen), and the nucleotide sequence was determined.
DNase I hypersensitivity assay.
DNase I hypersensitivity assays were performed as described previously (17). Briefly, Molt-3 cells were activated with 10 ng/ml of phorbol 12-myristate 13-acetate (PMA) plus 1 μg/ml of ionomycin for 4 h at 37°C. The nuclear pellet of the cells was digested with DNase I. The purified genomic DNA was digested with NdeI (−7617 and +6809) and BglI (−8118 and +3147) and Southern blotted (Nytran Plus, Schleicher & Schuell, Keene, NH) using random primer-labeled probe 1 (+6331 to +6754) and probe 2 (+1051 to +1352) and Quickhyb solution (Stratagene, La Jolla, CA).
Restriction enzyme accessibility assay.
A restriction enzyme accessibility assay was performed as described previously (8). Briefly, human PB T cells were activated with plate-bound anti-CD3 (2 μg/ml) and 1 μg/ml of anti-CD28 for 4 h, followed by digestion with 5 U of BstEII for 10 min at 37°C. Two micrograms of the DNA was digested with 10 U of StuI for 2 h at 37°C. Following blunting of the DNA ends with Klenow, the DNA was ligated to a universal linker (34). The cleavage sites were detected by PCR (34 cycles) with linker primer and a downstream antisense GSP (from +58 to +80, 5′-GAGCTTACGGTCACTCAG CAGAG-3′). 32P-labeled downstream antisense primer (from +39 to +62, 5′-CAGAGAGACGCCAGTGGGTCTGTC-3′) was added to the PCR for the last two cycles, and the PCR products were separated on polyacrylamide gels, visualized by autoradiography, and quantitated using a PhosphorImager.
IL-21R luciferase reporter assays.
The 5′ regulatory region of the IL21R gene from −2300 to +350 was subcloned between the BglII and XhoI polylinker sites in the pREP4 luciferase reporter vector (27). Site-directed mutagenesis or deletion of the IL-21R −61 to −32 region were performed using a QuickChange kit (Stratagene).
Molt-3 cells were transfected using DEAE-dextran with 10 μg of the reporter construct plasmid and 40 ng of pREP7 renilla luciferase vector as a transfection efficiency control. After 24 h, cells were stimulated with medium alone or PMA and ionomycin (PI). Luciferase activity was measured (18) after 18 h.
Transient transfection of normal murine splenic T cells was performed by electroporation, as described previously (18). Preactivated murine splenic T cells were mixed with 30 μg of the IL-21R promoter reporter plasmid and 500 ng of pREP7 renilla luciferase vector. After electroporation, cells were cultured for 3 h, and then half of the cells were stimulated with anti-CD3 plus anti-CD28. After 18 h, the cells were harvested and analyzed for luciferase activity.
EMSAs and Western blotting.
Nuclear extracts were prepared as described previously (25) from untreated naïve T cells or PI-activated T cells, and 10 μg of nuclear extracts and 20,000 cpm of 32P-labeled probe were used in electrophoretic mobility shift assays (EMSAs). For competition and supershift assays, before adding the labeled probe, either excess unlabeled oligonucleotides or antibodies to Sp1 or Sp3 (Upstate Biotechnology, Lake Placid, NY) or Sp2 or Sp4 (Santa Cruz Biotechnology, Santa Cruz, CA) were preincubated with the nuclear extracts. Recombinant Sp1 was from Sigma-Aldrich (St. Louis, MO), and Sp3 was from Upstate Biotechnology.
Nuclear extracts (15 μg) were resolved on 8% Novex Tris-glycine gel or 8% NuPAGE Tris-acetate gel (Invitrogen) and blotted onto Immobilon-P membranes (Millipore Corporation, Bedford, MA). The membranes were then blotted with anti-γ-tubulin (Santa Cruz Biotechnology) as a control or with anti-Sp1 and anti-Sp3 and developed by SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
DNA affinity chromatography and mass spectrometry.
DNA affinity chromatography was performed as described previously (55). Briefly, the protein binding site was concatemerized by a self-priming PCR technique using a wild-type sequence (5′-bio-CCCAGCTGCGGGTGGGCGGGGCTGGCGGGG-3′) or a similar mutant oligonucleotide (5′-biotin-CCCAGCTGCGGGGTTGCGGGG CTGGCGGGG-3′). Six milligrams of nuclear extract from PI-activated human PB T cells was incubated with streptavidin-conjugated biotinylated PCR product. The bound proteins were eluted with binding buffer containing 600 mM KCl and then dialyzed at 4°C overnight. The final eluate was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver stained. The bands enriched with wild-type oligonucleotides were identified by liquid chromatography mass spectrometry (LC-MS) and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) (Lerner Research Institute, Mass Spectrometry Laboratory for Protein Sequencing, the Cleveland Clinic Foundation, Cleveland, Ohio).
Small interfering RNA (siRNA)-based inhibition.
Purified human PB T cells were transfected using the human T cell Nucleofector kit (Amaxa, Gaithersburg, MD) with 100 nM siSp1, siSp3 (siGENOME SMARTpool siRNA, Dharmacon, Lafayette, CO), or siControl (Dharmacon). Since the transfection efficiency in human primary T cells was only 30 to 40%, we cotransfected the cells with pEYFP-N1 (BD Clontech, Franklin Lakes, NJ), which expresses enhanced yellow fluorescent protein (YFP). After culturing in antibiotic-free medium for 36 h, cells positive for YFP were isolated by cell sorting and cultured in medium alone or stimulated with anti-CD3 plus anti-CD28. After 2 h of incubation, total RNA was extracted and analyzed by quantitative real-time PCR.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (27). Human PB T cells were activated with anti-CD3 plus anti-CD28 for 2 h, followed by cross-linking with formaldehyde. Immunoprecipitations were performed with 6 μg of anti-Sp1 (Upstate Biotechnology) or normal rabbit IgG (Santa Cruz Biotechnology) conjugated to Dynabeads protein A (Dynal Biotech, Brown Deer, WI). Immunoprecipitated DNA samples were analyzed by real-time PCR for IL-21R promoter or β-actin as a control. The sequences of the primers and TaqMan probes were as follows: IL-21R promoter forward primer, 5′-GGTCCCTAAGAGGGAAGTGTCA-3′; reverse primer, 5′-GCACCCACTGTCACCAAAGG-3′; probe, 5′-FAM-CCCGATGGCCCC AAATGTCTTACTTG-TAMRA-3′; β-actin forward primer, 5′-TCCACCTTCCAGCAGATGTG-3′; reverse primer, 5′-GCAACTAAGTCATAGTCCGCC TAGA-3′; and probe, 5′-FAM-AGCAGGAGTATGACGAGTCCGGCCC- TAMRA-3′.
RESULTS
IL-21R expression is induced by TCR stimulation.
