Abstract
p53 binding protein 1 (53BP1) is a putative DNA damage sensor that accumulates at sites of double-strand breaks (DSBs) in a manner dependent on histone H2AX. Here we show that the loss of one or both copies of 53BP1 greatly accelerates lymphomagenesis in a p53-null background, suggesting that 53BP1 and p53 cooperate in tumor suppression. A subset of 53BP1−/− p53−/− lymphomas, like those in H2AX−/− p53−/− mice, were diploid and harbored clonal translocations involving antigen receptor loci, indicating misrepair of DSBs during V(D)J recombination as one cause of oncogenic transformation. Loss of a single 53BP1 allele compromised genomic stability and DSB repair, which could explain the susceptibility of 53BP1+/− mice to tumorigenesis. In addition to structural aberrations, there were high rates of chromosomal missegregation and accumulation of aneuploid cells in 53BP1−/− p53+/+ and 53BP1−/− p53−/− tumors as well as in primary 53BP1−/− splenocytes. We conclude that 53BP1 functions as a dosage-dependent caretaker that promotes genomic stability by a mechanism that preserves chromosome structure and number.
p53 binding protein 1 (53BP1) was identified in a yeast two-hybrid screen as a protein that binds to the central DNA binding domain of the tumor suppressor protein p53 (15). It contains two tightly packed tudor domains and a C-terminal tandem BRCT motif (9, 19). BRCT domains are thought to be protein-protein interaction domains and are found in many DNA damage response proteins (5, 19). The observation that, upon exposure of cells to ionizing radiation (IR), 53BP1 rapidly localizes in a dose-dependent manner to discrete nuclear foci supported the proposed role of 53BP1 in the DNA damage response (2, 16, 25, 29, 36). Moreover, following irradiation, 53BP1 interacts with and becomes phosphorylated by ataxia telangiectasia mutated (ATM), a central kinase in the DNA double-strand break (DSB) response (2, 25, 36). Colocalization of 53BP1 foci with those of phosphorylated histone H2AX and the Mre-11/Nbs1/Rad50 complex mapped the IR-induced accumulation of 53BP1 to regions of DNA DSBs (2, 25, 29). Subsequent studies using small interfering RNAs indicated that 53BP1 is involved in DNA damage checkpoint control by facilitating the phosphorylation of ATM substrates like Chk2 or SMC1 (12, 31). Moreover, 53BP1 was reported to be required for p53 accumulation in response to IR (31).
Defects in DNA damage checkpoint control were also observed in cells derived from 53BP1 knockout mice (13); however, the more striking observations obtained with these mice were of sensitivity towards ionizing radiation and greatly impaired class switch recombination (CSR) combined with immunodeficiency and an increased lymphoma predisposition (23, 24, 33, 34). Since CSR involves the generation and rejoining of DNA DSBs, a function of 53BP1 in DSB repair has been proposed (23, 34). It is well established that defects in the joining of DSBs can lead to chromosome translocations and gene amplifications that promote tumorigenesis, particularly in the context in which p53-dependent apoptosis is abrogated (11, 37). In this study, we address how 53BP1 contributes to genomic stability by analyzing tumorigenesis in p53-null mice that express various amounts of 53BP1.
MATERIALS AND METHODS
Mice.
53BP1−/− mice (33) were bred with p53−/− mice of the same mixed 129Sv × C57BL/6 background to generate the 53BP1+/− p53+/− offspring, which were used for subsequent intercrosses. Mice were observed daily for signs of poor health, and moribund mice were sacrificed and screened for tumors.
Characterization of tumors.
Tissues from solid tumors were analyzed by standard histological analysis. Lymphomas were characterized by flow cytometry using anti-CD3, -CD4, -CD8, -CD19, -B220, -Thy1.2, and -immunoglobulin M (IgM) antibodies (Pharmingen).
SKY and FISH.
Spectral karyotype analysis (SKY) was performed as described previously (22). For fluorescence in situ hybridization (FISH) analyses, metaphases were hybridized with bacterial artificial chromosome probes specific for IgHcα, c-myc, TCRcα, and TCRvα together with chromosome-specific probes (11).
Centrosome staining.
Immortalized p53−/− mouse embryonic fibroblasts (MEFs) as well as primary 53BP1+/+ and 53BP1−/− MEFs were grown on coverslips, fixed with ice-cold methanol, and stained with antibodies against γ-tubulin and α-tubulin (Sigma). Similarly, cells derived from a 53BP1−/− p53+/+ thymoma and a 53BP1+/+ p53−/− thymoma were fixed, cytospun on slides, and stained with the anti-γ-tubulin and anti-α-tubulin antibodies. Mitotic cells were identified by DAPI (4′,6′-diamidino-2-phenylindole) staining on a Nikon Eclipse E800 microscope (100× objective) equipped with a SPOT RT digital camera (Diagnostic Instruments, Inc.).
DNA repair and genomic instability assays.
