Abstract
The chromatin accessibility complex (CHRAC) is an abundant, evolutionarily conserved nucleosome remodeling machinery able to catalyze histone octamer sliding on DNA. CHRAC differs from the related ACF complex by the presence of two subunits with molecular masses of 14 and 16 kDa, whose structure and function were not known. We determined the structure of Drosophila melanogaster CHRAC14-CHRAC16 by X-ray crystallography at 2.4-Å resolution and found that they dimerize via a variant histone fold in a typical handshake structure. In further analogy to histones, CHRAC14-16 contain unstructured N- and C-terminal tail domains that protrude from the handshake structure. A dimer of CHRAC14-16 can associate with the N terminus of ACF1, thereby completing CHRAC. Low-affinity interactions of CHRAC14-16 with DNA significantly improve the efficiency of nucleosome mobilization by limiting amounts of ACF. Deletion of the negatively charged C terminus of CHRAC16 enhances DNA binding 25-fold but leads to inhibition of nucleosome sliding, in striking analogy to the effect of the DNA chaperone HMGB1 on nucleosome sliding. The presence of a surface compatible with DNA interaction and the geometry of an H2A-H2B heterodimer may provide a transient acceptor site for DNA dislocated from the histone surface and therefore facilitate the nucleosome remodeling process.
ATP-dependent nucleosome remodeling endows chromatin with dynamic properties that permit adjustment of structure and gene function in response to developmental or environmental cues (3, 37). SNF2-type ATPases that catalyze transitions of canonical nucleosome structure commonly reside in large protein complexes (14). The function of the other factors associated with the ATPase is unclear in most cases, but regulatory or targeting roles are presumed. The nucleosome remodeling ATPase ISWI is an integral part of several distinct complexes with diverse functions, including chromatin assembly, chromosome replication, and gene transcription (12, 30, 36). Because most ISWI complexes known are built of a relatively small number of subunits, they lend themselves to biochemical and mechanistic analysis. The most simple physiological chromatin remodeling factors are made of ISWI and one member of the BAZ/WAL family of proteins that are characterized by several evolutionary conserved structures, including PHD fingers and bromodomains. Prominent examples are ACF, NoRC, and WICH/WCRF complexes (5, 7, 27, 43). Judged by the presence of conserved sequence elements and domains, BAZ/WAL proteins are multifunctional, but only a few functions have been described. Some surfaces may integrate the remodeling function into physiological processes, as is clear in the case of Tip5, which targets SNF2H, the vertebrate homologue of ISWI, to rRNA gene promoters (42, 43). Likewise, NURF301 can be recruited to promoters by transcription activation domains (1, 48). However, our analysis of ACF1 revealed that BAZ/WAL proteins are not just involved in targeting but are active components of the nucleosome remodeling mechanism. Association of ACF1 with ISWI (constituting the ACF complex) improves the energy efficiency of catalysis by an order of magnitude and modulates the precise outcome of nucleosome mobilization (15). Nucleosome sliding requires interaction of the SLIDE domain of ISWI with nucleosomal DNA (22). We recently found that contacts of the PHD fingers of ACF1 with the histone body contribute a crucial anchor point on the nucleosome substrate that enables efficient conversion of the force generated by ATP hydrolysis into disruption of DNA-histone interactions (17).
The metazoan chromatin accessibility complex (CHRAC) is formed by association of two small proteins of 14 and 16 kDa with ACF1 and ISWI (11, 40). The yeast ISW2 complex also contains a pair of histone fold proteins and thus appears to be related to CHRAC (26, 35).
The histone folds in CHRAC14 and CHRAC16 have been predicted by sequence analysis, but they so far have not been characterized in any detail. In the present work, we determined the crystal structure of a CHRAC14-16 heterodimer from Drosophila melanogaster, which indeed revealed two variant histone folds that interact in a histone-like handshake manner. Histone fold motifs are novel structures in a nucleosome remodeling machinery. Related structures are used for heterodimerization in the transcription regulators NFYB-NFYC (CBFA-CBFC) (49), HAP3-HAP5 (2), and NC2α-NC2β (21). Histone folds are abundant in the basal transcription factor TFIID, where they mediate dimerization of several TBP-associated factors (4, 20). Recently, Kukimoto and colleagues described the interaction of the human CHRAC14-CHRAC16 homologues with hACF1 and showed that the presence of the two proteins facilitated nucleosome sliding in vitro (29). We show here that these functional interactions are conserved in the Drosophila complex. Our detailed analysis of the DNA binding properties of the CHRAC14-16 heterodimer suggests that these proteins function as DNA chaperones in striking analogy to nucleosome sliding enhancement previously observed for HMGB1 (6).