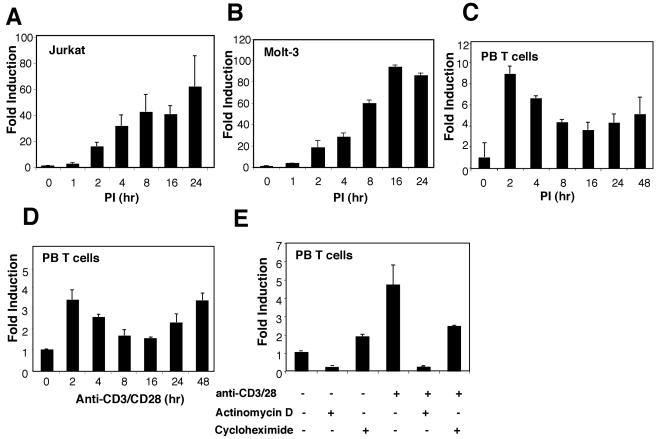
We initially investigated the regulation of IL-21R expression in human T cells using real-time PCR. IL-21R mRNA was not expressed in unstimulated Jurkat or Molt-3 cells, but levels gradually increased following stimulation with PI (Fig. 1A and B). IL-21R mRNA levels in primary human peripheral T cells were also induced by PI (Fig. 1C) or anti-CD3 plus anti-CD28 (Fig. 1D) within 2 h, then declined to a nadir at 16 h, and then again increased (Fig. 1C and D), reproducibly exhibiting a biphasic induction pattern. The induction of IL-21R mRNA expression after T-cell receptor (TCR) stimulation was primarily at the level of transcription, as the transcription inhibitor, actinomycin D, markedly diminished the inducibility (Fig. 1E). Interestingly, although the protein synthesis inhibitor, cycloheximide, moderately increased IL-21R mRNA in resting cells, it almost abolished the induction of IL-21R mRNA upon TCR activation (Fig. 1E), indicating that de novo protein synthesis is required for TCR-induced transcription of IL-21R.
FIG. 1.
TCR-induced IL-21R expression in human T lymphocytes. Jurkat cells (A), Molt-3 cells (B), or purified human PB T cells (C) were cultured in the presence of 10 ng/ml PMA plus 1 μg/ml ionomycin for the indicated times. Human PB T cells were also cultured in the presence of 2 μg/ml anti-CD3 plus 1 μg/ml anti-CD28 antibody, as indicated (D). Human PB T cells were also incubated for 2 h with actinomycin D (10 μg/ml) or cycloheximide (10 μg/ml) with or without anti-CD3/anti-CD28 (E). Total RNA was isolated from cells, and first-strand cDNA was synthesized. IL-21R mRNA levels were quantitated by real-time PCR and normalized to the level of 18S rRNA. For each sample, the bar represents induction (n-fold) compared with no treatment. Values are means ± standard errors of the mean (SEM) of results from three experiments.
Identification of the IL-21R major transcription initiation site.
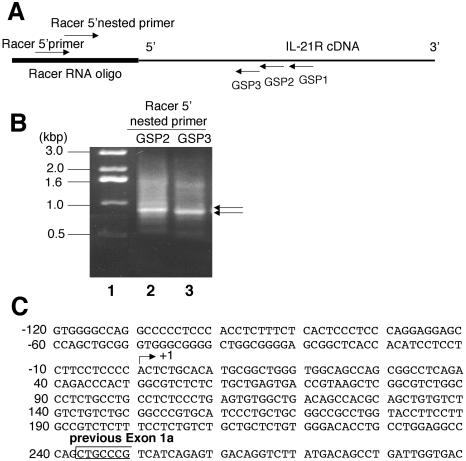
The human IL21R gene contains nine exons, with exons 1a and 1b coding for the 5′ untranslated region (35). Exons 1a and 1b were defined based on the 5′ ends of cDNA clones that have not been proven to correspond to transcription initiation sites. We used 5′ RACE to identify the IL-21R transcription initiation site(s) in human PB T cells activated with anti-CD3 plus anti-CD28. In this method, a “RACER RNA oligonucleotide” was ligated to the 5′ end of the mRNA. Three antisense IL21R gene-specific primers, GSP1, GSP2, and GSP3, targeting different regions of the receptor mRNA (at +805 to +828, +647 to +670, and +607 to +630, respectively), were paired with the “RACER 5′ primer” complementary to the “RACER RNA oligonucleotide” to amplify the 5′ end of the IL-21R mRNA (Fig. 2A). Since multiple weak PCR products were observed when using either GSP1 or GSP2, we performed nested PCR by using the PCR product from the GSP1 amplification paired with GSP2 and “RACER 5′ nested primer” (Fig. 2B, lane 2) or the PCR product from the GSP2 amplification paired with GSP3 and “RACER 5′ nested primer” (Fig. 2B, lane 3). Each yielded a major band of approximately 900 bp, but as expected, the level was slightly higher in the GSP2 reaction (Fig. 2B, lane 2). Subcloning and sequencing of this PCR product showed that the major transcription initiation site for IL-21R is 243 bp upstream of the original exon 1a (Fig. 2C) (13 of 19 clones yielded this position, and the other clones had a 5′ end 205 nucleotides upstream of exon 1a, which may be a minor transcription initiation site). Thus, we redefined the boundary of exon 1 as the major transcription initiation site.
FIG. 2.
Mapping of a major human IL-21R transcription initiation site by 5′ RACE. (A) Locations of primers used in the 5′ RACE. IL21R GSPs 1, 2, and 3 are complementary to the regions from +805 to +828, +647 to +670, or +607 to +630, respectively (relative to the boundary of IL-21R 5′ cDNA) (35). RACER 5′ primer and RACER 5′ nested primer complementary to the RACER RNA oligonucleotide were provided by the GeneRACER kit. (B) Total RNA was isolated from human PB T cells treated with 2 μg/ml of anti-CD3 plus 1 μg/ml of anti-CD28 antibody for 2 h. The 5′ end of IL-21R was PCR amplified by RACER 5′ primer with GSP1 (product 1) or GSP2 (product 2). The nested PCR was performed by using nested primers and PCR product 1 (lane 2) and product 2 (lane 3). Lane 1 shows the molecular weight marker. Arrows indicate the bands cut from the gel for subcloning and sequencing. (C) Partial sequence of the 5′ regulatory region of the IL21R gene showing the major transcription initiation site (arrow) 243 nucleotides 5′ of exon 1a (open box).
Identification of a DNase I-hypersensitive site approximately 100 bp 5′ of the transcription initiation site.