For the DNA repair assay, primary MEFs or stable MEF cell lines were plated onto coverslips. Once the cells had reached confluence and stopped dividing, they were irradiated with 2 Gy and allowed to recover for 27 h prior to immunostaining with anti-phospho H2AX antibodies (26). For the genomic instability assay, splenocytes were stimulated for 48 h with 100 U/ml interleukin 2 (Research Diagnostics, Inc.) and 4 μg/ml concanavalin A (Pharmacia) prior to exposure to 0 or 1 Gy of IR. After a recovery period of 30 min, Colcemid was added and metaphase spreads were prepared 3 h later according to a standard protocol.
Western blot analysis.
Thymocytes were isolated from 53BP1+/+ and 53BP1−/− littermates and cultured overnight in Dulbecco modified Eagle medium containing 100 U/ml interleukin 2 (Research Diagnostics, Inc.) and 4 μg/ml concanavalin A (Pharmacia). Aliquots of the cell suspensions were irradiated with either 0, 0.5, 2, or 5 Gy and lysed 2 h later in TNG buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 50 mM β-glycerophosphate, 1% Tween 20, 0.2% Nonidet NP-40) containing protease inhibitors. Equal amounts of protein were separated on a 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, transferred to a polyvinylidene difluoride membrane, and blotted with antibodies against p53 (FL-393-G; Santa Cruz), phosphorylated ATM (Ser1981) (10H11.E12; Cell Signaling), Chk2 (33), and β-actin (Sigma).
Real-time PCR analysis.
Total RNA was isolated from untreated or irradiated (10-Gy) splenocytes using Trizol (Invitrogen). Following treatment with RNase-free DNase (Invitrogen), RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed on a ABI PRISM 7700 sequence detection system (Applied Biosystems) with SYBR green PCR master mix (Applied Biosystems) and the following primer sets: 5′ p21, CCTGGTGATGTCCGACCTGTT; 3′ p21, GAGCGGCCTGAAGATTCCCC; 5′ mdm2 CAGCTTCGGAACAAGAGACTC; and 3′ mdm2, CTGCTCTCA CTCAGCGATGT. PCR conditions consisted of a 2-min 50°C cycle followed by 10 min at 95°C and 40 cycles of 95°C for 15 s and 55°C for 1 min. The average threshold cycle for each gene was determined from triplicate reactions, and the levels of gene expression were determined relative to that of β-actin (5′ β-actin, TGGGCCGCTCTAGGCACCA; 3′ β-actin, CCCCTGAACCCTAAGGCCA).
RESULTS
Increased tumor incidence in 53BP1-deficient mice.
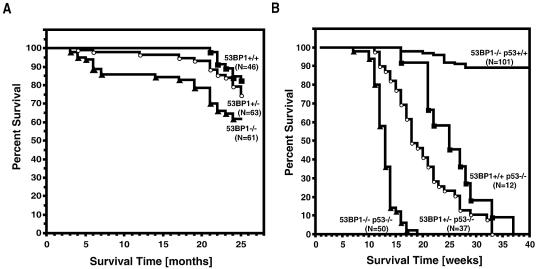
53BP1-deficient mice have an increased propensity to develop malignant lymphoma early in life (33). To assess whether the absence of functional 53BP1 also contributes to tumor susceptibility later in life, we analyzed the viability of a colony of 53BP1+/+ (n = 46), 53BP1+/− (n = 63), and 53BP1−/− (n = 61) mice over a period of 2 years (Fig. 1A). Only 11% of the 53BP1+/+ mice succumbed to malignant tumors during this time period compared to 29% of 53BP1−/− mice. Interestingly, 53BP1+/− mice were also approximately two times more likely to develop spontaneous tumors (19%) than wild-type mice of the same mixed background. All of the tumors that evolved during the first year of life were thymomas. Histopathological analysis of the tumors in 12- to 15-month-old animals also revealed leukemia/lymphoma as the most prevalent cancer in 53BP1+/− and 53BP1−/− mice (Table 1). The second most common tumor type in older 53BP1−/− mice was pulmonary carcinoma, while 53BP1+/− mice were more prone to tumors of vascular origin (hemangioma/hemangiosarcoma), which preferentially developed in liver or spleen (Table 1).
FIG. 1.
Survival of 53BP1-deficient mice in a p53 wild-type or p53-null background. (A) Survival of a cohort of 53BP1+/+ 53BP1+/− and 53BP1−/− mice observed for 25 months. (B) 53BP1 and p53 synergize in tumor suppression. Survival analysis of 53BP1+/+ p53−/−, 53BP1+/− p53−/−, 53BP1−/− p53−/−, and 53BP1−/− p53+/+ mice. Percent survival is plotted as a function of time as indicated. N represents the number of mice analyzed.
TABLE 1.
