Abstract
The expression of beta interferon genes from humans and mice is under the immediate control of a virus-responsive element (VRE) that terminates 110 bp upstream from the transcriptional start site. Whereas a wealth of information is available for the enhanceosome that is formed on the VRE upon the signals generated by viral infection, early observations indicating the existence of other far-upstream control elements have so far remained without a molecular fundament. Guided by a computational analysis of DNA structures, we could locate three as-yet-unknown transcription factor-binding regions at −0.5, −2, and −3 kb. Our present study delineates the interplay of factors YY1 and YY2 as it occurs at the sites at −3 kb and −2 kb (otherwise called HS1 and HS2), consistent with the idea that the novel factor YY2 antagonizes the negative actions exerted by YY1. Differences between the human and murine control regions will be described.
The expression of specific genes in response to extracellular signals requires not only the activation of distinct sets of transcription factors but also a mechanism that integrates their information. A well-known paradigm is the beta interferon (IFN-β) gene, which remains silent throughout the cycle of a differentiated cell unless it is activated by viral or bacterial infection. Together with its proximal promoter/enhancer, this gene is sometimes regarded the best-understood transcription unit of higher eukaryotes. Various signal transduction pathways have been elucidated that lead to the assembly of a multicomponent enhancer complex, the so-called enhanceosome (24).
For humans and mice, the regulation of the IFN-β gene has been elucidated in considerable detail (1, 2, 24, 32). In both cases, the single-copy genes are members of the respective IFN-α/β gene clusters; according to early reports, their induction appeared to obey the same rules. Both promoters can be dissected into several positive regulatory domains (PRDs) and negative regulatory domains (NRDs), which accommodate the respective factors, permit their interaction, and together form the virus-responsive element (VRE). Evidence has been presented for the existence of a NRD I element (−37 to −60) that overlaps PRD II, starting from the transcriptional start site. This is followed by PRD III/I, which in turn partially overlaps an element PRD IV that extends to position −110. A more recent addition is an NRD II element between positions −110 and −220 (32).
Virus infection results in the coordinate activation of three sets of factors, p50/p65 (on PRD II), interferon regulatory factors 3 and 7 (IRF3/7; on PRD I/III), and activating transcription factor 2 (ATF-2)/c-Jun (on PRD IV) which, together with the architectural high-mobility group protein I(Y) [HMG-I(Y)] protein, form the enhanceosome, essentially a platform for recruiting the transcriptional apparatus (the long and short forms of the latter protein are otherwise called HMGA1a and HMGA1b, respectively) (33). The silent state prior to induction is characterized by two positioned nucleosomes: nucleosome II has boundaries at +132 and −15 covering the transcriptional start site; nucleosome I is localized between positions −118 and −268, overlapping the NRD II domain. In addition, some authors have discussed factors interfering with PRD II (the NF-κB-repressing factor binding to NRE) (29) and with PRD I/III (factor IRF-2 and Blimp-1) (17, 22). Interference with this repressed state, for instance by a trichostatin A-mediated local acetylation of histone H4 tails, has been shown to derepress the IFN-promoter even in the absence of virus induction (32).
Chromatin immunoprecipitation experiments have elucidated the order of events subsequent to enhanceosome formation: transient recruitment of GCN5, a histone acetyltransferase (HAT), reaches a maximum 4 to 5 h postinfection. This event coincides with the acetylation of the nucleosome core and of HMG I(Y). These early acetylation steps strengthen interactions within the enhanceosome and protect HMG I(Y) from a subsequent acetylation mediated by CBP (CREB-binding protein, a factor accommodating the histone acetyltransferase p300), which would otherwise disassemble the enhanceosome (24).
Acetylation steps alone do not suffice for nucleosome remodeling, which also requires the recruitment of CBP, and is maximal at 9 to 12 h, a time when histone acetylation at the promoter starts to decline. CBP and p300 associate with IRF-3 and IRF-7 in a step that is followed by the recruitment of a chromatin-remodeling complex (SWI/SNF), TFIID recruitment, and TBP, as well as acetylation-dependent nucleosome sliding. TFIID is also required for the association of other preinitiation complex components such as TFIIB and TFIIA. Nucleosome II remains remodeled during the entire transcription process, even after the dissociation of SWI/SNF.
Related events appear to happen at the NRD II region: a virus-induced DNase I-hypersensitive site arises in this highly A/T-rich stretch of DNA that is otherwise characterized by an interaction with linker histone H1. In mouse cells, HMG I(Y) associates at position −130 between the VRE and NRD II, whereby H1 is displaced followed by derepression of the promoter (29).
Until recently, no information was available about details of the process by which GCN5 is recruited to the enhanceosome to initiate this extended series of modification/remodeling steps. This situation changed significantly after Weill et al. (32) demonstrated the participation of factor Yin Yang 1 (YY1) in the induction of IFN-β for murine cells; prior to infection, YY1 appears to be bound to a −90 site where it participates in promoter repression through an associated histone deacetylase (HDAC) that maintains the core histones of nucleosome I in a deacetylated state. It is suggested that shortly after infection, two molecules of YY1 bound at positions −90 and −122 mediate the recruitment of GCN5 and initiate nucleosome sliding via histone hyperacetylation. However, while there exist two proximal YY1 sites in the murine regulatory region, no corresponding functional sites were found for the human counterpart (19). The detection of two far-upstream YY1 sites for both the human IFN-β (huIFN-β) gene (positions −2000 and −3000) and the murine IFN-β (muIFN-β) gene (positions −2700 and −4700) has further complicated this situation (19).
Present work assigns a significant function not only to the ubiquitously expressed transcription factor (YY1), but also to its recently discovered homologue YY2 (26), which both associate with the sites far upstream from the human IFN-β gene. Both binding sites coincide with a DNase I hypersensitive site (HS2 at −2 kb and HS1 at −3 kb) and they represent the only YY1/YY2 sites within 4.5 kb of DNA. We examine the relevance of these interactions for regulating IFN-β expression by systematic target site mutagenesis and overexpression studies. Our results show the existence of as-yet-unknown remote control mechanisms acting on IFN-β transcription during virus induction.
MATERIALS AND METHODS
SIDD calculations.
Sites with secondary structure-forming potential can be localized in genomic sequences by the analysis of stress-induced duplex destabilization (SIDD) properties. SIDD profiles have been calculated according to an algorithm developed by Benham et al. (5) as described previously (18). In a preceding study (19), we screened sequences far upstream from the beta interferon gene for candidate sites with regulatory potential. The coincidence of these signals with DNase I hypersensitive sites suggested that stress-induced secondary DNA structures have a signal function. That this prediction is borne out is demonstrated by the biochemical studies in the present contribution.