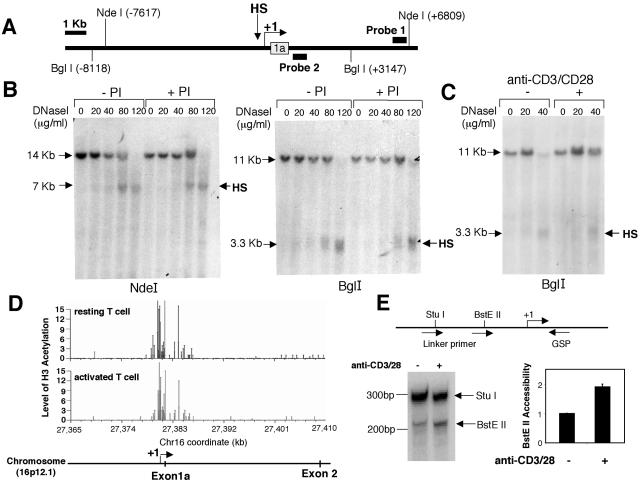
To identify potential regulatory elements that mediate human IL-21R induction, we examined the sequence flanking the transcription initiation site for DNase I-hypersensitive sites. We used NdeI, which cleaves at −7.6 kb and +6.8 kb relative to the transcription initiation site (Fig. 3A). Nuclei from untreated or PI-activated Molt-3 cells were subjected to DNase I hypersensitivity assays. Southern blotting of NdeI-digested genomic DNA and hybridization with probe 1 revealed a hypersensitive site approximately 100 bp 5′ to the transcription initiation site in both PI-stimulated and unstimulated cells (Fig. 3B, left panel). Digestion of DNase I-treated genomic DNA with BglI and subsequent hybridization with probe 2 revealed the same hypersensitive site (Fig. 3B, right panel). A similarly positioned hypersensitive site was also detected in human primary T cells stimulated with anti-CD3 plus anti-CD28 or not stimulated (Fig. 3C). The fact that the same DNase I-hypersensitive site was observed in both unstimulated and stimulated cells indicates that this site is accessible under either basal or induced conditions.
FIG. 3.
Characterization of a DNase I-hypersensitive site in the human IL21R gene. (A) Schematic showing the location of a DNase I-hypersensitive site in the human IL21R gene. The thick bars represent the probes; the open box represents exon 1a. The major transcription initiation site mapped by 5′ RACE is indicated by the arrow. (B) Nuclei were isolated from Molt-3 cells cultured for 4 h with or without PI and then digested with DNase I as indicated. DNA was extracted, digested with either NdeI or BglI, and analyzed on a 0.6% agarose gel. The hypersensitive site was mapped by hybridizing NdeI-digested DNA with probe 1 (+6331 to +6754) and BglI-digested DNA with probe 2 (+1051 to +1352). (C) Nuclei isolated from human PB T cells cultured with or without anti-CD3 and anti-CD28 were digested with DNase I. The hypersensitive site was mapped by hybridizing BglI-digested DNA with probe 2. (D) Acetylation levels of the IL21R gene on chromosome 16 in resting primary human T cells (upper panel) or anti-CD3 and anti-CD28 antibody-activated (24 h) primary T cells (lower panel) were analyzed by GMAT. The genome-wide data are posted at the website http://dir.nhlbi.nih.gov/labs/lmi/zhao/epigenome/G&D2005.htm. The retrieved region includes the promoter and the first intron of the IL21R gene, as indicated by the scheme at the bottom. (E) Nuclei isolated from human PB T cells treated with anti-CD3 and anti-CD28 for 4 h or not treated were briefly digested with BstEII. The purified genomic DNA was digested to completion with StuI. The cleavage sites were detected by linker ligation-mediated PCR with the antisense IL-21R GSP. The data were quantified by PhosphorImager. The intensity of the BstEII band was normalized for the intensity of the StuI band. The bars represent induction of BstEII accessibility (n-fold) compared with no treatment. Values are means ± SEM of results from three experiments. Locations of restriction enzymes and primers for PCR are shown at the top.
Recently, a genome-wide mapping technique (GMAT) of histone H3 acetylation in resting and activated human T cells was established (44). This method combines chromatin immunoprecipitation assays using an antibody to K9/K14 diacetylated histone H3 and serial analysis of gene expression technique (SAGE) to generate a GMAT library wherein the frequency of each SAGE tag reflects the level of histone modification at the corresponding gene locus. The level of H3 hyperacetylation is correlated with chromatin accessibility of promoters and regulatory elements. Using the human genome acetylation database (http://dir.nhlbi.nih.gov/labs/lmi/zhao/epigenome/G&D2005.htm), we analyzed H3 acetylation throughout the IL21R gene and found the promoter region around the DNase I-hypersensitive site was highly acetylated in activated as well as resting T cells (Fig. 3D). To confirm that this region is under chromatin structure remodeling upon activation, we performed a restriction enzyme accessibility assay, which has been extensively used to determine subtle cleavages in nucleosomal structure (8, 12, 53). We briefly digested the nuclei of human PB T cells with BstEII, which has a cleavage site at −140 relative to the transcription initiation site. Following complete digestion of the purified DNA with StuI (cleavage site at −214), which serves as an endogenous control, the cleavage sites were detected by linker ligation-mediated PCR with a universal linker primer and a downstream antisense GSP. Cells treated with anti-CD3 plus anti-CD28 manifested a twofold increase in BstEII accessibility (Fig. 3E), demonstrating that the IL-21R promoter is remodeled to a more open structure.
The TGGGCG motif in the promoter region is important for IL-21R expression.
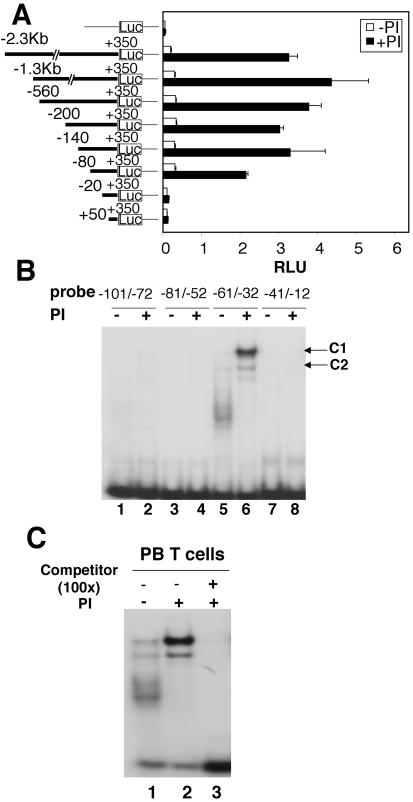
To identify the sequences critical for IL-21R promoter activity, the −2300 to +350 region was cloned in a luciferase reporter vector. Transient transfection of Molt-3 cells with this construct showed a potent increase in PI-induced activity (Fig. 4A). Analysis of a series of 5′ deletion constructs revealed that potent reporter activity was present in a construct extending to −80 but not to −20 (Fig. 4A), indicating a critical role for the −80 to −20 region in PI-induced IL-21R promoter activity.
FIG. 4.