Spectrum of spontaneous tumors in 53BP1+/+, 53BP1+/−, and 53BP1−/− mice between 12 months and 25 months of life
| Type of tumor | No. of tumors
|
||
|---|---|---|---|
| 53BP1+/+ mice | 53BP1+/− mice | 53BP1−/− mice | |
| Hemangioma/hemangiosarcoma | 0 | 4 | 0 |
| Leukemia | 2 | 2 | 2 |
| Lymphoma | 1 | 3 | 4 |
| Pulmonary carcinoma | 0 | 1 | 4 |
| Adenocarcinoma | 1 | 1 | 0 |
| Hepatoma | 1 | 1 | 2 |
| Othera | 1 | 1 | 3 |
Histopathology not determined.
53BP1 and p53 synergize in tumorigenesis.
The 53BP1 N-terminal BRCT motif, together with the inter-BRCT linker region, binds to the central DNA binding site of p53, a domain that is frequently mutated in cancer (10, 17). To determine whether 53BP1 acts as an upstream modulator of p53 in tumorigenesis, we assessed the tumor latency of 53BP1+/+, 53BP1+/−, and 53BP1−/− mice in a p53-null background.
Loss of both alleles of 53BP1 greatly accelerated the tumor development of p53−/− mice (53BP1−/− p53−/− mice versus 53BP1+/+ p53−/− mice; P < 0.0001) (Fig. 1B). While 53BP1+/+ p53−/− mice (n = 12) had an average life span of 26 weeks before they became moribund with lymphoma or other types of tumors, the median survival time of 53BP1−/− p53−/− mice (n = 50) was only 12 weeks. Double-knockout mice succumbed to lymphomas as early as 7 weeks, and within 18 weeks all animals had died. Of the 50 53BP1−/− p53−/− mice analyzed, 46 developed thymic lymphomas, while 3 mice succumbed to B-lineage lymphomas, and 1 had both a thymic lymphoma and a pro-B cell lymphoma. Flow cytometric analysis revealed that the thymic lymphomas were of immature T-cell origin, with the majority of tumors being CD3 negative and either CD4/CD8 double positive or CD8 single positive. Three of the B-lineage lymphomas were B220+ CD19+/IgM−, while one was B220+ CD19+/IgM+.
Loss of only one allele of 53BP1 also significantly accelerated the tumor onset in p53-null mice, albeit to a lesser degree than complete 53BP1 deficiency (53BP1+/− p53−/− versus 53BP1+/+ p53−/− mice; P < 0.0001) (Fig. 1B). 53BP1+/− p53−/− mice (n = 37) became moribund with tumors as early as 11 weeks, with a median survival time of 17.8 weeks and a maximum survival time of 33 weeks. The tumor spectrum in these mice was broader than in the double-knockout mice, with only 26 of the animals developing lymphomas and 11 animals developing solid tumors, including five sarcomas. Two of the 22 lymphomas analyzed were B220/CD19+/IgM+ B-lineage lymphomas, while the 20 others were thymic lymphomas that expressed various amounts of CD3, CD4, and/or CD8. The increase in tumor predisposition observed in 53BP1−/− p53−/− and 53BP1+/− p53−/− mice indicates that loss of 53BP1 synergizes with p53 inactivation in tumorigenesis.
Normal p53 activation in 53BP1−/− p53+/+ thymocytes.
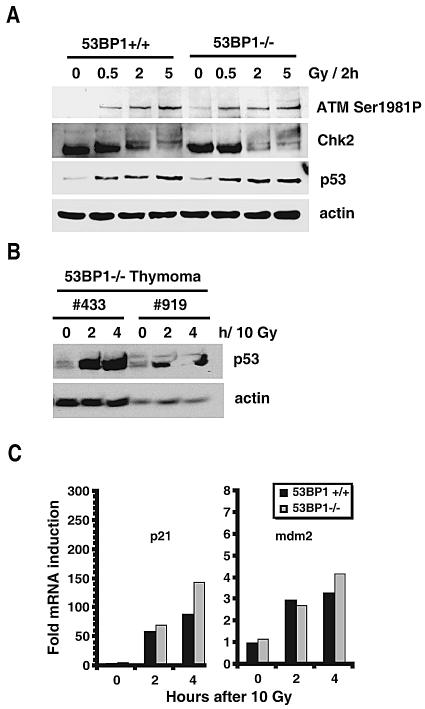
The observed synergism of 53BP1 and p53 suggests that they have different functions in tumor suppression. To test this hypothesis, we assessed p53 accumulation in 53BP1-deficient thymocytes 2 h after exposure of cells to 0.5 Gy, 2 Gy, or 5 Gy of IR. Compared to wild-type controls, 53BP1−/− thymocytes showed normal p53 stabilization in response to low-dose as well as high-dose irradiation (Fig. 2A). Moreover, levels of phosphorylation of ATM at Ser 1981 were very similar in wild-type and mutant cells, although we did see slightly impaired Chk2 phosphorylation upon low-dose IR in the 53BP1−/− thymocytes, as reported previously (33) (Fig. 2A). Consistent with these findings, p53 accumulation was readily observed in two thymoma samples derived from 53BP1-null mice (Fig. 2B). Since 53BP1 has been proposed to act as a transcriptional coactivator of p53, we also quantified the expression of the p53-dependent target genes mdm2 and p21 using real-time PCR. As shown in Fig. 2C, induction of mdm2 and p21 mRNA in response to IR was normal or somewhat increased in 53BP1-deficient thymocytes, suggesting that 53BP1 is not essential for p53 transactivation activity in murine thymocytes.