Basically, the SIDD algorithm (http://genomics.ucdavis.edu/benham/sidd/index.php) predicts the propensity of a DNA base pair to undergo strand separation under superhelical tension as it arises, due to tracking proteins or the loss of nucleosomes (19). To this end, the incremental free energy [G(x)] needed to separate the base pair at each position x is computed in the context of a defined stretch of DNA. A value of G(x) near or below zero indicates an essentially completely destabilized base pair with a high probability to denature at equilibrium. SIDD profiles, i.e., plots of G(x) versus x, visualize regions of the sequence where superhelical stress will destabilize the duplex (Fig. 1).
FIG. 1.
Localization of all identified factor binding sites in the IFN-β upstream control region (SIDD profile). SIDD properties of the human IFN-β gene were calculated as part of the standard plasmid pTZ18R (5). The destabilized interferon beta promoter and terminator regions are marked PI and TI, respectively, whereas the coding sequence (IFN-β) is marked by a horizontal arrow. The strongly destabilized main upstream peaks of interest are termed HS1, HS2, and HS3. Transcription factor-binding sites identified by EMSAs are indicated as follows: arrowheads, YY1/YY2; dashed arrow, PURα, PURβ, and UF complex; small black arrows, Oct-1.
Reporter gene constructs.
pILGTkneo (P), a virus-inducible human IFN-β promoter fragment (representing position 0 to −281) drives the transcription of a firefly luciferase gene; neomycin phosphotransferase serves as a selection marker. pILGTkneoEcoC (P-C) contains a 4.5-kb control sequence of the genomic 5′ region, in addition. pILGTkneoEcoCMutHS1 (P-CHS1mut) is the same as P-C, but with a mutated YY1/YY2 site within the promoter-distal destabilized region of HS1 (primer pair SDM1/SDM2 was applied for mutagenesis). pILGTkneoEcoCMutHS2 (P-CHS2mut)is the same as P-C, but with a mutated YY1/YY2-binding site within destabilized region HS2 (primer pair SDM3/SDM4 applied for mutagenesis). pILGTkneoEcoCMutHS1/HS2 (P-CHS1/HS2mut): as “P-C,” but with mutated YY1/YY2-binding sites within both destabilized regions HS1 and HS2 (primer pairs SDM1/SDM2 and SDM3/SDM4 were applied for mutagenesis). pILGTkneoEcoCMutHS3 (P-CHS3mut) is the same as P-C, but with a mutated binding site for an undetermined factor (UF complex) within promoter-proximal destabilized region HS3 (the primer pair SDM5/SDM6 was applied for mutagenesis).
Expression vectors.
pcDNA3.1(−)FLAGYY1 was cloned with a pCEP4FLAGYY1 (Ed Seto, Tampa, Fla.)-derived EcoRI/XbaI FLAGYY1 fragment, containing human YY1. pcDNA3.1FLAGYY2 from Ed Seto, Tampa, Fla. (26) encodes human YY2, whereas pcDNA3.1(−)muYY2 possesses the murine orthologue without a FLAG tag. The empty pcDNA3.1(−) vector was used as a control plasmid.
Identification and cDNA cloning of murine YY2 (muYY2).
The NCBI BLAST programs were used to search for a protein with homology to human YY2. A good candidate was the cDNA clone NM_178266, encoding a protein called membrane-bound transcription factor protease, site 2 (NP_839997). To amplify the corresponding cDNA, a set of different primers was designed (see “Oligonucleotides for cDNA amplification,” below). For reverse transcription (RT), RNA was isolated from murine NIH 3T3 cells using the RNeasy Mini kit from QIAGEN according to the manufacturer's suggestions. Subsequently, 5 μg of RNA was incubated with 10 mM deoxynucleoside triphosphate mixture and 0.5 μg of oligo(dT) primer in a total volume of 10 μl for 5 min at 65°C. After 1 min on ice, the mixture was pooled with 9 μl of RT reaction mixture (2 μl 10× RT buffer, 100 mM MgCl2, 200 mM dithiothreitol, and 40 U of RNase Out) in a PCR tube. Prior to the RT-PCR run, the mixture was incubated at 42°C for 2 min and 50 U of SuperScript II Reverse Transcriptase (Invitrogen) was added. RT-PCR was performed at 42°C for 50 min, followed by 15 min at 70°C. To amplify the double-stranded cDNA of interest, 500 ng of single-stranded cDNA was mixed with corresponding primer pairs (see above; 15 pmol each), 10 mM deoxynucleoside triphosphate mixture, 5 μl 10× buffer 2, and 2.5 U of Pol mixture (Expand Long Template PCR System; Roche) in a total volume of 50 μl. The PCR was performed at 95°C for 120 s and 30 cycles, each cycle consisting of at 95°C for 30 s, 65°C for 60 s, and 68°C for 90 s. The last step was 68°C for 10 min. The final full-length PCR product was purified, subcloned into a pCR2.1 TA-cloning vector (Invitrogen), and verified by DNA sequencing.
Oligonucleotides used with the site-directed mutagenesis kit (Stratagene).
The oligonucleotides were as follows: SDM1, 5′-CTTCACCTGTTTCTTTTCCTCTTTCAATACTCGAGCTAGAGCTTTTTAAATTGC-3′; SDM2, 5′-GCAATTTAAAAAGCTCTAGCTCGAGTATTGAAAGAGGAAAAGAAACAGGTGAAG-3′; SDM3, 5′-GCCAAATCAAGCCACTATTAAAATCTCGAGTTACTTCCTTTTATTAATTTTCTC-3′; SDM4, 5′-GAGAAAATTAATAAAAGGAAGTAACTCGAGATTTTAATAGTGGCTTGATTTGGC-3′; SDM5, 5′-CAAAGAAGATTGGTTCTAGGACCACGAATTCGCTGCCTCCACAGATACCAAAATC-3′; and SDM6, 5′-GATTTTGGTATCTGTGGAGGCAGCGAATTCGTGGTCCTAGAACCAATCTTCTTTG-3′.
Oligonucleotides for electrophoretic mobility shift assay (EMSA) studies.
The oligonucleotides were as follows: sE7fw, 5′-CAAGCCACTATTAAAATGGTGGTTTACTTC-3′; sE7re, 5′-GAAGTAAACCACCATTTTAATAGTGGCTTG-3′; sE26r, 5′-TGTGGAGGCAGCGGGATGGTGGTCCTAGAA-3′; E0cf, 5′-TCACCTGTTTCTTTTCCTCTTTCAATATGGCTACTAGAGCTTTTTAAATT-3′; E0cr, 5′-AATTTAAAAAGCTCTAGTAGCCATATTGAAAGAGGAAAAGAAACAGGTGA-3′; M4fw, 5′-CAAGCCACTATTAAAATCTCGAGTTACTTC-3′; M4re, 5′-GAAGTAACTCGAGATTTTAATAGTGGCTTG-3′; Pmufw, 5′-GGCCTTTTCCTCTGTCATTTTCTCTTGATC-3′; Pmure, 5′-GATCAAGAGAAAATGACAGAGGAAAAGGCC-3′; Phufw, 5′-GAAACTACTAAAATGTAAATGACATAGGAA-3′; and Phure, 5′-TTCCTATGTCATTTACATTTTAGTAGTTTC-3′.