MATERIALS AND METHODS
Cloning of bicistronic p14-p16 expression vectors.
The CHRAC16 coding sequence was amplified by PCR from the plasmid pET24d-CHRAC16 (11), with a 5′ primer introducing a linker including an NdeI site and an internal ribosomal entry site upstream of the CHRAC16 start codon and the following sequence: 5′-GGGGGTCTCGAATTCAATAATTTTGTTTAACTTTAAGAAGGAGAT ATACATATGGGCGAACCAAGGAGCCAA-3′ (modified after reference 34). The ribosomal entry site is underlined, the CHRAC16 CDS is shown in italics. The 3′ primer introduced an XbaI site downstream of the CHRAC16 stop codon (5′-GCCGGTCTCGAATTCTAGACTATTCATCAGACTCCGATTC-3′; CHRAC16 CDS is shown italics.). The ends of the PCR fragment included two EcoRI sites masked by BsaI sites. The BsaI-digested PCR fragment was cloned into the EcoRI site of the CHRAC14 expression plasmid pGEX2T-CHRAC14 (11). In a second step, the NdeI site was used to insert a 57-bp DNA fragment (5′-TAT GGTTAACCATCATCACCATCACCACCATCACGAGAATTTGTATTT TCAGGG-3′) encoding an N-terminal eight-histidine tag and a TEV cleavage site. The deletion mutants CHRAC14ΔN (Δ2 to 8), CHRAC14ΔC (1 to 108), CHRAC16ΔN (Δ2 to 18), and CHRAC16ΔC (1 to 117) were produced by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) on the bicistronic expression plasmid. All constructs were verified by sequencing.
Expression and purification of p14-p16 in Escherichia coli.
Full-length Drosophila p14-p16 and deletion variants were expressed from the bicistronic plasmids in E. coli BL21(DE3)(pLysS). Colonies were grown in LB medium containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C to an optical density (at 600 nm) of 0.8 and induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 3 h. Bacterial cell pellets were resuspended in phosphate-buffered saline (PBS) containing protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 0.7 μg/ml pepstatin, 1 μg/ml leupeptin). Cells were lysed by sonication, and the cell extract was passed over glutathione-Sepharose 4B beads (Amersham) equilibrated in PBS-0.05% NP-40. Beads were washed in 20 column volumes PBS500 (PBS with 500 mM NaCl, 0.05% NP-40) and 10 column volumes PBS-0.05% NP-40. The protein was either eluted from the beads with 100 mM Tris, pH 8.0, 50 mM KCl, 30 mM glutathione (Sigma) or cleaved off the glutathione S-transferase (GST) tag with thrombin (Amersham) according to the manufacturer's protocol. The protein was then passed over an Ni2+-loaded 1-ml Hi-Trap chelating fast protein liquid chromatography column (Amersham) and eluted with an imidazole gradient (0 to 500 mM) in HEMG500 buffer (25 mM HEPES, pH 7.6, 500 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol). The heterodimer eluted at approximately 170 mM imidazole. For crystallization, the CHRAC16 C-terminal His tag was cleaved off by TEV protease. After cleavage, the protein was passed over the Ni2+ column again and collected from the flowthrough. Finally, the p14-p16 heterodimer was applied to a Superdex 200 or Superdex 75 gel filtration column, respectively (depending on the presence or absence of the GST tag). Peak fractions were pooled and concentrated. The protein concentration was determined using the Bio-Rad protein assay.
Crystallization and structure determination.