FIG. 2.
p53 stabilization and p53-dependent transcription in primary 53BP1+/+ and 53BP1−/− cells. (A) Levels of p53, phosphorylated ATM, and Chk2 in normal 53BP1+/+ and 53BP1−/− thymocytes before and 2 h after irradiation with 0.5, 2, and 5 Gy. (B) p53 levels in 53BP1−/− tumor cells derived from two thymic lymphomas. The time points after treatment with 10 Gy are indicated at the top. (C) Quantitative RT-PCR analysis of p21 and mdm2 mRNA levels in 53BP1+/+ and 53BP1−/− splenocytes before and after irradiation with 10 Gy. The mRNA levels in the untreated 53BP1+/+ cells are set to 1 after standardization of all values relative to the mRNA levels of actin.
Chromosomal aberrations in 53BP1−/− p53−/− lymphomas.
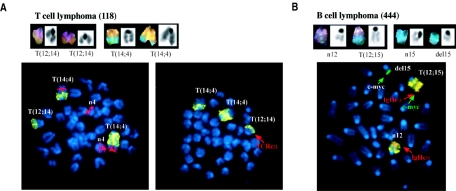
To understand how the loss of 53BP1 contributes to tumorigenesis, we examined metaphases from 53BP1−/− p53−/− lymphomas by SKY. Unlike p53−/− lymphomas, which have been reported to rarely harbor chromosome translocations (21), four out of seven 53BP1−/− p53−/− tumors analyzed showed structural aberrations, including clonal translocations and deletions (Fig. 3 and Table 2). Some of the aberrations found in 53BP1−/− p53−/− lymphomas mapped to chromosome 12 or 14, which contain, respectively, the IgH or T-cell receptor (TCR) alpha locus. To assess whether translocations arise in the context of aberrant V(D)J recombination, we analyzed metaphase spreads by FISH. One out of seven of the rearrangements observed in thymic lymphomas involved the TCRα locus (Fig. 3A). In addition, one of one IgM+ B-lineage lymphoma analyzed contained a T(12;15) translocation that juxtaposed the distal part of the c-myc oncogene on chromosome 15 to the IgHc alpha locus on chromosome 12 (Fig. 3B). Therefore, it is likely that some of the thymic lymphomas as well as B-lineage lymphomas in 53BP1−/− p53−/− mice arise during antigen receptor rearrangements.
FIG. 3.
Chromosomal translocations involving antigen receptor loci in 53BP1−/− p53−/− lymphomas. (A) SKY (top panel) and FISH (bottom panel) of a thymic lymphoma harboring two T(14;4) translocations and a T(12;14) translocation in which the breakpoint was near the T-cell receptor alpha locus (TCRcα; red, indicated by an arrow). Chromosome 14 and 4 painting probes were used for FISH analysis together with a TCRcα bacterial artificial chromosome probe. The TCRvα locus is deleted in this tumor (not shown). (B) SKY (top panel) and FISH (bottom panel) analyses of a B-cell lymphoma harboring a T(12;15) translocation that juxtaposes IgHcα (red) and c-myc (green) (indicated by arrows).
TABLE 2.
Karyotype of 53BP1−/− p53+/+ and 53BP1−/− p53−/− lymphomas observed by Sky and FISH
| Tumor | Genotype | Phenotype | Age (wk) | Composite karyotype | Locus involvement
|
|
|---|---|---|---|---|---|---|
| TCR | IgH | |||||
| 259 | 53BP1−/− p53+/+ | Thymic | 24 | 43-45 (2n), XY, +4, +15, +15 | No | No |
| 56-57 (3n), XYY, +15, −17, −18 | ||||||
| 118 | 53BP1−/− p53−/− | Thymic | 13.9 | 40-41 (2n), XY, +6, Rb(11.16), T(12;14), T(14;4)(two), +15 | Yes | No |
| 136 | 53BP1−/− p53−/− | Thymic | 13.9 | Tetraploid, aneuploid | No | No |
| 423 | 53BP1−/− p53−/− | Thymic | 13.6 | 44-46 (2n), XYY, +5, +10, −13, +14, +15, +Y | No | No |
| 592 | 53BP1−/− p53−/− | Thymic | 9.9 | 37-41 (2n), XXX, Rb(4.4), +4, +5, Rb(X.X), +X | No | No |
| 705 | 53BP1−/− p53−/− | Thymic | 11.3 | 76 (4n), XXXX, +1, T(4;16), Rb(4.4)X2, +4, T(7;13), −9, −12, −12, T(14;4)(two), +16, −17a | No | No |
| 708 | 53BP1−/− p53−/− | Thymic | 12.4 | 61-62 (3n), XXXXYY, −2, −3, +4, +5, −6, −7, −8, −9, −10, −11, −13, +del(14)X5, +15, +16, −18, +X, −Y | No | No |
| 444 | 53BP1−/− p53−/− | B | 11.4 | 33-42 (2n), X, −3, +5, −6, T(12;15), −13, +15, −17, −X | No | Yes |
This karyotype represents only one clone; the tumor is highly heterogeneous and polyploid/aneuploid.