Oligonucleotides for cDNA amplification.
The oligonucleotides were as follows: mYY2fw, 5′-ATGGCCTCTGAGACAGAGAAACTTCTGTG-3′; mYY2r1, 5′-GAGAGCATGCTATAGGTCTTGGAAAGTCTC-3′; mYY2r2, 5′-CTCGGCCCATGGATGTGCAAGTGCTTTCTC-3′; mYY2r3, 5′-CTCCAGTATGGATTCGCACATGTGTGCGCA-3′; and mYY2r4, 5′-TTACTGGTCATTCTTGTTCTTAACATGGG-3′.
Cell culture and transfection.
Mouse LMTk− cells and human MG63 cells were cultivated at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Indicated amounts of expression plasmid DNA were transiently transfected using lipofection reagent Metafectene (Biontex) according to the manufacturer's protocol. If possible, transient expression was verified by Western blot analysis.
Stable clone mixtures.
Murine LMTk− cells were electroporated with appropriate amounts of linearized reporter gene constructs to generate a multiplicity of single-clones (≥50/construct) with a low number of stably integrated copies (4). At 24 h after transfection, the medium was exchanged for G418-containing selection medium (800 to 1,000 μg/ml). Dead cells were removed every 2 days by the addition of fresh selection medium. After 7 to 10 days, single clones were counted, trypsinized, pooled, and plated onto one suitable tissue culture plate to obtain the stable clone mixture. These cells were subjected to virus induction experiments as soon as possible to ensure homogeneity of the clone mixture.
Virus induction.
Experiments were carried out with Newcastle disease virus (NDV). A total of 2.5 × 105 cells were seeded onto six-well plates. After 48 h, the cells were washed three times with phosphate-buffered saline and three times with serum-free DMEM. After removal of the medium, 1 ml of NDV suspension (1:1,600 in serum-free DMEM) was added to the cells, which were then incubated for 1 h at 37°C. Virus induction was terminated by three washes with serum-containing DMEM. After the final washing step, cells were incubated in fresh serum-supplemented DMEM for an additional 24 h before virus-inducible IFN-β promoter activity was measured.
Luciferase assay.
Firefly luciferase activity was determined by using the luciferase assay system and 5× passive lysis buffer (Promega) according to the manufacturer's instructions and normalized for total protein content by bicinchoninic acid reaction.
IFN test.
Human osteosarcoma MG63-cells (6) were treated with NDV (see above) to induce IFN-β production. At 24 h after virus induction, the supernatants were harvested to determine the titer of secreted IFN-β. A microtiter plate with 103 Vero cells/well was cultivated for 24 h to form a confluent monolayer. Supernatants to be tested and an IFN-β standard (500 U/ml) were added to the first row. Subsequent rows contained 1:1 dilutions. After 24 h, the medium was replaced by 100 μl vesicular stomatitis virus (VSV) suspension (DMEM plus 5% fetal calf serum and VSV, 1:30,000). The next day lysis of unprotected cells was monitored under the microscope. A subsequent staining with crystal violet (50-g/liter crystal violet, 8.5-g/liter NaCl, 143 ml 37% formaldehyde, 500 ml ethanol) was used to detect living cells. The units of secreted IFN-β within the supernatants were calculated relative to an IFN-β standard.
Yeast one-hybrid system; construction of target reporters and yeast reporter strains.
The YY1/YY2 target site reporter constructs were prepared by insertion of a monomer of the sE7 motif into the pLacZi vector (Clontech). Two antiparallel oligonucleotides were synthesized, one representing the sense strand and the other representing its antisense complement. The oligonucleotides were annealed at 94°C for 5 min, followed by slow cooling to room temperature. The annealed fragments were fused to the pLacZi vector linearized with SmaI. The structure of the fusion construct was confirmed by sequencing. To prepare a reporter yeast strain after linearization with NcoI (motif sE7 monomer-pLacZi), the target reporter constructs were integrated into the ura3 gene of yeast strain YM4271 (Saccharomyces cerevisiae) using the Matchmaker one-hybrid system protocol (Clontech) and Yeast Protocols handbook (Clontech). Colonies that grew onto SD/uracil plates, were selected and replated at least twice onto fresh SD/uracil plates. To check the background expression of the constructed reporter strains, a β-galactosidase filter assay was performed. If the lifted colony turns blue within 15 min, the background lacZ expression is too high. However, all colonies of the strains tested turned weakly blue after 6 h, indicating that background lacZ expression in the constructed reporter strains was low. This reporter strain was termed YM4271LacZisE7.
(i) Verification of DNA-protein interactions.
To test sE7-YY1 and YY2 interactions, both cDNAs were cloned into the pGAD424 vector (YY1, SalI/BglII; YY2, EcoRI/SalI), encoding YY1 and YY2 activation domain fusion proteins. These constructs were subsequently transiently transformed into the reporter strain YM4271LacZisE7 using the Matchmaker one-hybrid system protocol (Clontech) and the Yeast Protocols handbook (Clontech). After approximately 3 days, the grown yeast clones were verified by a β-galactosidase colony lift filter assay.
(ii) β-Galactosidase colony lift filter assay.
Yeast clones were grown for 3 days and assayed for β-galactosidase activity. The yeast cells were lifted onto a Porablot NCL membrane (Macherey-Nagel, Dueren, Germany), permeabilized by immersion in liquid nitrogen for 20 s, thawed at room temperature for several minutes, and placed onto a sheet of filter paper (Macherey-Nagel, Dueren, Germany) that was soaked with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol) containing X-Gal (5-bromo-4 chloro-3-indolyl-d-galatopyranoside; 1 mg/ml). Afterwards, the filter was incubated at 30°C until colonies turned blue.
In vitro translation.
Human FLAGYY2 and muYY2 has been in vitro translated by using the TNT Coupled Transcription/Translation system (Promega) according the manufacturer's protocol. The corresponding expression plasmids pcDNA3.1(−)FLAGYY2 and pcDNA3.1(−)muYY2 used in this protocol utilizes a T7 promoter.
Nuclear extract.
Nuclear extracts from human MG63 cells were prepared using the Nuc Buster protein extraction kit from Novagen (Madison, Wis.) according to the manufacturer's protocols.
EMSA.
Synthetic single-stranded oligonucleotides were radioactively end labeled with T4 polynucleotide kinase and [γ-32P]ATP and subsequently purified via microspin G-50 columns (Amersham Biosciences, United Kingdom). Double-stranded probes were prepared by annealing equal amounts of the complementary single strands. Nuclear extracts (each, 10 μg) or in vitro-translated proteins were added to 25-μl reaction mixtures in binding buffer containing 10 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM KCl, 0.005% xylene cyanol (wt/vol), 0.005% bromophenol blue (wt/vol), 2% Ficoll (wt/vol), 1 μg poly(dI-dC) (D-68305; Roche Diagnostics GmbH Mannheim) in the presence of a 200× molar excess of a nonspecific single-strand competitor and incubated with 50 fmol 32P-end-labeled single- and double-stranded probe mixtures at 37°C for 30 min.