Crystallization experiments were performed using a Cartesian crystallization robot (Genomic Solutions) with drop sizes of 0.2 μl protein solution mixed with 0.2 μl reservoir solution at a concentration of 35 mg/ml heterodimer. Crystals grew under several conditions of the Hampton Index screen. The largest crystals grew as regular rhomboids with sizes of 350 by 200 by 100 μm3 above a reservoir containing 0.1 M citric acid, pH 3.5, 2 M ammonium sulfate (condition 1, Index screen). The space group of these crystals is P3221: a, 76.0 Å; c, 166.1 Å; γ, 120°. A second crystal form yielding crystals sufficiently large for data collection was obtained with 0.1 M HEPES, pH 7.5, 12% (wt/vol) polyethylene glycol 3350, 5 mM CoCl2, 5 mM NiCl2, 5 mM CdCl2, 5 mM MgCl2 (condition 64, Index screen) as the reservoir solution. Those crystals grew as cubes of 100 by 100 by 100 μm3 and possess space group P4212: a, 130.5 Å; c, 59.7 Å. An initial data set of crystal form I, space group P3221 was collected at ESRF beam line ID14-4 to 2.4 Å resolution (Table 1). Attempts to solve the structure of this crystal form by molecular replacement failed. Therefore, we used the second crystal form (space group P4212) to solve the structure by single isomorphous replacement combined with anomalous scattering using data from a native crystal and a methylmercury acetate derivative collected at ESRF beam line ID29 (Table 1). Two major heavy-atom sites were located and refined with the program SOLVE (45), and the initial single isomorphous replacement combined with anomalous scattering map was further improved by solvent flattening and histogram matching using the program RESOLVE (44). The two mercury atoms were bound to Cys49 of CHRAC16 in both heterodimers present in the asymmetric unit and served as starting points for model building. The structure was manually built using the program O (28) and refined with the program CNS (8) against native data of this crystal form. The refined model was subsequently used to locate both heterodimers of the asymmetric unit in crystal form I (space group P3221) using the program AMORE (10) followed by refinement using the program CNS. The models in both crystal forms possess excellent stereochemistry. The final refinement statistics in both crystal forms are given in Table 1.
TABLE 1.
Data collection, structure solution, and refinement
| Parameter | Result for:
|
||
|---|---|---|---|
| Crystal form I | Crystal form II
|
||
| Native data | Hg data | ||
| Data collection | |||
| Space group | P3221 | P4212 | P4212 |
| Cell dimensions (Å) | a = 76.0, c = 166.1 | a = 130.5, c = 60.1 | a = 130.5, c = 59.7 |
| Wavelength (Å) | 0.9393 | 0.9795 | 0.9795 |
| ESRF beamline | ID14-4 | ID29 | ID29 |
| Resolution (Å)f | 24.0-2.4 (2.5-2.4) | 30.0-2.8 (2.95-2.80) | 35.0-3.0 (3.2-3.0) |
| No. of measurements | 130,593 (18,682) | 186,423 (27,241) | 142,145 (20,824) |
| No. of unique reflections | 22,490 (3,257) | 13,325 (1,902) | 10,828 (1,536) |
| Completeness (%) | 99.9 (99.9) | 99.9 (99.9) | 99.9 (99.9) |
| I/σ (I) | 4.4 (3.0) | 5.6 (1.6) | 5.3 (2.1) |
| Rmeas (%)a | 6.9 (26.4) | 12.8 (47.5) | 13.2 (36.8) |
| Structure determination | |||
| No. of Hg sites (found/total) | 2/2 | ||
| Riso (%)b | 21.3 | ||
| Z-scorec | 11.5 | ||
| Figure of merit (before/after solvent flattening) | 0.33/0.78 | ||
| Refinement | |||
| Resolution (Å) | 20-2.4 | 30-2.8 | |
| Total no. of atoms | 2,519 | 2,348 | |
| No. of protein atoms | 2,465 | 2,324 | |
| No. of water molecules | 39 | 21 | |
| Bound ions | 3 sulfate | 3 Cd2+ | |
| RMSD from ideal geometry | |||
| Bond length (Å) | 0.008 | 0.007 | |
| Bond angles (°) | 1.261 | 1.193 | |
| Rcryst (%)d (no. of reflections) | 23.7 (20,214) | 21.7 (11,861) | |
| Rfree (%)e (no. of reflections) | 27.8 (2,231) | 27.1 (1,363) | |
Rmeas = Σh [nh/(nh − 1)]1/2 ΣhΣi|Ii(h) − <I(h)>|/ΣhΣiI(h,i), where <I(h)> is the mean of the I observations of reflection h and n is the multiplicity of reflection h.
Riso = Σ|FPH − FP|/ΣFP, where FPH and FP are the derivative and native structure factor amplitudes, respectively.
Calculated according to the program SOLVE.
Rcryst = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated for crystal forms I and II using 9.9% and 10.3% of the reflections, respectively.
Values for the highest-resolution shell are given in parentheses.
FLAG affinity purification from Sf9 cell extract.
Sf9 cells were coinfected with baculovirus vectors carrying p14FLAG-HISp16 (pFASTBACDual; Invitrogen) and ACF1-myc full-length and deletion constructs (pFASTBAC; Invitrogen) (16) and grown for 48 h at 26°C. Cells were washed in PBS containing protease inhibitors, resuspended in EX100 buffer containing protease inhibitors (20 mM HEPES, pH 7.6, 100 mM KCl, 1.5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.05% NP-40), and lysed by two freeze-thaw cycles and mild sonication. The extract was incubated with 10 μl FLAG beads (Sigma) per 12 × 106 cells while rotating for 2 to 3 h at 4°C. Beads were washed once with EX100, three times with EX500 (500 mM KCl), and once again with EX100 buffer. Bound protein was eluted with FLAG peptide overnight at 4°C. ACF1-myc constructs were detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using the 9E10 anti-myc antibody (Sigma).