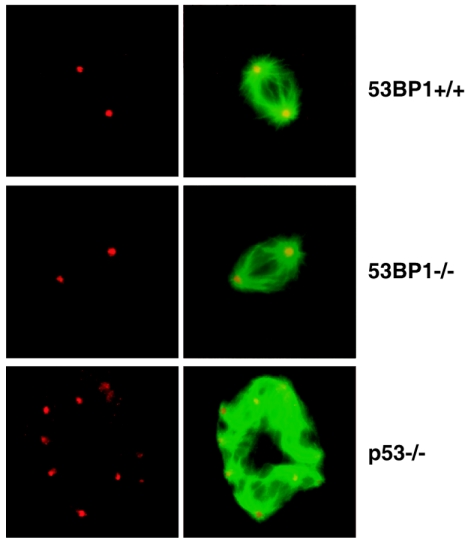
Thymomas in p53-deficient mice develop independently of V(D)J recombination, with aneuploidy as a characteristic chromosomal aberration (21). Of the five tumors that did not show TCR or IgH involvement, three of them as well as one 53BP1−/− p53+/+ thymoma harbored numerical chromosome aberrations (Table 2). These tumors arose within 14 weeks of age, which is considerably earlier than tumors arising in p53−/− mice. Aneuploidy in p53-deficient cells has been linked to centrosome hyperamplification (30), and we readily observed centrosome abnormalities in MEFs lacking p53 (38% [n = 100]). In contrast, aneuploidy in immortalized or primary 53BP1−/− p53+/+ MEFs was usually not accompanied by centrosome hyperamplification (Fig. 4). Only 4% of immortalized 53BP1−/− p53+/+ MEFs (n = 100) and 10% of primary 53BP1−/− p53+/+ MEFs (n = 100) showed centrosome hyperamplification, a frequency very similar to that found in wild-type 53BP1+/+ p53+/+ MEFs (2% in immortalized MEFs and 13% in primary MEFs [n = 100]). Moreover, centrosome hyperamplification was clearly less common in aneuploid 53BP1−/− p53+/+ thymomas (24% [n = 100]) than in a similar aneuploid 53BP1+/+ p53−/− thymomas (56% [n = 100]). Together, these data imply that 53BP1 may suppress aneuploidy through a mechanism distinct from that of p53.
FIG. 4.
Normal centrosome duplication in 53BP1-deficient cells. 53BP1+/+, 53BP1−/−, and p53−/− MEFs were immunostained for γ-tubulin, a component of centrosomes (red), and α-tubulin, a component of microtubules (green, here shown merged with red). Mitotic p53-deficient MEFs, but not 53BP1-deficient MEFs, frequently show centrosome hyperamplification and abnormal mitotic spindle.
Impaired DNA DSB repair and genomic stability in 53BP1+/− cells.
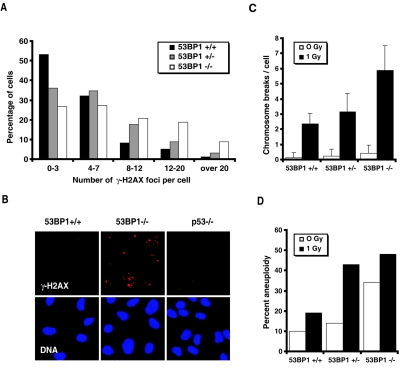
One of the mechanisms by which the loss of 53BP1 promotes tumorigenesis is through a defect in the proper rejoining of DNA DSBs that arise during lymphocyte development. We had shown earlier that 53BP1+/− cells express approximately half the amount of 53BP1 protein expressed by 53BP1+/+ cells, and Western blot analyses of 53BP1+/− p53−/− thymomas confirmed that the tumor cells had not lost heterozygosity (33; data not shown). To investigate whether the loss of only one allele of 53BP1 is sufficient to impair DNA DSBs repair, we estimated the number of unrepaired DSBs in irradiated 53BP1+/+, 53BP1+/−, and 53BP1−/− MEFs using phospho-H2AX (γ-H2AX) focus analysis. Nonreplicating G0/G1 53BP1 wild-type cells showed an average of 4.4 residual γ-H2AX foci 27 h after 2 Gy of IR, with the majority of cells having 0 to 3 foci (Fig. 5A). In contrast, 53BP1−/− MEFs exhibited a significantly higher number of unrepaired DSBs (on average, 9.5 foci per cell), while irradiated 53BP1+/− MEFs showed an intermediate phenotype, with an average of 6.5 residual γ-H2AX foci per cell (53BP1+/+ versus 53BP1+/− cells, P < 0.001; 53BP1+/− versus 53BP1−/− cells, P < 0.001) (Fig. 5A). No repair defect was detected in 53BP1+/+ p53−/− MEFs (Fig. 5B).