The −80 to −20 region is important for IL-21R promoter activity in response to TCR activation and forms complexes with nuclear factors. (A) Luciferase (Luc) reporter construct schematics are on the left. These were transfected into Molt-3 cells that were not stimulated or stimulated with PI. Luciferase activity was normalized with renilla luciferase activity (relative light units, RLU). Shown are means ± SEM of results from three experiments. (B) Nuclear extracts from PI-treated (4 h) or untreated human PB T cells were incubated with the indicated IL-21R promoter probes and subjected to EMSA. Two DNA-binding protein complexes, C1 and C2, are indicated. (C) EMSA with the −61 to −32 probe and nuclear extract from untreated or PI-treated human PB T cells. In lane 3, a 100-fold molar excess of unlabeled −61 to −32 oligonucleotide was preincubated with nuclear extract before adding the probe.
To identify binding sites for the transcription factor(s), we next performed EMSAs using nested overlapping oligonucleotide probes that together span the −101 to −12 region (−101 to −72, −81 to −52, −61 to −32, or −41 to −12) and nuclear extracts from resting or PI-treated human PB T cells. Two complexes, C1 and C2, were formed only with the −61 to −32 probe (Fig. 4B, lanes 5 and 6). C1 was potently induced after PI stimulation, whereas C2 was only slightly induced (Fig. 4B, lanes 5 and 6, and C, lanes 1 and 2). The formation of the complexes was eliminated by competition with unlabeled wild-type oligonucleotide (Fig. 4C, lane 3). The “smear-like” DNA-binding protein complex seen with extracts from resting T cells (Fig. 4B, lane 5, and C, lane 1) was consistently observed in extracts isolated from different donors, minimizing the possibility that the smear represents protein degradation. Furthermore, when the same extracts were incubated with an IL-7R promoter probe spanning an Ets binding motif for GA binding protein (55), the appropriate band was identified without a “smear,” indicating that general protein degradation had not occurred (data not shown).
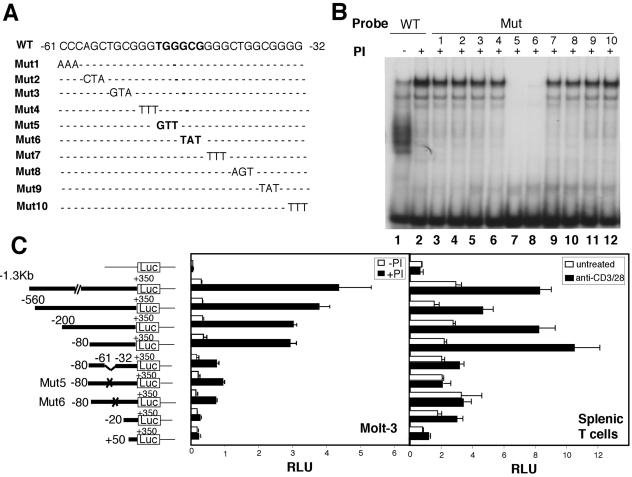
To further define the factor binding sites, we made a series of mutant oligonucleotide probes by changing every three nucleotides within the −61 to −32 region (Fig. 5A) and incubated these probes with nuclear extracts from PI-activated human PB T cells. Mutation of either TGG to GTT (Mut5) or GCG to TAT (Mut6) greatly diminished DNA-protein complex formation (Fig. 5B, lanes 7 and 8). Similar findings were observed in Molt-3 cells (data not shown).
FIG. 5.
Characterization of the TGGGCG motif in the 5′ regulatory region of IL-21R responsible for TCR-induced activity. (A) Sequence of the wild-type (WT) and mutant (Mut) −61 to −32 IL-21R probes used in EMSAs. (B) EMSAs with WT or Mut probes and nuclear extract from human PB T cells treated with PI for 4 h or not treated. (C) Transient transfection of IL-21R promoter reporter constructs in Molt-3 cells and mouse splenic T cells. Luciferase (Luc) reporter construct schematics are on the left. Mut5 and Mut6 are shown in panel A. The luciferase constructs were transfected into Molt-3 cells followed by no stimulation or stimulation with PI or transfected into preactivated mouse splenic T cells that were then not treated or stimulated with anti-CD3/CD28. Luciferase activity was measured and normalized with Renilla luciferase activity (relative light units, RLU). Values are means ± SEM of results from three experiments.
Consistent with the importance of these complexes, deletion of the −61 to −32 region from the −80 to +350 construct greatly decreased PI-induced IL-21R promoter activity in Molt-3 cells, and when the TGG (Mut5) and GCG (Mut6) mutations were made in the −80 to +350 construct, the induced IL-21R promoter activity was also decreased (Fig. 5C, left panel). Because we could not achieve efficient transfection in human T cells, we transiently transfected these constructs into primary murine splenic T cells and found activation-induced IL-21R promoter activity (Fig. 5C, right panel) similar to the findings in Molt-3 cells. These data indicate a critical role for the TGGGCG region for IL-21R promoter activity.
Identification of Sp1 and Sp3 binding to the GC box in the IL-21R promoter in vitro.
To characterize factor(s) binding to the TGGGCG region, we used DNA affinity chromatography (55). PCR-amplified concatemers spanning the entire −61 to −32 region (containing the TGGGCG motif) were biotinylated and tethered to streptavidin paramagnetic particles. A parallel control purification was performed in which TGG GCG was mutated to GTTGCG (Mut5). Nuclear extracts from PI-activated human PB T cells were separately incubated with each affinity resin and washed to remove the nonspecific binding, and eluates were resolved by SDS-PAGE and silver stained. Several bands were more enriched in the eluate from wild-type resin than in that from mutant resin. The major band of approximately 95 kDa that was present only in the wide-type resin eluate (Fig. 6A) was analyzed by LC-MS and MALDI-TOF mass spectrometry and identified as Sp1. The p80 band also contained Sp1 peptides and perhaps represents a degradation product of Sp1. No peptides were detected from the p40 band, so the identity of p40 is completely unclear. The inability to recover peptides may have been due to protein degradation or a low abundance of the material.
FIG. 6.
Sp1 and Sp3 bind to the GC motif in the IL-21R promoter in vitro. (A) Proteins purified by DNA affinity chromatography. Nuclear extracts from PI-stimulated (4 h) human PB T cells were subjected to DNA affinity chromatography. Eluates from WT or mutant (Mut5, as in Fig. 5A) DNA-conjugated beads were resolved by SDS-PAGE and silver stained. LC-MS and MALDI-TOF mass spectrometry were conducted on the 95-, 80-, and 40-kDa bands (arrows). The 95-kDa band is Sp1. (B) Sequence comparison of human and mouse IL-21R promoter-proximal region showed a conserved GGGCGGGGC motif. (C) EMSA using WT probe and recombinant Sp1, Sp3, or nuclear extracts (NE) from human PB T cells treated with PI or not treated. Antibodies to Sp1, Sp2, Sp3, Sp4, or unlabeled oligonucleotides containing the Sp1 consensus sequence at 25-, 50-, or 100-fold molar excess were preincubated with nuclear extract before adding WT probe, as indicated. (D) Nuclear extracts (15 μg) from anti-CD3-plus-anti-CD28-activated human PB T cells or mouse splenic T cells were blotted with anti-Sp1 or γ-tubulin (control). (E) Nuclear extracts (15 μg) were blotted with anti-Sp3 or γ-tubulin.