In vitro translation.
ACF1 derivatives were translated in vitro using the TNT system (Promega). Constructs were either expressed from pSPORT (Invitrogen) or pING14A (23) vectors and labeled with [35S]methionine and [35S]cysteine (ratio, 70% to 30%) during the reaction.
GST pull-down assays.
Glutathione-Sepharose 4B beads (Amersham) were equilibrated with EX250 buffer (250 mM KCl). The beads were loaded with GST, GSTp14, GSTp14-p16, and deletion constructs with a final concentration of 0.75 mg of protein/ml of beads by rotating overnight at 4°C. Beads were washed twice with EX250. To 25 μl of these beads, in vitro-translated ACF1 constructs were bound by rotating in a total volume of 100 μl (EX250 buffer) for 3 h at 4°C. Beads were washed once with EX250, three times with EX500 buffer, once with EX250, and once with EX100 buffer. Protein on the beads was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and signals were enhanced by incubation in Amplify solution (Amersham) for 30 min before drying. Bound in vitro translation constructs were detected by exposure of the gel to X-ray film.
Electrophoretic mobility shift assays (EMSA).
Band shifts were either performed with 0.5 nM radiolabeled 35-bp DNA (5′-CCCTATAACCCCTGCATTGAATTCCAGTCTGATAA-3′), 72-bp DNA (linker sequence for cloning of the bicistronic p14-p16 expression vector, see above), and 248-bp rRNA genes (32) (see Fig. S5 in the supplemental material) or with 6 nM radiolabeled 248-bp rRNA genes (see Fig. 6B) with the protein concentrations given in the figure legends. Protein was allowed to bind to DNA for 10 min at room temperature. DNA-protein complex formation was tested on 4.5% to 6.5% polyacrylamide gels in 0.4× Tris-borate-EDTA. Gels were run for 3 to 5 h at 100 V.
FIG. 6.
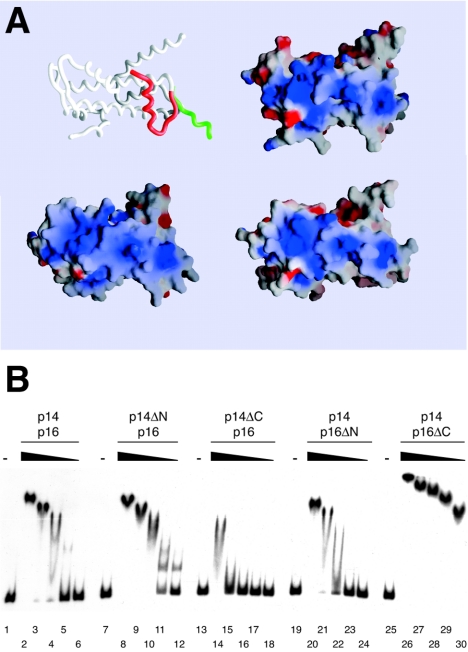
DNA binding by CHRAC p14-p16. (A) Surface charge distribution of p14-p16 compared to that of NFYB-NFYC and H2A-H2B heterodimers calculated with GRASP (38). Negative and positive potentials (±15 kBT [kB, Boltzmann constant; T, temperature]) are depicted in red and blue, respectively. The orientation of the three protein pairs is identical and corresponds to that shown for the p14-p16 worm model. Helix α1 of CHRAC16 was modeled onto NFYC and is depicted in red (compare also to Fig. S1 in the supplemental material). (B) EMSA with wt p14-p16 and p14-p16 deletion mutants. Six-nanomolar concentrations of radiolabeled 248-bp DNA were incubated with 20, 5, 2, 0.5, and 0.2 μM of the respective p14-p16 derivatives before complexes were resolved on a native polyacrylamide gel. An autoradiography of the dried gel is shown. −, absent.
Nucleosome sliding assays.
Positioned nucleosomes were generated and used for nucleosome mobility shift assays as described previously (16). Six-nanomolar end-positioned mononucleosomes were incubated with purified wild-type (wt) ACF or ACF ΔWAC (approximately 30 to 300 pM) (15) in EX50 buffer containing 1 mM ATP and 0.2 mg/ml bovine serum albumin for 45 min at room temperature. Where indicated, full-length and truncated p14-p16 heterodimers were added to the sliding reaction mixtures in various concentrations as indicated in the figure legends before the start of the reaction.