FIG. 5.
Genomic instability in 53BP1−/− in 53BP1+/− cells. (A) Numbers of γ-H2AX foci/cell in primary 53BP1+/+, 53BP1+/−, and 53BP1−/− MEFs 27 h following 2 Gy. (B) Immunofluorescence analysis of residual γ-H2AX foci in 53BP1+/+, 53BP1−/−, and p53−/− MEF lines 27 h after 2 Gy. (C) Average number of chromosome breaks per cell in 53BP1+/+, 53BP1+/−, and 53BP1−/− splenocytes before and after irradiation with 1 Gy. The error bars represent the standard deviations from at least 30 metaphase spreads. (D) Percentages of splenocytes (n = 30) with gains or losses of chromosomes before and after irradiation with 1 Gy.
To further investigate whether 53BP1 haploinsufficiency compromises genomic stability, we assessed the number of chromosome breaks in 53BP1+/+, 53BP1+/−, and 53BP1−/− mouse splenocytes before and 3 h after exposure to 1 Gy of IR. 53BP1−/− splenocytes exhibited an increased number of spontaneous chromosome breaks per cell (0.45 ± 0.55 [mean ± standard deviation]) relative to 53BP1+/+ cells (0.15 ± 0.35; 53BP1−/− versus 53BP1+/+ cells, P < 0.005) and 53BP1+/− cells (0.25 ± 0.46; P ≥ 0.2). Upon treatment with 1 Gy of IR, the average number of chromosome breaks in 53BP1+/− splenocytes (3.2 ± 1.2) was intermediate between that found in 53BP1+/+ cells (2.4 ± 0.69; 53BP1+/− versus 53BP1+/+ cells, P < 0.005) and 53BP1−/− cells (5.9 ± 1.66; 53BP1+/− versus 53BP1−/− cells, P < 0.001) (Fig. 5C). Moreover, irradiated 53BP1+/− splenocytes showed a high degree of aneuploidy (43%) that was more similar to that of irradiated 53BP1−/− cells (49%) than of 53BP1+/+ cells (19%) (Fig. 5D). A slight increase in aneuploidy was also found in nonirradiated 53BP1+/− splenocytes, with 14.3% of the cells showing losses or gains of chromosomes compared to 10.3% in 53BP1+/+ cells and 34.5% in 53BP1−/− cells (Fig. 5D). Together these findings demonstrate that a reduction in 53BP1 protein levels is sufficient to compromise the maintenance of chromosomal structure and number.
DISCUSSION
In the present study we have shown that the loss of 53BP1 synergizes with p53 deficiency in tumorigenesis. While the suppression of lymphomas in p53-deficient mice is thought to rely mainly on activation of the apoptotic pathway (28), 53BP1 probably functions at a different step in tumor suppression by participating in the repair of DNA DSBs and the suppression of aneuploidy. These findings are consistent with our earlier observations that 53BP1-deficient thymocytes show increased radiation sensitivity while thymocytes from p53-null mice are known to be resistant to apoptosis (34).
We have shown that 53BP1 is not essential for p53 stabilization and the induction of mdm2 and p21 expression in irradiated mouse cells. In fact, the absence of 53BP1 rather resulted in an increase in the p53 transactivation activity, suggesting that 53BP1 may negatively modify p53 function. Such a model is contrary to initial studies, which implied that 53BP1 may stimulate p53-mediated transcriptional activation, based on the ability of overexpressed 53BP1 to increase the expression of a p53-dependent chloramphenicol acetyltransferase reporter gene and of endogenous p21 (16). However, later structural analyses suggest that p53 may not be able to bind simultaneously to 53BP1 and p53 DNA binding sites (10, 17). Thus, it appears unlikely that 53BP1 could directly facilitate p53-mediated transcription. The increase in gene expression observed in the early study might be a consequence of artificial 53BP1 overexpression.
An involvement of 53BP1 in DNA DSB repair has been proposed previously based on the increased radiation sensitivity and impaired CSR observed in 53BP1-deficient mice (24, 33, 34). Moreover, an approximately 50% decrease in the production of mature T cells in the absence of 53BP1 suggested a defect in the rearrangements of the TCR locus required for normal T-cell development (33). Normally, cells which fail to successfully rejoin the rearranged gene segments undergo cell cycle arrest or apoptosis. However, in some cells, misrepair of DNA DSBs can lead to oncogenic translocations and subsequent tumor formation (11, 37). The low but significant lymphoma incidence in 53BP1-deficient mice together with the dramatic acceleration of lymphomagenesis observed in 53BP1−/− p53−/− mice suggests a role of 53BP1 in suppressing oncogenic translocations. Indeed, the one 53BP1−/− p53−/− B-lineage lymphoma analyzed contained a clonal T(12;15) translocation, which juxtaposed the c-myc oncogene to the IgH antigen receptor locus and may have evolved by aberrant RAG-induced V(D)J recombination. Moreover, two out of five 53BP1−/− p53−/− thymic lymphomas harbored clonal translocations, one of which involved the TCR locus, suggesting that faulty TCRα/δ V(D)J recombination initiated the oncogenic transformation. Interestingly, thymomas that developed in ATM-deficient mice also harbor translocations in the TCR locus (20), supporting our previous conclusion that 53BP1 and ATM may be components of the same tumor suppression pathway (33).