Sp1 is a sequence-specific zinc finger transcription factor that recognizes GGGGCGGGGC motifs (GC box) and closely related sequences (5, 21). Sequence comparison showed that the GGGCGGGGC sequence is exactly conserved in the 5′-proximal region in the mouse Il21r gene even though the adjacent nucleotide diverges (Fig. 6B). We confirmed the binding of recombinant Sp1 protein to the IL-21R promoter by EMSAs, and this complex has similar migration to that of complex C1 from human PB T cells (Fig. 6C, lane 1 versus lane 6). Addition of anti-Sp1 also decreased the intensity of C1 (Fig. 6C, lane 7) but did not abrogate the complex the way it did the complex generated with recombinant Sp1 (Fig. 6C, lane 2), and it had no effect on complex C2 (Fig. 6C, lane 7). This might relate to a possible modification of Sp1 in the T-cell extract that might compromise the accessibility of the antibody to Sp1 or alternatively may represent the binding of a factor in addition to Sp1. We therefore tested the effect of antibodies to other Sp family proteins, as Sp3 and Sp4 act through the same GC (GGGGCGGGG) box while Sp2 mainly acts through a GT box (GGTGTGGGG) (4, 7). An antibody to Sp3 abolished the C2 complex (Fig. 6C, lane 9), analogous to its elimination of the complex formed with recombinant Sp3 (Fig. 6C, lane 4 versus lane 3). The recombinant Sp3 contains only the 100-kDa isoform, which may explain why its migration was different from C2, which potentially contains the 60- and 58-kDa isoforms of Sp3. As expected, when an unlabeled Sp1 consensus oligonucleotide containing the GC box was added at 25-, 50-, or 100-fold molar excess, the formation of both C1 and C2 was partially reduced (Fig. 6C, lanes 11 to 13). Antibodies to the other Sp family proteins, Sp2 and Sp4, had no effect (Fig. 6C, lanes 8 and 10). This is consistent with Sp4 being more specific for the brain (7) and the fact that in vitro-translated Sp2 could not bind to the IL-21R promoter (data not shown), making it less likely that Sp2 or Sp4 is involved in the regulation of IL-21R.
We next examined whether the expression levels of Sp1 and Sp3 in primary T cells were changed upon TCR stimulation. Interestingly, in human PB T cells, the basal expression of Sp1 was variable among different donors. In those donors with low basal expression, Sp1 was potently induced after stimulation with anti-CD3 plus anti-CD28 (Fig. 6D, donor 1), whereas in other donors where Sp1 was expressed at a high level in resting cells, no inducibility was observed (Fig. 6D, donor 2). It is possible that the altered basal levels of Sp1 expression might reflect recent exposure of some donors to antigenic stimuli. Consistent with this, the level of Sp1 was low in naïve resting mouse splenic T cells but consistently induced after TCR activation (Fig. 6D). Our data thus indicated that Sp1 bound to the IL-21R promoter in vitro and that TCR stimulation augmented levels of Sp1. In contrast, the expression level of Sp3 was relatively stable upon TCR activation, and even in the donors with induced Sp1 expression, no inducibility of Sp3 was observed (Fig. 6E).
Suppression of TCR-mediated IL-21R induction by siSp1.
To investigate the role of Sp1 and Sp3 in vivo for TCR-induced IL21R gene regulation, we used synthetic small interfering RNAs (21-nucleotide duplex siRNAs). As a control, we used an siRNA that, based on bioinformatic algorithms, was designed to have more than four mismatches with other known human genes. We transfected 25, 50, 100, 150, or 200 nM siSp1 or siSp3 into human primary T cells and found that 100 nM provided an optimal silencing effect by measuring the Sp1 or Sp3 mRNA levels using quantitative real-time PCR (data not shown), and we used this dose in subsequent experiments. In human PB T cells, when the Sp1 expression level was effectively silenced by siRNA transfection (Fig. 7B), the level of TCR-induced IL-21R expression was diminished by over 50% (Fig. 7A). However, when the expression level of Sp3 was effectively silenced by siSp3 (Fig. 7C), the TCR-induced IL-21R expression was at most minimally reduced. Moreover, the combination of siSp3 and siSp1 did not decrease the expression of IL-21R more than transfection of only siSp1 (Fig. 7A). The control siRNA did not affect Sp1 (Fig. 7B), Sp3 (Fig. 7C), or IL-21R (Fig. 7A) expression. These results demonstrate that although both Sp1 and Sp3 bind to the IL-21R promoter, Sp1 plays a more critical role in regulating IL-21R expression in human T cells.
FIG. 7.
Binding of Sp1 to the IL-21R promoter is essential for TCR-induced IL-21R expression. Suppression of IL-21R expression in human primary T cells by siRNA targeting of Sp1 is also shown. Human PB T cells were transfected with 100 nM siRNAs along with pEYFP-N1 and were cultured for 36 h. YFP+ cells were sorted and stimulated with anti-CD3 and anti-CD28 for 2 h. Total RNA was extracted and analyzed for IL-21R (A), Sp1 (B), and Sp3 (C) expression by quantitative real-time PCR. IL-21R, Sp1, and Sp3 expressions were normalized to the level of expression of 18S rRNA. For each sample, the bar represents the induction (n-fold) compared to no TCR activation. Values are means ± SEM of results from three experiments. (D) In vivo binding of Sp1 to the IL-21R promoter in human primary T cells by chromatin immunoprecipitation. Human PB T cells were cultured with or without anti-CD3 and anti-CD28 for 2 h. Formaldehyde-cross-linked chromatin was immunoprecipitated with preimmune rabbit serum or with anti-Sp1. Real-time PCR was performed to quantitate the DNA fragment containing the Sp1 binding site in the IL-21R promoter. β-Actin was a control. Values are means ± SEM of results from three experiments.
Sp1 binds in vivo to the IL-21R promoter.
To determine if Sp1 bound to the IL-21R promoter in human primary T cells in vivo, we performed ChIP assays using an antibody to Sp1. Real-time PCR was used to quantitate the abundance of DNA fragment spanning the IL-21R promoter with Sp1 binding sites, with the β-actin gene as an indicator of the nonspecific background after immunoprecipitation. Upon TCR activation, the Sp1 antibody reproducibly resulted in a 2- to 2.5-fold increase in the IL-21R promoter ChIP signal compared to that seen with controls (Fig. 7D). These results demonstrate that Sp1 binds to the IL-21R promoter in vivo and that the binding increases after TCR activation.
TCR-induced IL-21R expression depends on the dephosphorylation of Sp1.