ATPase assay.
The ATPase assay was performed as described previously (13) with the following modifications. Standard reaction mixtures (15 μl) contained 50 mM Tris-HCl, pH 7.5, 50 mM KCl, 0.5 mM 2-mercaptoethanol, 0.1 g/liter bovine serum albumin, 20 μM ATP, 0.67 mM MgCl2, and 35 kBq [γ-32P]ATP (Amersham). The ATPase activity of 3 fmol ACF was determined in the presence of either 0.1 μg double-stranded DNA or the same amount of chromatinized DNA. Reaction mixtures were incubated at 26°C for 30 min.
Protein structure accession numbers.
The coordinates and structure factors of the CHRAC14-CHRAC16 heterodimer have been deposited with the PDB under accession numbers 2BYK and 2BYM for crystal forms I and II, respectively.
RESULTS
Crystallization and X-ray structure determination of a CHRAC14-CHRAC16 heterodimer.
Since the ratio of CHRAC14 (p14) and CHRAC16 (p16) in CHRAC is 1:1, we applied a coexpression strategy that yields stoichiometric amounts of p14-p16. Crystals were obtained with one batch of protein under several crystallization conditions; however, we realized that due to inadvertent proteolytic trimming during preparation, an estimated 2.5 kDa was removed from p16. Despite several attempts, crystallization could not be repeated with full-length protein. Crystallization yielded two crystal forms (Table 1). Initial attempts to solve the crystal structure by molecular replacement were unsuccessful. The structure of the p14-p16 heterodimer was therefore solved in crystal form II by single isomorphous replacement using a mercury derivative (Table 1). An initial model was built and refined in this crystal form at 2.8-Å resolution, which was subsequently used to solve crystal form I by molecular replacement and to refine the structure at 2.4-Å resolution (Fig. 1B).
FIG. 1.
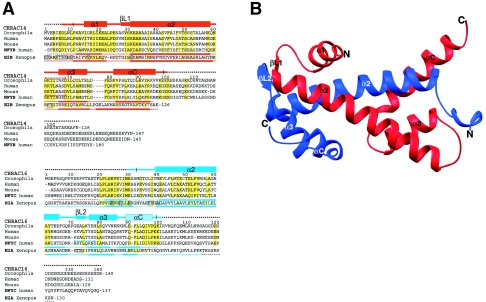
Structure of the CHRAC p14-p16 heterodimer. (A) Alignment of Drosophila CHRAC14 and CHRAC16 with human and mouse homologues, NFYB and NFYC, subunits of the human transcription factor NFY, and Xenopus histones H2A and H2B. Conserved or conservatively substituted residues within the p14-p16 families and compared to NFYB-NFYC are depicted with a yellow background. In addition, residues conserved or conservatively substituted in H2A-H2B compared to p14-p16 and NFYB-NFYC are also shown with a yellow background. Secondary structure elements in the p14 and p16 subunits were determined by manual inspection. The noncanonical p16 helix αC is shown in light blue. Secondary structure elements of NFYB-NFYC and H2A-H2B are depicted as published previously (33, 41). Disordered regions present in the initial constructs are represented by broken lines and are not included in the final model. H2A-H2B residues forming hydrogen bonds with nucleosomal DNA are shown in boxes. Intrachain salt bridges conserved between p14 and NFYB (red), but not between p16 and NFYC (blue), are indicated. (B) Ribbon representation of the p14-p16 heterodimer. CHRAC14 and CHRAC16 are depicted in red and blue, respectively. The color code for the two CHRAC subunits is kept throughout the figures. Figure 1B (see also Fig. S1 and S3 in the supplemental material) was produced with program Ribbons (9).
Structure of a CHRAC14-CHRAC16 heterodimer.
Our current model in crystal form I contains p14 residues 7 to 98 (second molecule in the asymmetric unit, residues 11 to 99), p16 residues 30 to 100 (second molecule in the asymmetric unit, residues 33 to 98), 39 water molecules, and 3 sulfate ions. In crystal form II, the structure of the heterodimer is very similar, although ordered N- and C-terminal extensions are slightly shorter. The overall structure of the p14-p16 heterodimer is depicted in Fig. 1B. The two core domains of each protein adopt a histone-like fold and pack head to tail against each other. In p14, the core histone motif helix α1-loop L1-helix α2-loop L2-helix α3 is extended by a long fourth helix, αC, characteristic for the H2B family. p16 also contains a C-terminal helical region following helix α3, similar to other H2A-related proteins. To allow comparison with other histone fold proteins, we designate the last helix αC, although its conformation is rather irregular compared to canonical α or 310 helices.