For supershift experiments, 1 μg of antibodies (anti-YY1, sc-281, sc-1703, anti-Oct-1, and sc-8024x) were added to the protein mixture and incubated at room temperature for 30 min prior to addition of the probe. Reaction mixtures were fractionated by electrophoresis on an 8% nondenaturing acrylamide gel at 140 V for 2 to 3 h in 0.25× Tris-buffered EDTA buffer at room temperature. Gels were dried and subjected to autoradiography on phosphorimager plates.
RESULTS
The human IFN-β gene domain extends over 14 kb on the telomeric end of a gene cluster with 26 IFN-α/β genes and pseudogenes on the short arm of chromosome 9 (9). The domain boundaries were first characterized biochemically by their scaffold/matrix association potentials (8). Subsequently, we could show that these scaffold/matrix attachment regions coincided with destabilized regions in the SIDD profile. An SIDD profile reflects the energy required to separate DNA strands at a given superhelical tension (5). In addition to these regions, three isolated, very restricted ∼250-bp-wide signals were detected within the huIFN-β domain, which coincide with prominent DNase I hypersensitive sites (HS1, HS2, and HS3) (Fig. 1) (21). One of these (HS3) marked a fragile genomic site next to the promoter (9). Sites HS2 and HS3 flanked a register of six nucleosomes that are positioned in the absence of gene activity. Site HS1 marks a site next to the region of increased scaffold/matrix association potential (19).
Before the onset of transcription, not only the proximal nucleosomes (II and I according to the above nomenclature) become mobilized but also the upstream member of the six-nucleosome array next to HS2. These observations and early findings from our laboratory, which have shown the influence of this region on the induction of IFN-β (20), led to the identification of HS1- to HS3-associating factors (19).
Factors associating with sequences at destabilized sites.
Systematic screens for DNA-binding proteins via EMSA revealed at least two double-stranded and three single-stranded DNA-binding proteins associating with HS1, HS2, and HS3. In our previous contribution (19), we demonstrated that a multifunctional transcription factor, YY1, has specific binding sites adjacent to minima in the SIDD profile, i.e., at the downstream flank of HS1 and the upstream flank of HS2, which both share an ATGG core recognition motif. We have found that these are the only strong YY1 sites within 5 kb of the huIFN-β upstream region, suggesting an evolutionary conservation of both the consensus sequence and the strand separation potential. This conservation may indicate that such a coincidence is a prerequisite for structural distortion and thereby for factor binding (25, 27).
During our studies, it became apparent that these properties may be of a more general relevance, since we have traced factors in addition to YY1 that comply with these criteria (19). Among these are the ubiquitous factor Oct-1 (Fig. 1) and at least three more factors that bind to a single strand probe (sE26re; see Materials and Methods). The similarity of this sequence with PUR (the purin-rich element-binding protein, which prefers a single purine-rich strand for binding), especially the repetitive occurrence of GGN (N = A, C, and T) tracts, led to the identification of PURα/β complexes by supershift analyses with specific antibodies (19).
We wanted to focus the present study on factors with obvious relevance for the induction process, and therefore we prepared the constructs depicted in Fig. 2 with mutations in the HS1 and/or HS2 recognition sequences for YY1, as well as in the HS3-associated binding site for the UF complex. To this end, we introduced five- to sixfold point mutations, which abolished binding according to an EMSA test (19). Figure 3A shows the effect of a 5-bp mutation in HS2 (ttaaaATctcgag in place of ttaaaATGGtggt; lowercase letters are base pairs around or substituted base pairs within the ATGG core motif) on the activity of a luciferase reporter at comparable copy number levels (Fig. 3B), i.e., it compares the induced level of gene activity for the stably expressed constructs P, P-C, and PCHS2mut. For construct P, the reporter was controlled by the minimal interferon promoter (up to the EcoRI site at position −284), for P-C by an intact 4.5-kb upstream control region and for P-CHS2mut by the same region containing the HS2 mutation. This experiment demonstrates a fivefold difference of induced expression levels between P-C and P-CHS2mut, hinting at a significant (positive) influence of the associating factor(s). It is remarkable that the effect of deleting the entire upstream sequence was reproducibly smaller, and it is concluded that in this range we have to expect positive as well as negative cis-acting elements.
FIG. 2.
Luciferase reporter constructs used to demonstrate the existence of far-upstream control mechanisms. Construct P is restricted to IFN-β promoter sequences down to position −281. All other constructs contain the ∼4.2-kb EcoRI-C upstream fragment in addition. Construct P-C comprises the intact upstream region, whereas the bottom four constructs contain base pair substitutions in the marked positions, which coincide with the SIDD minima HS1, HS2, and HS3. Mutants P-CHS1mut and P-CHS2mut represent single mutants of either of the upstream YY1/YY2-binding sites. P-CHS1/HS2mut is a construct with mutations at both positions. For P-CHS3mut, the site for the undetermined factor (UF complex) (Fig. 1) in the promoter-proximal site HS3 has been modified to elucidate its potential contribution to the induced expression level of IFN-β.
FIG. 3.
The influence of upstream sequences on huIFN-β promoter induction in stable clone mixtures. (A) Consequences of a deletion (P) and a mutation in HS2 (P-CHS2mut). The construct with the intact −4.5-kb control sequence (P-C) shows a maximum induced expression which is impaired significantly (fivefold) due to a 5-bp mutation (P-CHS2mut; ttaaaATctcgag in place of ttaaaATGGtggt). For the minimal construct (P), the induction level is reduced only twofold, demonstrating that the extended upstream region contains both positive and negative regulatory elements. All values have been normalized to the basal construct, which was set at a value of 100; this procedure was maintained for all subsequent measurements. Standard deviations of at least three independent experiments are indicated. (B) Copy number control (Southern blots) for stable integrants with a luciferase probe. All signals in lanes 1 to 6 refer to equal amounts of genomic, high-molecular-weight DNA. Lanes 4 (P), 5 (P-C), and 6 (P-CHS2mut) refer to genomic DNA from the stable clone mixtures tested under the experiments shown in panel A. Controls: lanes 1 and 2, genomic DNA from an independent experiment on cells previously transfected with either P or P-C, respectively; lane 3, genomic DNA from nontransfected cells; lane 7, plasmid DNA. The radioactive signals correspond to a Sca I-fragment of approximately 2.4 kb of the reportergene constructs. A DNA standard as well as relative densiometric values, as percentages of the integrated copies, are indicated.
In a more extended series of experiments, we investigated the relative importance of mutations in HS1, HS2, and HS3. Data shown in Fig. 4 reproduce the relative expression of P-CHS2mut and demonstrate that the same reduction of induced levels of gene activity is also found for P-CHS1mut and for a mutation in both HS1 and HS2 (construct P-CHS1/HS2mut). These results show that both sites together are required to constitute a strong (or stronger) activation of an IFN-β promoter. A mutation in HS3 was without major effects. We therefore dedicated further work to the factors associating with the former two sites.