Posttranslational modification of Sp1 is known to regulate its transcriptional activity. For example, lipopolysaccharide treatment reduced Sp1 DNA-binding activity and transcriptional activation by dephosphorylation of Sp1 at serine and threonine residues (56). In addition, it was reported that in human T lymphocytes treated with anti-CD2 plus anti-CD28, Sp1 was dephosphorylated, correlating with cell cycle progression. The dephosphorylation of Sp1, which was mediated by protein phosphatase 2A (PP2A), increased its transcriptional activity in a reporter assay in Kit225 cells (22). We thus examined the effect of calyculin A, a potent inhibitor of both PP2A and protein phosphatase 1 (PP1) that specifically inhibits dephosphorylation at serine/threonine residues (43), on TCR-induced IL-21R expression. When 10 nM calyculin A was incubated with human PB T cells stimulated with anti-CD3 plus anti-CD28 for 2 h, it blocked the IL-21R mRNA induction (Fig. 8A) without affecting viability or expression of 18S rRNA (data not shown). A second phosphatase inhibitor, okadaic acid, which binds to the catalytic subunits of PP1 and PP2A (with a 200-fold-higher affinity for PP2A) (22), caused a similar reduction of TCR-induced IL-21R expression (Fig. 8A). By EMSA, the DNA-binding activity of Sp1 was decreased by calyculin A or okadaic acid treatment in T cells activated with anti-CD3 plus anti-CD28 (Fig. 8B, lane 4 versus lane 3 and lane 8 versus lane 7). The decrease in DNA-binding activity did not result from protein degradation after cytotoxic calyculin A treatment, as Sp1 protein expression was if anything somewhat increased after calyculin A treatment, as detected by Western blotting (Fig. 8C, lane 2 versus lane 1 and lane 4 versus lane 3). By Western blotting, two Sp1 bands were detected in resting T cells (Fig. 8C, lane 1), with the lower-molecular-weight form most likely arising from dephosphorylation, given the increase in the upper band after calyculin A treatment (Fig. 8C, lane 2). TCR stimulation augmented Sp1 with a relative increase in the lower dephosphorylated band (Fig. 8C, lane 3), and the addition of calyculin A again increased the higher-molecular-weight phosphorylated form (Fig. 8C, lane 4). Similar effects on Sp1 expression level were observed in okadaic acid-treated T cells (Fig. 8C). Together, these results indicate that TCR stimulation induces Sp1 with a relative increase in the dephosphorylated form, and the dephosphorylation of Sp1 may be required for IL-21R promoter activity.
FIG. 8.
Dephosphorylation of Sp1 is essential for TCR activation-induced IL-21R expression. Human PB T cells were not stimulated or stimulated with anti-CD3 and anti-CD28 for 2 h. Half of the cells were coincubated with 10 nM calyculin A or 0.8 μM okadaic acid. Total RNA and nuclear extract were isolated from these cells. (A) Suppression of TCR-induced IL-21R mRNA expression by calyculin A or okadaic acid treatment. Total RNA was transcribed to first-strand cDNA, and IL-21R mRNA levels were quantitated by real-time PCR and normalized to the level of 18S rRNA. The bar represents the induction (n-fold) compared with no treatment. Values are means ± SEM of results from three experiments. (B) Calyculin A or okadaic acid treatment diminished the Sp1 DNA-binding activity. Nuclear extracts were incubated with the −61 to −32 probe of IL-21R promoter and subjected to EMSA. Different donors were used by the calyculin A and okadaic acid experiments, presumably explaining the difference in the levels of Sp1. (C) Calyculin A or okadaic acid treatment inhibited Sp1 dephosphorylation. Fifteen micrograms of the nuclear extract was subjected to SDS-PAGE on 8% Novex Tris-glycine gel or 8% NuPAGE Tris-acetate gel, transferred to an Immobilon-P membrane, and blotted with anti-Sp1 or γ-tubulin.
DISCUSSION
We have demonstrated that TCR-induced IL-21R expression is regulated at the level of transcription in human T cells and depends on the synthesis of new proteins. Interestingly, the induction pattern is biphasic in primary T cells. This pattern may be due to the involvement of different factors that regulate early/late induction. In the early induction phase, IL-21R mRNA induction requires the synthesis of new or activation of preexisting transcription factors. We hypothesize that the late induction of IL21R gene expression may result from cytokines produced by activated T cells. For example, IL-21 is known to up-regulate expression of its own receptor (57).
In examining the molecular mechanism underlying the transcriptional regulation of the IL21R gene, we found that Sp1 binds to a GC-rich motif in the proximal promoter and mediates IL-21R induction in activated human T cells. Sp1 is known to activate many vital genes, including genes involved in cell growth and development (5, 15, 21), and homozygous deletion of Sp1 in mice causes severe lethal embryonic malformation (30). Classically, Sp1 was viewed as a constitutive transcription factor that regulates the basal expression of many cellular genes (7, 14). However, it has now been found to be involved in tissue-specific gene expression and in the control of transcription following different stimuli (3, 40, 51).
In the immune system, Sp1 has been reported to control the expression of genes that mediate important cellular functions. For example, Sp1 regulates the expression of genes encoding IL-2Rβ (26), TCR Vα (19), and FasL (54) and plays a critical role in regulating the division of T lymphocytes after costimulation with anti-CD2 and anti-CD28 (22). We now show that Sp1 plays an essential role in regulating TCR-induced IL-21R expression. Mutation of the Sp1 binding sites in the IL-21R proximal promoter region diminished PI-induced reporter activity. Strikingly, Sp1 expression was significantly increased in primary T cells after TCR stimulation. The TCR-induced augmentation of Sp1 transcriptional activity correlated with induction of IL-21R mRNA, and transfection of Sp1 siRNAs into primary human T cells significantly decreased TCR-induced IL-21R mRNA expression. Unexpectedly, however, Sp1 expression is only slightly increased 2 h after TCR activation, while IL-21R mRNA levels were highly induced at this time point. We therefore hypothesized that posttranslational modification of Sp1 might play a critical role for IL-21R gene expression. Depending on cell type and stimuli, phosphorylation of Sp1 has been reported to either increase (41, 45) or decrease (1, 23, 32, 58) Sp1 transcriptional activity. We thus examined the effect of the phosphatase inhibitors calyculin A and okadaic acid, which inhibit PP1 and PP2A activity, and found that they decreased IL-21R expression. Moreover, the phosphatase inhibitors increased the phosphorylation of Sp1 and correspondingly decreased Sp1 DNA-binding activity for the IL-21R promoter. These results suggest that posttranslational modification of Sp1 is critical for TCR-induced IL-21R expression.