Probably as a result of partial proteolytic trimming, the predicted p16 helix α1 is missing, and presumably as a consequence, residues preceding helix α2 adopt a conformation different from other histone fold proteins. In both crystal forms, two p14-p16 heterodimers interact through the same extensive interface, where p14 helix α2 of a neighboring heterodimer inserts into a groove, which in a classical histone fold would be occupied by p16 helix α1. We suppose that, under physiological conditions, the N-terminal helix α1 in CHRAC16 occupies this position and that it was only displaced under our experimental conditions, which allowed crystallization. The interface between the two heterodimers is probably not physiologically relevant, which is further corroborated by the observation that, in ultracentrifugation studies, p14-p16 behave as heterodimers (K. Hartlepp, N. Mücke, and J. Langowski, unpublished observations).
Comparison with other histone-like proteins.
The overall structure of the heterodimer closely resembles other histone-like protein pairs like the NFYB-NFYC heterodimer (41) of the trimeric transcription factor NFY (root mean square deviation [RMSD] = 1.68 Å, 143 Cα atoms) and histone pairs H2A-H2B (33) (RMSD = 1.87 Å, 127 Cα atoms). Main differences to the NFYB-NFYC and H2A-H2B heterodimers are in the C-terminal end of p16 helix α2 and in the conformation of the following loop L2 (see Fig. S1 in the supplemental material). Compared to NFYC, helix α2 of p16 is shorter by two residues, whereas loop L2 has one additional residue inserted. As a result, loop L2 adopts a different conformation and p16 residues 72 to 74 are able to form a short two-stranded sheet with βL1 residues 28 to 30 of p14. Additional differences between p14-p16 and H2A-H2B are a longer loop between helices α3 and αC in p14 than in H2B and differently positioned helices α3 and αC in p16 compared to H2A (see below). Thus, the histone folds of p14-p16 and NFYB-NFYC are more similar than those of the H2A-H2B heterodimer. Indeed, histone core regions of CHRAC14 and CHRAC16 share 32% and 27% identical residues with NFYB and NFYC, respectively, whereas the core regions of CHRAC14 and CHRAC16 share only 9% and 15% identical amino acids with H2B and H2A, respectively (Fig. 1A). Despite the better conservation of the histone cores between p14-p16 and NFYB-NFYC, the N- and C-terminal extensions are not conserved, either. Accordingly, these extensions either are disordered in the crystal structure (p14-p16) or were essentially omitted from the crystallized constructs (NFYB-NFYC).
CHRAC14-CHRAC16 interact with the N terminus of ACF1.
p14 and p16 bind to the ISWI ATPase, but this interaction is disrupted by moderate salt concentrations (11). Since the association of p14-p16 with CHRAC resists salt washes of up to 1 M KCl (A. Eberharter, unpublished observation), we tested for binding of the small subunits to ACF1 after coexpression in insect cells. Sf9 cells were coinfected with a tandem baculovirus expression vector encoding p14FLAG and His6-tagged p16 and viruses coding for myc-tagged ACF1 or variants lacking parts of the protein (Fig. 2A). The FLAG tag on p14 was used to purify interacting proteins from the whole-cell extracts, and ACF1 was detected by Western blotting with anti-myc antibody. By this assay, we observed binding of full-length ACF1 and the N-terminal 1,064 amino acids (aa) of ACF1 to p14-p16, but a C-terminal fragment containing aa 497 to 1,476 did not bind (Fig. 2B). Interestingly, in this experiment, several N-terminal degradation products that could be detected by the anti-myc antibody in the input (Fig. 2B, lanes 2 to 4) did not bind to the FLAG beads, which also points to an ACF1 N-terminal binding site for p14-p16.
FIG. 2.