FIG. 4.
Relative influence of sites HS1, HS2, and HS3 on the induction process. All experiments refer to a series of transfections similar to but independent from the results shown in Fig. 3. The three left-hand experiments reproduce the above results. The subsequent two experiments demonstrate an effect similar to P-CHS2mut either for mutations in HS1 (P-CHS1mut) or for a simultaneous double mutation in HS1 and HS2 (P-CHS1/HS2mut). Construct P-CHS3mut demonstrates that the factor underlying the UF-complex in Fig. 1 is not essential for the induction of the IFN-β promoter.
Hypersensitive sites HS1 and HS2 can accommodate a YY1-related factor, YY2.
YY1 has been described as a factor with both positive and negative effects on transcription. This diversity of actions has mostly been ascribed to the recruitment of proteins involved in the turnover of histone acetyl groups, i.e., HATs and HDACs (32), through the YY1 C-terminal Zn finger domain (34). An activating function on the muIFN-β promoter became apparent in case both sites at positions −90 and −122 were intact (32). In contrast, mutations at either site (mut122 or mut90) displayed reduced virus-induced activities starting shortly after infection.
Following this concept, a straightforward explanation for the repression of induced activity levels of P-CHS1/HS2mut constructs in Fig. 4 would be the simultaneous binding of YY1 by HS1 and HS2 sequences. An alternative explanation emerged when Nguyen et al. (26) reported the identification of a human cDNA encoding a protein called YY2 with a significant homology (86.4%) to YY1 in its zinc finger region. Zn fingers form the protein domain by which both HATs and HDACs are recruited to YY1. They also direct YY1 to their DNA-binding site, which explains the observation that YY2 can bind to some (although not all) promoters that have been reported to be subject to regulation by YY1 (26). Both factors may therefore compete for the enzymes involved in the turnover of lysine-linked acetyl groups. Figure 5A shows that common polyclonal anti-YY1 antibodies do in fact cross-interact with YY2 (lanes 5 and 6), making it possible that various actions that have so far been ascribed to YY1 are actually the consequence of YY2 binding.
FIG. 5.

Interaction of YY2 with HS1 and HS2: EMSA and yeast one-hybrid experiments. (A) The effect of in vitro-translated FLAGYY2 on HS1- and HS2-derived DNA sequences (E0c and sE7,) in an EMSA. Lanes 2 and 4 show a distinct DNA-FLAGYY2 complex, whereas lanes 1 and 3 (negative control; in vitro-translated luciferase) are blank. Lanes 5 and 6 show blocking/supershift assays with common polyclonal anti-YY1 antibodies sc-281 (Santa Cruz) (5) and sc-1703 (6) in experiments corresponding to the results shown in lane 4. The SIDD profile marks the positions of the EMSA probes (arrowheads), which are seen to coincide with the flanks of the destabilized regions. (B) Yeast cells were stably transformed with the sE7 30mer (YY1/YY2-binding site of HS2). YY2 or YY1 binding was tested by transient transformation with pGAD424YY2 (a) and pGAD424YY1 (b). (c) Negative control (pGAD424). The figure reflects the results of a lift filter assay where the β-galactosidase signals (dark dots) in panels a and b indicate interaction of the 30mer with both YY proteins.
These experiments have been extended in Fig. 5 by EMSA and one-hybrid experiments using human FLAGYY2 produced in an in vitro translation system. The results clearly demonstrate that the sites at HS1 and HS2 can both accommodate YY2 in place of YY1; this is supported by the analyses shown in Fig. 5B, which demonstrate activity of either YY1 or YY2 derivatives in a yeast one-hybrid approach. We therefore decided to study the relative effects of both factors on reporter genes that have been linked to the huIFN-β control region.
Opposing actions of YY1 and YY2 in a stable reporter gene assay.
By destroying the ATGG tracts in HS1 and HS2, induction of the luciferase reporter via the huIFN-β promoter was severely impaired (Fig. 3 and 4). In principle, this would be in line with the loss of a positive regulatory factor. The experiments shown in Fig. 6A and 6B, however, are explained more easily by the YY1/YY2 competition model introduced above.
FIG. 6.
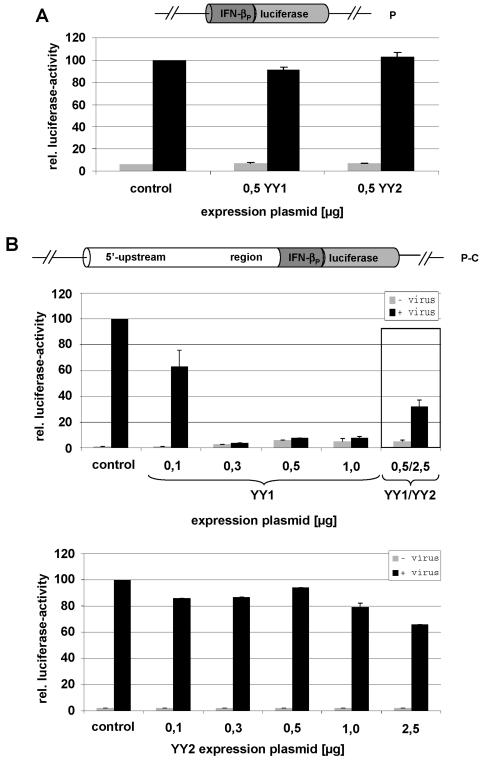
Effects of YY1 and YY2 overexpression on the induction of minimal (P) and extended (P-C) constructs stably transfected into LMTk− cells. (A) Both factors do not act upon the minimal promoter. Clone mixtures were supertransfected with the indicated amounts of either YY1 or YY2 expression plasmid, respectively. Neither YY1 nor YY2 overexpression influences the promoter activity of the minimal construct. All values have been normalized to the control experiment, which was set at a value of 100; this procedure was maintained for all subsequent measurements. Standard deviations of at least three independent experiments are indicated. (B) Mutual influences of YY1 and YY2 on the induction of reporter genes under the control of an extended upstream region (P-C). While the maximum effect of YY1 is already attained at 0.3 μg of transgene, YY2 alone appears to be without an effect throughout the concentration range (0.1 to 1 μg or 2.5 μg, respectively, expression plasmid). In a coexpression situation (YY1 plus YY2) (experiment shown in a separate box), however, YY2 clearly reduces the negative effect of YY1, indicating the competition of both factors for a single binding site.