Sp1 has been reported to exert its transcriptional activation with the help of other comodulators or cofactors, such as NF-κB (20), STAT proteins (6, 28), Smad family factors (33), Egr family factors (26, 50), NFAT family proteins (54), or other members of the Sp family (9, 46). Complex C2, as detected in EMSA, appears to represent Sp3, since anti-Sp3 diminished the formation of C2. However, the DNA-binding activity of complex C2 was only slightly induced by PI activation, in contrast to Sp1, whose binding activity was markedly induced. In addition, siSp3 had little effect on TCR-induced IL-21R expression, and the cotransfection of siSp1 and siSp3 was not more potent than siSp1 transfection alone. Thus, unlike Sp1, Sp3 plays only a minor role in regulating IL-21R repression. So far, we do not yet have definitive data that support factors other than Sp1 as critically contributing to the regulation of IL-21R. Since PI activation is associated with protein kinase C activation and calcium/calcineurin pathway, it is conceivable that other factors, such as AP-1, NF-κB, or NFAT (10, 13, 42, 49), might be involved in IL-21R regulation, an area for future investigation.
Taken together, our results reveal that Sp1 is indeed a critical regulator of IL21R gene expression, a finding with implications as to how the expression of this critical receptor can be controlled. Moreover, the observation that Sp1 is potently induced and dephosphorylated in response to anti-CD3 plus anti-CD28 has much broader implications for the regulation of Sp1-dependent genes in antigen-activated T cells. Further studies are needed to clarify how TCR activation induces the dephosphorylation of Sp1 and to identify the relevant target residues. Such studies will not only further clarify the basis for IL-21R regulation but also help us to understand how Sp1-regulated genes in general are broadly controlled in the immune system.
Acknowledgments
We thank Jian-Xin Lin, Howard Young, and Maria Berg (all at NIH) for valuable suggestions and/or critical comments.
This research was supported by the Intramural Research Program of the NHLBI, NIH.
REFERENCES
- 1.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 2.Asao, H., C. Okuyama, S. Kumaki, N. Ishii, S. Tsuchiya, D. Foster, and K. Sugamura. 2001. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J. Immunol. 167:1-5. [DOI] [PubMed] [Google Scholar]
- 3.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 4.Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 195:27-38. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, M. R., J. T. Kadonaga, S. P. Bell, and R. Tjian. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234:47-52. [DOI] [PubMed] [Google Scholar]
- 6.Cantwell, C. A., E. Sterneck, and P. F. Johnson. 1998. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol. Cell. Biol. 18:2108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, T., B. Gebelein, and R. Urrutia. 1999. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 880:94-102. [DOI] [PubMed] [Google Scholar]
- 8.Cui, K., P. Tailor, H. Liu, X. Chen, K. Ozato, and K. Zhao. 2004. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell. Biol. 24:4476-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, S., T. Kojima, M. Shiraiwa, M. C. Mechin, S. Chavanas, G. Serre, M. Simon, A. Kawada, and H. Takahara. 2005. Regulation of the expression of peptidylarginine deiminase type II gene (PADI2) in human keratinocytes involves Sp1 and Sp3 transcription factors. J. Investig. Dermatol. 124:1026-1033. [DOI] [PubMed] [Google Scholar]
- 10.Favero, J., and V. Lafont. 1998. Effector pathways regulating T cell activation. Biochem. Pharmacol. 56:1539-1547. [DOI] [PubMed] [Google Scholar]
- 11.Habib, T., S. Senadheera, K. Weinberg, and K. Kaushansky. 2002. The common gamma chain (gamma c) is a required signaling component of the IL-21 receptor and supports IL-21-induced cell proliferation via JAK3. Biochemistry 41:8725-8731. [DOI] [PubMed] [Google Scholar]
- 12.Hempel, W. M., and P. Ferrier. 2004. Restriction endonuclease accessibility as a determinant of altered chromatin structure. Methods Mol. Biol. 287:53-63. [DOI] [PubMed] [Google Scholar]
- 13.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 14.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 16.Kasaian, M. T., M. J. Whitters, L. L. Carter, L. D. Lowe, J. M. Jussif, B. Deng, K. A. Johnson, J. S. Witek, M. Senices, R. F. Konz, A. L. Wurster, D. D. Donaldson, M. Collins, D. A. Young, and M. J. Grusby. 2002. IL-21 limits NK cell responses and promotes antigen-specific T cell activation: a mediator of the transition from innate to adaptive immunity. Immunity 16:559-569. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. P., J. Kelly, and W. J. Leonard. 2001. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity 15:159-172. [DOI] [PubMed] [Google Scholar]
- 18.Kim, H. P., and W. J. Leonard. 2002. The basis for TCR-mediated regulation of the IL-2 receptor alpha chain gene: role of widely separated regulatory elements. EMBO J. 21:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsley, C., and A. Winoto. 1992. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 12:4251-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krehan, A., H. Ansuini, O. Bocher, S. Grein, U. Wirkner, and W. Pyerin. 2000. Transcription factors ets1, NF-kappa B, and Sp1 are major determinants of the promoter activity of the human protein kinase CK2alpha gene. J. Biol. Chem. 275:18327-18336. [DOI] [PubMed] [Google Scholar]
- 21.Kriwacki, R. W., S. C. Schultz, T. A. Steitz, and J. P. Caradonna. 1992. Sequence-specific recognition of DNA by zinc-finger peptides derived from the transcription factor Sp1. Proc. Natl. Acad. Sci. USA 89:9759-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacroix, I., C. Lipcey, J. Imbert, and B. Kahn-Perles. 2002. Sp1 transcriptional activity is up-regulated by phosphatase 2A in dividing T lymphocytes. J. Biol. Chem. 277:9598-9605. [DOI] [PubMed] [Google Scholar]
- 23.Leggett, R. W., S. A. Armstrong, D. Barry, and C. R. Mueller. 1995. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J. Biol. Chem. 270:25879-25884. [DOI] [PubMed] [Google Scholar]
- 24.Leonard, W. J. 2001. Cytokines and immunodeficiency diseases. Nat. Rev. Immunol. 1:200-208. [DOI] [PubMed] [Google Scholar]
- 25.Lin, J.-X., N. K. Bhat, S. John, W. S. Queale, and W. J. Leonard. 1993. Characterization of the human interleukin-2 receptor β-chain gene promoter: regulation of promoter activity by ets gene products. Mol. Cell. Biol. 13:6201-6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, J. X., and W. J. Leonard. 1997. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor beta-chain promoter through noncanonical Egr and Sp1 binding sites. Mol. Cell. Biol. 17:3714-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 28.Look, D. C., M. R. Pelletier, R. M. Tidwell, W. T. Roswit, and M. J. Holtzman. 1995. Stat1 depends on transcriptional synergy with Sp1. J. Biol. Chem. 270: 30264-30267. [DOI] [PubMed] [Google Scholar]