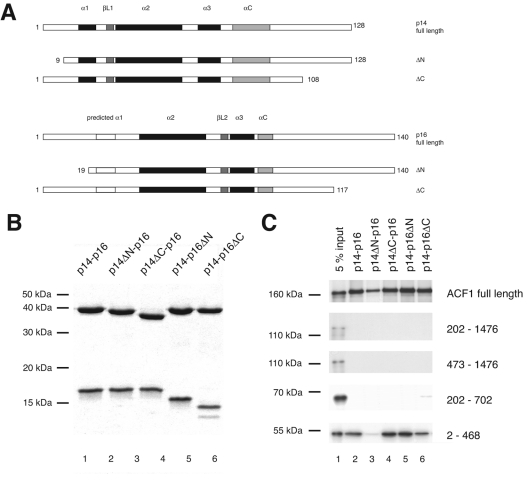
p14-p16 interacts with the N terminus of ACF1. (A) ACF1 derivatives used in this study. Top panel: baculovirus-encoded, myc-tagged ACF1 derivatives; bottom panel: in vitro-translated ACF1 derivatives. Interaction with p14-p16, as determined by the results shown in panels B and D, is indicated to the right: +, interaction; −, no interaction. (B) FLAG affinity purification of complexes from whole-cell extracts of SF9 cells coinfected with p14FLAG-HISp16 and myc-tagged ACF1 variants (shown in panel A). The Western blot was probed with anti-myc antibody. In the mock infection (lane 1), the cells were transfected with p14FLAG-HISp16 alone. Bands corresponding to the ACF1 constructs in the input are marked with asterisks. (C) Coomassie-stained 15% polyacrylamide gel of glutathione-Sepharose beads loaded with purified GST, GSTp14, and GSTp14-HISp16 (3.75 μg/lane). (D) GST pull down of in vitro-translated ACF1 constructs (shown in panel A). Top panel: 5% of input; bottom panel: pull down with recombinant GSTp14-HISp16 heterodimer.
To map the site of p14-p16 interaction on ACF1 more precisely, we immobilized GST-tagged p14-p16 expressed in E. coli (Fig. 2C) on glutathione-Sepharose beads and assayed for the interaction of in vitro-translated ACF1 and various deletion derivatives in pull down assays. The p14-p16 heterodimer interacted with full-length ACF1 and with derivatives containing the N-terminal 201 amino acids (Fig. 2D, lanes 1 to 3). This interaction was direct, since it resisted DNase and RNase treatment (see Fig. S2 in the supplemental material). p14 alone was unable to bind to ACF1, suggesting that p14-p16 form a functional unit (see Fig. S2 in the supplemental material). The N terminus of ACF1 contains a so-called WAC motif (for WSTF, ACF1, and cbp146), a sequence similarity found in other members of the BAZ/WAL family (27, 43, 48). Deletion of the first 202 aa from ACF1 abolished p14-p16 binding, suggesting that the WAC motif is part of a binding determinant. We could not determine whether the WAC domain was sufficient for the interaction, since an in vitro translation construct consisting of ACF1 aa 2 to 201 yielded variable results, possibly due to improper folding of the small fragment. From our data, we cannot formally exclude a contribution of flanking sequences, such as the conserved DDT motif, to p14-p16 binding. However, the human homologues of p14 and p16, human CHRAC17 (hCHRAC17) and hCHRAC15, have recently been shown to interact with the WAC domain of hACF1 (29).
CHRAC14-CHRAC16 enhance ACF-mediated nucleosome sliding.
Previously, we have shown that ACF and CHRAC can catalyze the sliding of histone octamers on short DNA fragments (15, 32). The nucleosome sliding assay exploits the different electrophoretic mobilities of nucleosomes, which are situated either centrally or at the end of a 248-bp DNA fragment. ACF is able to slide a histone octamer from the end to the center of a DNA fragment in an ATP-dependent reaction (15).
We monitored the effect of p14-p16 on ACF-induced nucleosome sliding by titration of increasing amounts of the heterodimer into sliding reactions (Fig. 3). At high ACF concentrations, no difference in sliding activity could be detected in the absence or presence of p14-p16 (lanes 1 to 6). However, at limiting ACF concentrations, nucleosome relocation was significantly enhanced by the presence of p14-p16 (Fig. 3, compare lane 16 with lanes 17 and 18 and lane 22 with lanes 23 and 24). This effect was ATP-dependent (lanes 8 to 13), indicating that the difference in nucleosome migration was caused by nucleosome repositioning and not by interaction of p14-p16 with the nucleosome (lanes 7 and 14).
FIG. 3.
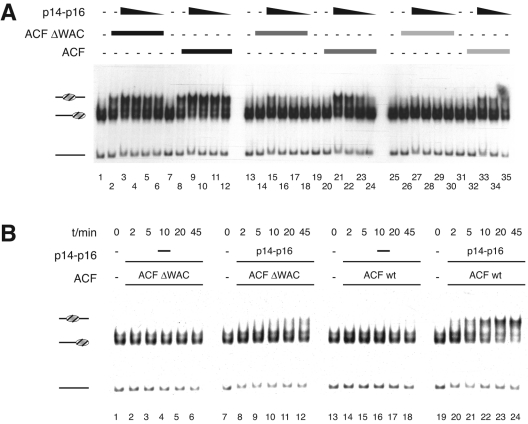
ACF-catalyzed nucleosome sliding is enhanced by p14-p16. Six-nanomolar radiolabeled end-positioned nucleosomes were incubated with approximately 300 pM, 90 pM, and 30 pM ACF, and decreasing amounts of p14-p16 (8, 4, 2, and 1 μM) were added to the reaction mixtures. With 300 pM ACF, sliding is maximal and no further enhancement due to p14-p16 is seen (lanes 2 to 6). Nucleosomes do not slide in the absence of ATP (lanes 9 to 13). Migration of the nucleosomes is not affected by p14-p16 alone (lane 7 and 14). +, present; −, absent. The bands correspond to (from the top) centrally positioned nucleosome, end-positioned nucleosome, and free DNA fragment, as schematically indicated to the left.