We used murine cells that stably integrated either the minimal promoter construct (P), the functional (nonmutated) construct P-C, or the mutated variants P-CHS2mut and P-CHS1/HS2mut. For these constructs, we determined the effect of YY1- and YY2-overexpression on the induced activity of the luciferase reporter. While expression under the control of the minimal promoter (Fig. 6A) and for the mutants (not shown) remains unaffected in the presence of elevated levels of either YY1 or YY2, YY1 exerts a significant negative effect in the presence of the intact far-upstream region (Fig. 6B). This effect increased significantly if the concentration of the expression plasmid was raised from 0.1 to 1 μg, suggesting that the results of our mutagenesis study (Fig. 3 and 4) cannot be ascribed solely to YY1. Starting at 0.3 μg, higher cellular concentrations of YY1 even resulted in a slight derepression of the promoter, meaning that the base level of expression was elevated while, at the same time, a subsequent induction by virus became less and less effective.
If YY2 was overexpressed alone, base levels and induction factors were indistinguishable from those of the control (Fig. 6B). This could mean that endogenous levels of (murine) YY2 are sufficient to support the induction process and that its overexpression has no negative consequences. Finally, both factors were expressed simultaneously, and it was found that an excess of YY2 reduced the negative effect that was otherwise observed in the presence of YY1 (Fig. 6B, box).
While this interpretation appeared plausible, it still had to be proven that murine cells do, in fact, express an orthologue of the human YY2 protein. In fact, both humans (Xp22.1-p22.2) and mice (XF4) contain related YY2-encoding exons within the MBTPS2 coding region (accession numbers XP_355849 [human] and NP_839997 [mice], NCBI). We generated single-stranded cDNAs from the total RNA of murine NIH 3T3 cells by RT-PCR, which were used as templates to amplify YY2 specific PCR products of the expected extension (Fig. 7A). The longest of these fragments was used for in vitro translation of muYY2 in parallel to human FLAGYY2. Both proteins yielded comparable signals in EMSAs (Fig. 7B).
FIG. 7.
Cloning and functional verification of the murine YY2 protein. (A) PCR fragments of the murine YY2 cDNA with different sets of primer pairs. RT-PCR generated single-stranded cDNAs derived from total RNA of murine NIH 3T3 cells were used as templates to amplify YY2-specific PCR products. Four distinct combinations of primer pairs were analyzed to ensure the correct amplification of YY2 DNA sequences corresponding to the published data of a cDNA originally designated as membrane-bound transcription factor protease, site 2 (MBTPS2, accession no. NP_839997 or NM_178266). 5′ primer (mYY2fw) plus lane 2, 3′ primer mYY2r1; lane 3, mYY2r2; lane 4, mYY2r3; and lane 5 (full length), mYY2r4. Lane 1 contains a 1-kb DNA ladder (Invitrogen). (B) DNA-binding capacity of in vitro-translated murine YY2, tested by EMSA. A nuclear extract of human MG63 cells (lane 1) was tested for sE7 binding, together with in vitro-translated YY2 from humans (lane 2; FLAGYY2) and mice (lane 3). Lane 4 (negative control) contains the in vitro translation mixture with the empty pcDNA3.1(−) plasmid.
So far, experiments have relied on the induction of a reporter gene under the control of the human IFN-β promoter as it occurs in a murine cell line (LMTk−) and under the control of the murine transcription machinery. Figure 8 shows that the negative effect exerted by YY1 can also be observed in an established human IFN-β producing cell line, i.e., the MG63 osteosarcoma line (6), for which the intact gene is in its native location. In this case, the induction process has been followed by a cytotoxicity test with the virus-protective action of secreted human interferon on a primate indicator cell line (Vero) (13).
FIG. 8.
The negative effect of YY1 is also valid for the endogenous IFN-β gene. For human MG63 cells, the IFN-β gene is induced by NDV subsequent to transfection of 0, 0.3, and 0.5 μg of YY1 expression plasmid (right-hand columns). Interferon levels have been determined via the protection of an indicator cell line (Vero) from the cytopathogenic effects of VSV.
YY2 sites in the muIFN-β promoter.
Both the human and the murine promoters contain equivalent YY1-binding sites in remote upstream positions (19). The functions of these sites were compared in ongoing experiments. On the other hand, in the huIFN-β promoter the proximal −90 and −122 YY1-binding sites that are present at the muIFN-β promoter are not conserved (19, 32). Figure 9 explores the possibility that the novel factor YY2 might contribute to YY1/YY2 interplay at the unique promoter-proximal muIFN-β sites. By an EMSA, we compared the binding of FLAGYY2 to the murine −90 site with the HS2-associated sequence (ds7; primer pair sE7fw/sE7re), first analyzed in the experiments shown in Fig. 5. These results extended earlier analyses for YY1 (19): the murine promoter (Pmu) but not the human promoter (Phu) contains a proximal site that bound YY2 in place of YY1. However, in this case binding was also much weaker than in the sites that correlated with the strong SIDD signals in the upstream region.
FIG. 9.
Interaction of YY2 with an IFN-β promoter-associated binding site in the murine gene (32). A radioactively labeled murine IFN-β probe (Pmu) including the YY1-binding site around position −90 (32) binds FLAGYY2 (lane 4); its position in the SIDD profile has been marked by an arrow. If compared with the signal from the HS2-specific probe (lane 1), its affinity appears largely reduced. Lane 2 shows an experiment performed with the ds7-mutant M4; and lane 3 represents an EMSA analogous to the data shown in lane 4 but using the corresponding sequence from the human IFN-β promoter (Phu).
DISCUSSION
Viral infection triggers a series of well-defined events at the IFN-β promoter that culminate in the remodeling of a positioned nucleosome overlapping the transcriptional start site. Most current models fully rely on the events that occur in the proximal VRE, which for the human gene is thought to terminate with PRD IV around position −110. Somewhat further downstream, a negative regulatory domain has been localized that is also contained in the EcoRI fragment extending to position −284. Nevertheless, dating back almost 2 decades, there are observations also indicating the existence of remote control mechanisms for the human IFN-β gene (IFNB1) (7, 20). These data indicate the induction-dependent modulation of a nucleosome residing at the downstream border of HS2 (7).
We have demonstrated specific binding sites for YY1 (exemplified by the human IFN-β control region) at positions −2 kb and −3 kb that coincide with flanks of the HS2 and HS1 imprints in the SIDD profile, respectively (19). Both sites contain almost identical aaATGGt motifs (lowercase letters are base pairs around or substituted base pairs within the ATGG core motif), which over 4.5 kb occur only at these positions. If either or both of these sites were destroyed by mutagenesis, the induced level of IFN-β decreased fivefold, which seemed to indicate the loss of an activating factor DNA-binding site (Fig. 3A and 4). Although YY1 may have both activating and repressing influences depending on site occupancy (32), overexpression of YY1 led to a consistent decrease in induced IFN-β levels both for transfected reporter constructs (Fig. 6B) and for the endogenous IFN-β gene in human MG63 osteosarcoma cells (Fig. 8).
Differences between the IFN-β genes in humans and mice (huIFN-β versus muIFN-β).