- 29.Ma, H. L., M. J. Whitters, R. F. Konz, M. Senices, D. A. Young, M. J. Grusby, M. Collins, and K. Dunussi-Joannopoulos. 2003. IL-21 activates both innate and adaptive immunity to generate potent antitumor responses that require perforin but are independent of IFN-gamma. J. Immunol. 171:608-615. [DOI] [PubMed] [Google Scholar]
- 30.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89:619-628. [DOI] [PubMed] [Google Scholar]
- 31.Mehta, D. S., A. L. Wurster, M. J. Whitters, D. A. Young, M. Collins, and M. J. Grusby. 2003. IL-21 induces the apoptosis of resting and activated primary B cells. J. Immunol. 170:4111-4118. [DOI] [PubMed] [Google Scholar]
- 32.Mortensen, E. R., P. A. Marks, A. Shiotani, and J. L. Merchant. 1997. Epidermal growth factor and okadaic acid stimulate Sp1 proteolysis. J. Biol. Chem. 272:16540-16547. [DOI] [PubMed] [Google Scholar]
- 33.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 95:6733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 35.Ozaki, K., K. Kikly, D. Michalovich, P. R. Young, and W. J. Leonard. 2000. Cloning of a type I cytokine receptor most related to the IL-2 receptor beta chain. Proc. Natl. Acad. Sci. USA 97:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozaki, K., R. Spolski, R. Ettinger, H. P. Kim, G. Wang, C. F. Qi, P. Hwu, D. J. Shaffer, S. Akilesh, D. C. Roopenian, H. C. Morse III, P. E. Lipsky, and W. J. Leonard. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361-5371. [DOI] [PubMed] [Google Scholar]
- 37.Ozaki, K., R. Spolski, C. G. Feng, C. F. Qi, J. Cheng, A. Sher, H. C. Morse III, C. Liu, P. L. Schwartzberg, and W. J. Leonard. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science 298:1630-1634. [DOI] [PubMed] [Google Scholar]
- 38.Parrish-Novak, J., S. R. Dillon, A. Nelson, A. Hammond, C. Sprecher, J. A. Gross, J. Johnston, K. Madden, W. Xu, J. West, S. Schrader, S. Burkhead, M. Heipel, C. Brandt, J. L. Kuijper, J. Kramer, D. Conklin, S. R. Presnell, J. Berry, F. Shiota, S. Bort, K. Hambly, S. Mudri, C. Clegg, M. Moore, F. J. Grant, C. Lofton-Day, T. Gilbert, F. Rayond, A. Ching, L. Yao, D. Smith, P. Webster, T. Whitmore, M. Maurer, K. Kaushansky, R. D. Holly, and D. Foster. 2000. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature 408:57-63. [DOI] [PubMed] [Google Scholar]
- 39.Parrish-Novak, J., D. C. Foster, R. D. Holly, and C. H. Clegg. 2002. Interleukin-21 and the IL-21 receptor: novel effectors of NK and T cell responses. J. Leukoc. Biol. 72:856-863. [PubMed] [Google Scholar]
- 40.Philipsen, S., and G. Suske. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rafty, L. A., and L. M. Khachigian. 2001. Sp1 phosphorylation regulates inducible expression of platelet-derived growth factor B-chain gene via atypical protein kinase C-zeta. Nucleic Acids Res. 29:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 43.Resjo, S., A. Oknianska, S. Zolnierowicz, V. Manganiello, and E. Degerman. 1999. Phosphorylation and activation of phosphodiesterase type 3B (PDE3B) in adipocytes in response to serine/threonine phosphatase inhibitors: deactivation of PDE3B in vitro by protein phosphatase type 2A. Biochem. J. 341:839-845. [PMC free article] [PubMed] [Google Scholar]
- 44.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohlff, C., S. Ahmad, F. Borellini, J. Lei, and R. I. Glazer. 1997. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 272:21137-21141. [DOI] [PubMed] [Google Scholar]
- 46.Santini, M. P., C. Talora, T. Seki, L. Bolgan, and G. P. Dotto. 2001. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21(WAF1/CIP1) expression in keratinocyte differentiation. Proc. Natl. Acad. Sci. USA 98:9575-9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strengell, M., S. Matikainen, J. Siren, A. Lehtonen, D. Foster, I. Julkunen, and T. Sareneva. 2003. IL-21 in synergy with IL-15 or IL-18 enhances IFN-gamma production in human NK and T cells. J. Immunol. 170:5464-5469. [DOI] [PubMed] [Google Scholar]
- 48.Suto, A., H. Nakajima, K. Hirose, K. Suzuki, S. Kagami, Y. Seto, A. Hoshimoto, Y. Saito, D. C. Foster, and I. Iwamoto. 2002. Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line C(epsilon) transcription of IL-4-stimulated B cells. Blood 100:4565-4573. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, R. S., M. J. Tymms, L. H. McKinlay, M. F. Shannon, A. Seth, and I. Kola. 1997. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene 14:2845-2855. [DOI] [PubMed] [Google Scholar]
- 50.Trejo, S. R., W. E. Fahl, and L. Ratner. 1997. The tax protein of human T-cell leukemia virus type 1 mediates the transactivation of the c-sis/platelet-derived growth factor-B promoter through interactions with the zinc finger transcription factors Sp1 and NGFI-A/Egr-1. J. Biol. Chem. 272:27411-27421. [DOI] [PubMed] [Google Scholar]
- 51.Turner, J., and M. Crossley. 1999. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci. 24:236-240. [DOI] [PubMed] [Google Scholar]
- 52.Wang, G., M. Tschoi, R. Spolski, Y. Lou, K. Ozaki, C. Feng, G. Kim, W. J. Leonard, and P. Hwu. 2003. In vivo antitumor activity of interleukin 21 mediated by natural killer cells. Cancer Res. 63:9016-9022. [PubMed] [Google Scholar]
- 53.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, S., K. Matsui, A. Fine, B. Zhu, A. Marshak-Rothstein, R. L. Widom, and S. T. Ju. 1999. FasL promoter activation by IL-2 through SP1 and NFAT but not Egr-2 and Egr-3. Eur. J. Immunol. 29:3456-3465. [DOI] [PubMed] [Google Scholar]
- 55.Xue, H. H., J. Bollenbacher, V. Rovella, R. Tripuraneni, Y. B. Du, C. Y. Liu, A. Williams, J. P. McCoy, and W. J. Leonard. 2004. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat. Immunol. 5:1036-1044. [DOI] [PubMed] [Google Scholar]
- 56.Ye, X., and S. F. Liu. 2002. Lipopolysaccharide down-regulates Sp1 binding activity by promoting Sp1 protein dephosphorylation and degradation. J. Biol. Chem. 277:31863-31870. [DOI] [PubMed] [Google Scholar]
- 57.Zeng, R., R. Spolski, S. E. Finkelstein, S. Oh, P. E. Kovanen, C. S. Hinrichs, C. A. Pise-Masison, M. F. Radonovich, J. N. Brady, N. P. Restifo, J. A. Berzofsky, and W. J. Leonard. 2005. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J. Exp. Med. 201:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, Q., and K. Liao. 2000. Differential expression of the adipocyte amino acid transporter is transactivated by SP1 and SP3 during the 3T3-L1 preadipocyte differentiation process. Biochem. Biophys. Res. Commun. 271:100-106. [DOI] [PubMed] [Google Scholar]