Some of the 53BP1−/− p53−/− lymphomas as well as the 53BP1−/− p53+/+ thymoma analyzed showed neither translocations nor deletions but exhibited abnormal numbers of chromosomes. Aneuploidy is a well-known hallmark or causal factor of cancers and may arise from centrosome malfunction or defects in mitotic segregation (6, 30). Unlike p53-deficient tumor cells, aneuploid 53BP1-deficient tumor cells showed only a moderate amount of centrosome hyperamplification, suggesting a difference in the mechanisms by which p53 and 53BP1 suppress aneuploidy. There are several possibilities regarding how the loss of 53BP1 may lead to numerical chromosomal alterations. For instance, 53BP1 has been shown to localize to the kinetochore in mitotic cells and 53BP1 accumulation appears to be the highest at the kinetochore of unaligned chromosomes (18), suggesting that 53BP1 may participate directly in sensing chromosome alignment during mitosis. Moreover, 53BP1 interacts with SNM1, a protein that is involved in an early mitotic checkpoint induced by spindle stress (1). However, we did not observe a defect in the ability of 53BP1-deficient MEFs to transiently arrest in mitosis in the presence of nocodazole, a microtubule-disrupting agent (data not shown). This suggests that mitotic spindle checkpoints are grossly intact in 53BP1-deficient cells, although we cannot rule out subtle defects. Another explanation for chromosome missegregation in 53BP1-deficient tumors could be reduced cohesion fidelity in the absence of 53BP1. 53BP1 is involved in the S-phase checkpoint. Recent data from Saccharomyces cerevisiae suggest that S-phase checkpoint proteins contribute to the establishment of chromatin configurations permissive for sister chromatid cohesion (35).
Interestingly, the loss of one allele of 53BP1 also significantly accelerated tumorigenesis in a p53-deficient background. The tumors appeared later than in 53BP1−/− mice and exhibited a broader spectrum, including lymphoma, sarcoma, and carcinoma. Moreover, 53BP1+/− p53+/+ mice also had a higher cancer predisposition than wild-type animals, further indicating that 53BP1 haploinsufficiency compromises genomic stability. The high percentage of solid tumors in these animals points to aneuploidy as a causal factor, while the observed hematological malignancies likely evolved from the misrepaired juxtaposition of genes at the sites of translocations. In support of this hypothesis, we observed a decreased ability to repair IR-induced DNA DSBs in 53BP1+/− cells as well as a marked increase in aneuploidy in 53BP1+/− splenocytes upon irradiation.
The high rate of chromosomal missegregation in some of the 53BP1−/− p53−/− and 53BP1−/− p53+/+ tumors distinguishes the tumor suppressor function of 53BP1 from that observed for H2AX. H2AX is a histone variant that becomes phosphorylated at megabase regions surrounding the sites of DNA strand breaks (27). 53BP1 interacts with phosphorylated H2AX in response to DNA damage, and H2AX is required for 53BP1 accumulation at the damage sites (8, 32). The loss of both alleles or only one H2AX allele has been shown to enhance tumor susceptibility in a dosage-dependent manner by promoting structural chromosomal anomalies, including translocations, deletions, and duplications (4, 7). Thus, while 53BP1 and H2AX act together in facilitating the repair of DNA DSBs, 53BP1 appears to have an additional role in suppressing numerical chromosomal aberrations. The fact that 53BP1−/− or 53BP1+/− mice show a higher tumor risk than H2AX−/− or H2AX+/− mice further supports the hypothesis of a dual role of 53BP1 in tumor suppression.
Given the impact of 53BP1 on tumorigenesis in our mouse model, it is not surprising that a 53BP1 or 53BP1-associated DNA damage response has also been implicated in human disease. Two recent studies describe the activation of the DNA damage response network in human precancerous lesions as an important barrier to tumorigenesis (3, 14). More directly, in a small number of lung tumors and melanomas tested, the progression to carcinoma or malignant melanoma was associated with a reduction in 53BP1 level (14). Thus, 53BP1 deficiency or haploinsufficiency may play a causal role in human cancer development.
Acknowledgments
This work was supported by a grant from the National Institutes of Health to J. Chen (CA100109). J. Chen is a recipient of a Department of Defense (DOD) breast cancer career development award (DAMD17-02-1-0472). I. Ward is supported by a postdoctoral fellowship from the DOD Breast Cancer Research Program (DAMD17-01-1-0317).