For both the human and the murine IFN-β genes, the enhanceosome has to accommodate the three mentioned sets of transcription factors, p50/p65 (NF-κB), IRF3/7, and ATF-2/c-Jun (AP1). In case of the human enhancer, the architectural protein HMG I(Y) is also involved (28): two molecules, each using two out of its three AT hooks for DNA binding, cause modifications enabling the cooperative association of transcriptional activators within the enhanceosome (35). There is no HMG I(Y) binding to the VRE region of the murine promoter, however (10). Instead, a specific HMG I(Y)-binding site was found at position −130 between the VRE and NRD II.
The murine VRE contains a potential YY1/YY2-binding site in its PRD IV part and another site at position −122 close to the HMG I(Y) site (32), which are not present in the human case (Fig. 10) (19). Whether or not binding of YY1/YY2 is possible in addition to ATF-2 is an open question. Similarly, in human VRE there is an overlap of ATF-2- and Oct-1-binding sites (Fig. 10) (19).
FIG. 10.
The established proximal IFN-β promoters from both humans and mice contain additional (but different) factor-binding sites in the PRD IV element (base pairs are indicated relative to the transcriptional start site). For the murine promoter, the ATF-2/c-Jun DNA element overlaps with a YY1/YY2-binding site (for a discussion of the underlined letters, see reference 32) For the human promoter, the corresponding ATF-2/c-Jun element overlaps with an Oct-1 site (underlined letters). The presence of a functional Oct-1 site within the human IFN-β promoter sequence has been confirmed by EMSA-supershift analysis (see small inset image) using Phu as a probe. +, the addition of an appropriate monoclonal anti-Oct-1 antibody (sc-8024; Santa Cruz); the arrow indicates the Oct-1-specific shift; the asterisk marks the corresponding supershifted DNA-protein complex.
An activating role was originally suggested for YY1, since one of its actions is the recruitment of GCN5, by which it may function as a transcriptional activator shortly after infection. A functional switch has been correlated with CBP/p300, which induces sliding of nucleosome II enabling formation of the preinitiation complex and gene transcription, before a shutoff is triggered by the CBP/p300-mediated acetylation of YY1. This modification leaves its DNA-binding capacity intact but restores its interaction with HDACs.
The discovery of YY2 introduced an additional level of complexity to the interpretation of YY1 actions. Since antibodies, thus far considered to be specific for YY1, cross-interact with YY2 (Fig. 5A), more attention will have to be devoted to the relative role of these factors. Overexpression of YY2 increases p53 as well as c-FOS promoter activity and c-MYC or CXCR4 promoter activity, in case it is expressed at moderate levels (26). Together with the finding that small interfering RNA knockdown of endogenous YY2 reduces the activity of all four promoters, these observations suggest that, in essence, YY2 is a transcriptional activator (26; E. Seto, personal communication). Our data add IFN-β to the series of candidate genes whose expression depends on the interplay between YY2 (activation) and YY1 (shutoff).
Similar to YY1, YY2 contains both transcriptional activation and repression domains. However, since YY2 does not contain an acidic domain, the mechanism must be different for both proteins. For YY1, the HAT association region has been localized to the C-terminal Zn finger region (34), which is highly conserved between YY1 and YY2. Future studies will therefore address the question of to what extent the activating role originally assigned to YY1 can be ascribed to YY2.
The actions of YY1 and YY2 on the huIFN-β promoter require the intact upstream region including HS1 and HS2 (construct P-C) (Fig. 3A, 4, and 6B). This is suggested by the fact that the minimal construct P remains unaffected by overexpression of either factor (Fig. 6A). Yet we had to prove that there are no other upstream sites for which YY1 or YY2 have an indirect effect, for instance, by titrating away (“squelching”) factors with a different specificity. The observation that expression from constructs P-CHS1/HS2mut and P-CHS2mut (Fig. 2) also remains unaffected by elevated levels of either YY1 or YY2 demonstrates that, in fact, HS1 and HS2 are the only sites that are involved in this aspect of regulation (data not shown). EMSAs clearly demonstrate that the same sequences that accommodate YY1 interact equally with YY2 (Fig. 5A and 7) in support of the YY1/YY2 competition model. Finally, it must be noted that overexpression of YY2 alone leaves the properties of the endogenous situation unchanged both regarding the repressed state (basal level of expression) and the induction factor after viral infection (Fig. 6B). In contrast, YY1 interferes at higher levels with both the repressed state and the induction process as demonstrated in the data shown in Fig. 6B. It is concluded that both factors act by distinct mechanisms and that the postulated activation by endogenous YY2 is mechanistically different from the derepression (and loss of virus inducibility) that becomes visible at higher concentrations of YY1.
YY1 was originally described as nuclear matrix protein 1 (NMP-1) by Guo et al. (12), i.e., a nuclear matrix-associated DNA-binding factor with sequence-specific recognition of a regulatory element next to a histone H4 gene. The finding that NMP-1 is YY1 suggests that this transcriptional regulator may mediate gene-matrix interactions. McNeil et al. (23) have shown that the C-terminal domain (amino acids 201 to 414) is necessary for this interaction, while the N terminus (amino acids 1 to 256) has at most a weak contribution. Investigations by Bushmeyer et al. (11) localized a nuclear matrix targeting signal in the segment of amino acids 256 to 340 within the Zn finger region, a domain already known to mediate binding to DNA, HAT, and HDAC (both enzymes being components of the nuclear matrix (14, 15). It appears likely that at least some of these interactions are mutually exclusive; therefore, it will be of interest to determine how the association of YY1 with its binding partners is regulated and what role YY2 plays in this circuitry.
The precise colocalization of factor-binding sites with sites of DNA strand-unpairing potential (SIDD minima) (Fig. 1) deserves particular attention, since it may indicate the conservation of this property together with a potent binding motif (19). While PURα/β are single-strand binding proteins that will prefer such a situation for obvious reasons, YY1 (and likewise YY2, Oct-1, and as-yet-unidentified factors) requires both DNA strands for binding. They may also profit from a flexible DNA backbone, since for YY1 there are intriguing observations that relate at least some actions of YY1 to its DNA-bending potential (25, 27). The direction of the bend, in turn, may determine whether YY1 acts as an activator or repressor (30). Both parameters, DNA-bending potential (3, 16, 31) and strand separation potential (5) are not only functionally related but are also properties that have been associated with scaffold/matrix attachment regions. Since it is assumed that at least 7% of all vertebrate promoters have YY1 sites, it will be rewarding to assign certain or all these regulatory aspects to their function.
Acknowledgments
We are grateful to Ed Seto (University of South Florida/Tampa) for materials, encouragement, and advice and to Peter Mueller (GBF) for his help with database researches. Particular thanks go to Craig Benham and A. K. Prashanth (UC Davis Genome Center) for continuous discussions on DNA structure/function relationships and for performing the relevant calculations (19).