REFERENCES
- 1.Akhter, S., C. T. Richie, J. M. Deng, E. Brey, X. Zhang, C. Patrick, Jr., R. R. Behringer, and R. J. Legerski. 2004. Deficiency in SNM1 abolishes an early mitotic checkpoint induced by spindle stress. Mol. Cell. Biol. 24:10448-10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 4.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114: 359-370. [DOI] [PubMed] [Google Scholar]
- 5.Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68-76. [PubMed] [Google Scholar]
- 6.Cahill, D. P., C. Lengauer, J. Yu, G. J. Riggins, J. K. Willson, S. D. Markowitz, K. W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392:300-303. [DOI] [PubMed] [Google Scholar]
- 7.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez- Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 9.Charier, G., J. Couprie, B. Alpha-Bazin, V. Meyer, E. Quemeneur, R. Guerois, I. Callebaut, B. Gilquin, and S. Zinn-Justin. 2004. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure (Cambridge) 12:1551-1562. [DOI] [PubMed] [Google Scholar]
- 10.Derbyshire, D. J., B. P. Basu, L. C. Serpell, W. S. Joo, T. Date, K. Iwabuchi, and A. J. Doherty. 2002. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 21:3863-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Difilippantonio, M. J., S. Petersen, H. T. Chen, R. Johnson, M. Jasin, R. Kanaar, T. Ried, and A. Nussenzweig. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196:469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiTullio, R. A., T. A. Mochan, M. Venere, J. Bartkova, M. Sehested, J. Bartek, and T. D. Halazonetis. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998-1002. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 14.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 15.Iwabuchi, K., P. L. Bartel, B. Li, R. Marraccino, and S. Fields. 1994. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. USA 91:6098-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwabuchi, K., B. Li, H. F. Massa, B. J. Trask, T. Date, and S. Fields. 1998. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J. Biol. Chem. 273:26061-26068. [DOI] [PubMed] [Google Scholar]
- 17.Joo, W. S., P. D. Jeffrey, S. B. Cantor, M. S. Finnin, D. M. Livingston, and N. P. Pavletich. 2002. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 16:583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jullien, D., P. Vagnarelli, W. C. Earnshaw, and Y. Adachi. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115:71-79. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V., S. F. Altschul, and P. Bork. 1996. BRCA1 protein products. Functional motifs. Nat. Genet. 13:266-268. (Letter.) [DOI] [PubMed] [Google Scholar]
- 20.Liao, M. J., and T. Van Dyke. 1999. Critical role for Atm in suppressing V(D)J recombination-driven thymic lymphoma. Genes Dev. 13:1246-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao, M. J., X. X. Zhang, R. Hill, J. Gao, M. B. Qumsiyeh, W. Nichols, and T. Van Dyke. 1998. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol. Cell. Biol. 18:3495-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liyanage, M., A. Coleman, S. du Manoir, T. Veldman, S. McCormack, R. B. Dickson, C. Barlow, A. Wynshaw-Boris, S. Janz, J. Wienberg, M. A. Ferguson-Smith, E. Schrock, and T. Ried. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14:312-315. [DOI] [PubMed] [Google Scholar]
- 23.Manis, J. P., J. C. Morales, Z. Xia, J. L. Kutok, F. W. Alt, and P. B. Carpenter. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481-487. [DOI] [PubMed] [Google Scholar]
- 24.Morales, J. C., Z. Xia, T. Lu, M. B. Aldrich, B. Wang, C. Rosales, R. E. Kellems, W. N. Hittelman, S. J. Elledge, and P. B. Carpenter. 2003. Role for the BRCT protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 10:10. [DOI] [PubMed] [Google Scholar]
- 25.Rappold, I., K. Iwabuchi, T. Date, and J. Chen. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G. C. Smith, M. J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, A. R. Parker, S. P. Jackson, A. Gennery, P. A. Jeggo, and M. Lobrich. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell 16:715-724. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt, C. A., J. S. Fridman, M. Yang, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1:289-298. [DOI] [PubMed] [Google Scholar]
- 29.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarapore, P., and K. Fukasawa. 2002. Loss of p53 and centrosome hyperamplification. Oncogene 21:6234-6240. [DOI] [PubMed] [Google Scholar]
- 31.Wang, B., S. Matsuoka, P. B. Carpenter, and S. J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298:1435-1438. [DOI] [PubMed] [Google Scholar]
- 32.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 33.Ward, I. M., K. Minn, J. Van Deursen, and J. Chen. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, I. M., B. Reina-San-Martin, A. Olaru, K. Minn, K. Tamada, J. S. Lau, M. Cascalho, L. Chen, A. Nussenzweig, F. Livak, M. C. Nussenzweig, and J. Chen. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren, C. D., D. M. Eckley, M. S. Lee, J. S. Hanna, A. Hughes, B. Peyser, C. Jie, R. Irizarry, and F. A. Spencer. 2004. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell 15:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, Z., J. C. Morales, W. G. Dunphy, and P. B. Carpenter. 2001. Negative cell cycle regulation and DNA-damage inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem. 276:2708-2718. [DOI] [PubMed] [Google Scholar]
- 37.Zhu, C., K. D. Mills, D. O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C. C. Morton, F. W. Alt, S. Banin, L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109: 811-821. [DOI] [PubMed] [Google Scholar]