This work was supported by grants from Deutsche Forschungsgemeinschaft (BO 419-6), BMBF (DHGP I/II), and INTAS (011-0279).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. N. 1986. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 21:8513-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, A., D. Schübeler, and J. Bode. 2000. Transcriptional properties of genomic transgene integration sites marked by electroporation or retroviral infection. Biochemistry 39:7041-7049. [DOI] [PubMed] [Google Scholar]
- 5.Benham, C., T. Kohwi-Shigematsu, and J. Bode. 1997. Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J. Mol. Biol. 272:181-196. [DOI] [PubMed] [Google Scholar]
- 6.Billiau, A., V. G. Edy, H. Hermans, J. van Damme, J. Desmyter, J. Georgiades, and P. de Somer. 1977. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob. Agents Chemother. 12:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode, J., H. J. Pucher, and K. Maaβ. 1986. Chromatin structure and induction-dependent conformational changes of human interferon-β genes in a mouse host cell. Eur. J. Biochem. 158:393-401. [DOI] [PubMed] [Google Scholar]
- 8.Bode, J., and K. Maaβ. 1988. The chromatin domain surrounding the human interferon-β gene as defined by scaffold-attached regions (SARs). Biochemistry 27:4706-4711. [DOI] [PubMed] [Google Scholar]
- 9.Bode, J., C. Benham, E. Ernst, A. Knopp, R. Marschalek, R. Strick, and P. Strissel. 2000. Fatal connections: when DNA ends meet on the nuclear matrix. J. Cell. Biochem. Suppl. 35:3-22. [DOI] [PubMed] [Google Scholar]
- 10.Bonnefoy, E., M-T. Bandu, and J. Doly. 1999. Specific binding of high mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol. 19:2803-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushmeyer, S. M., and M. L. Atchison. 1998. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem. 68:484-499. [PubMed] [Google Scholar]
- 12.Guo, B., P. R. Odgren, A. J. van Wijnen, T. J. Last, J. Nickerson, S. Penman, J. B. Lian, J. L. Stein, and G. S. Stein. 1995. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc. Natl. Acad. Sci. USA 92:10526-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser, H., G. Gross, W. Bruns, H. K. Hochkeppel, U. Mayr, and J. Collins. 1982. Inducibility of human β-interferon gene in mouse L-cell clones. Nature 297:650-654. [DOI] [PubMed] [Google Scholar]
- 14.Hendzel, M. J., G. P. Delcuve, and J. R. Davie. 1991. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 266:21936-21942. [PubMed] [Google Scholar]
- 15.Hendzel, M. J., J. M. Sun, H. Y. Chen, J. B. Rattner, and J. R. Davie. 1994. Histone acetyltransferase is associated with the nuclear matrix. J. Biol. Chem. 269:22894-22901. [PubMed] [Google Scholar]
- 16.Homberger, H. P. 1989. Bent DNA is a structural feature of scaffold-attached regions in Drosophila melanogaster interphase nuclei. Chromosoma 98:99-104. [DOI] [PubMed] [Google Scholar]
- 17.Huang, S. 1994. Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF1. Cell 78:9. [DOI] [PubMed] [Google Scholar]
- 18.Klar, M. 2005. Strukturgebende Signale im eukaryontischen Genom—Vorhersage und Verifizierung. Dissertation. Technical University of Braunschweig, Braunschweig, Germany. [Online.] http://opus.tu-bs.de/opus/volltexte/2005/713/pdf/DissMK.pdf.
- 19.Klar, M., E. Stellamanns, P. A. K., A. Gluch, and J. Bode. Dominant genomic structures: detection and potential signal functions in the interferon-beta domain. Gene, in press. [DOI] [PubMed]
- 20.Klehr, D., K. Maaβ, and J. Bode. 1991. Scaffold-attached regions from the human IFN-β domain can be used to enhance the expression of genes under the control of various promoters. Biochemistry 30:1264-1270. [DOI] [PubMed] [Google Scholar]
- 21.Knopp, A. 2001. Studien zur Struktur von S/MARs am Beispiel einer genomischen Domäne aus dem humanen Interferon-Gencluster. Dissertation. Technical University Braunschweig, Braunschweig, Germany. [Online.] http://opus.tu-bs.de/opus/volltexte/2001/200/pdf/Dissertati.PDF.
- 22.Lopez, L., R. Reeves, M. L. Island, M. T. Bandu, N. Christeff, J. Doly, and S. Navarro. 1997. Silencer activity in the interferon-A gene promoters. J. Biol. Chem. 272:22788-22799. [DOI] [PubMed] [Google Scholar]
- 23.McNeil, S., B. Guo, J. L. Stein, J. B. Lian, S. Bushmeyer, E. Seto, M. L. Atchison, S. Penman, A. J. van Wijnen, and G. S. Stein. 1998. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking, J. Cell. Biochem. 68:500-510. [PubMed] [Google Scholar]
- 24.Munshi, N., J. Yie, M. Merika, K. Senger, S. Lomvardas, T. Agalioti, and D. Thanos. 1999. The IFN-β enhancer: a paradigm for understanding activation and repression of inducible gene expression. Cold Spring Harb. Symp. Quant. Biol. 64:149-159. [DOI] [PubMed] [Google Scholar]
- 25.Natesan, S., and M. Z. Gilman. 1993. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 7:2497-2509. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, N., X. Zhang, N. Olashaw, and E. Seto. 2004. Molecular cloning and functional characterization of the transcription factor YY2. J. Biol. Chem. 279:25927-25934. [DOI] [PubMed] [Google Scholar]
- 27.Rothman-Denes, L. B., X. Dai, E. Davydova, R. Carter, and K. Kazmiercak. 1998. Transcriptional regulation by DNA structural transitions and single-stranded DNA bending motifs. Cold Spring Harbor Symp. Quant. Biol. 63:63-73. [DOI] [PubMed] [Google Scholar]
- 28.Schwanbeck, R., G. Manfioletti, and J. Wisniewski. 2000. Architecture of high mobility group protein I-C · DNA complex and its perturbation upon phosphorylation by Cdc2 kinase. J. Biol. Chem. 275:1793-1801. [DOI] [PubMed] [Google Scholar]
- 29.Shestakova, E., M. T. Bandu, J. Doly, and E. Bonnefoy. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 75:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 31.von Kries, J. P., L. Phi-Van, S. Diekmann, and W. H. Strätling. 1990. A non-curved chicken lysozyme 5′ matrix attachment site is 3′ followed by a strongly curved DNA sequence. Nucleic Acids Res. 18:3881-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill, L., E. Shestakova, and E. Bonnefoy. 2003. Transcription factor YY1 binds to the murine beta interferon promoter and regulates its transcriptional capacity with a dual activator/repressor role. J. Virol. 77:2903-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski, J. R., and R. Schwanbeck. 2000. High mobility group I/Y: multifunctional chromosomal proteins causally involved in tumor progression and malignant transformation. Int. J. Mol. Med. 6:409-419. [DOI] [PubMed] [Google Scholar]
- 34.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMG I(Y) in the assembly and function of the IFN-β enhanceosome. EMBO J. 18:3074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]