Since p14-p16 interact with the N terminus of ACF1, we reconstituted an ACF complex lacking part of the WAC motif of the ACF1 subunit (Δ4 to 111, ACFΔWAC) (19) and examined whether the effect of p14-p16 was dependent on interaction with ACF (Fig. 4). The N-terminal deletion did not affect the ATPase and nucleosome sliding activities of ACF (data not shown). The ATPase activity and concentration of the two ACF complexes were carefully matched, which allowed proper monitoring of the p14-p16 effect. Full sliding enhancement by p14-p16 could only be observed with intact ACF but not with the ACFΔWAC complex (Fig. 4A, compare lanes 14 and 15 with lanes 20 and 21 and lanes 26 and 27 with lanes 32 and 33). The dependence of the p14-p16-mediated enhancement on an intact N terminus of ACF1 was also obvious in a time course of nucleosome sliding (Fig. 4B). Together, these results show that p14-p16 are able to stimulate the activity of ACF and that this effect requires interaction with the N terminus of ACF1, consistent with similar observations made recently for human CHRAC/ACF (29).
FIG. 4.
The p14-p16 sliding enhancement is mediated by the ACF1 WAC domain. (A) Protein titrations. Nucleosome sliding is catalyzed by 300, 75, and 37.5 pM concentrations of either ACF or ACFΔWAC (decreasing shading of bar) in the presence of decreasing amounts of p14-p16 (8, 4, 2, and 1 μM) as indicated. (B) Time course. Reactions were performed with concentrations of ACF and ACFΔWAC complex (approximately 180 pM) which do not suffice to slide nucleosomes in the absence or presence of p14-p16 (approximately 8 μM). Time points are taken after 0, 2, 5, 10, 20, and 45 min. −, absent. Labeling is as described for Fig. 3.
The CHRAC14 N terminus is involved in ACF1 binding.
We created and purified heterodimers bearing deletions of individual N- and C-terminal tails of p14 and p16 (Fig. 5A and B) and tested their interactions with in vitro-translated ACF1 and deletion variants (Fig. 5C; Fig. 2C for ACF1 deletions). Interaction of all p14-p16 derivatives with ACF1 required N-terminal sequences as before (Fig. 2 and 5C). Deletion of 8 amino acid residues from the N terminus of p14, which is mostly disordered in the crystal structure, led to somewhat reduced binding to full-length ACF1 and essentially abolished binding to the N-terminal amino acids 2 to 468 of ACF1 (lane 3). This result suggests that the N terminus of p14 contributes to the interaction of the heterodimer with ACF1, but it may well not be sufficient. Interestingly, intact p14 alone does not interact with ACF1, suggesting a contribution of p16 sequences to ACF1 interaction (see Fig. S2 in the supplemental material).
FIG. 5.
Role of N- and C-terminal tails of p14-p16 for ACF1 binding. (A) Summary of p14-p16 derivatives. (B) Coomassie-stained 15% polyacrylamide gel of glutathione-Sepharose beads loaded with 3.75 μg of purified recombinant GSTp14-p16 derivatives as indicated. (C) GST pull down of in vitro-translated ACF1 constructs (shown in Fig. 1C). Lane 1, 5% of input. The p14-p16 derivatives used for the pull down are indicated above the lanes. The in vitro-translated ACF1 derivatives assayed for interaction are indicated to the right.
The C termini of CHRAC14 and CHRAC16 modulate the DNA binding properties of the heterodimer.
The calculated electrostatic surface potentials of the p14-p16, NFYB-NFYC, and H2A-H2B histone pairs are rather similar. Notably, the H2A-H2B surface that faces the DNA shows a similar basic overall charge in p14-p16 and NFYB-NFYC (Fig. 6A), while the opposite surface is rather negatively charged (data not shown). Binding of p14-p16 to DNA therefore probably involves a similar surface (see Fig. S3 in the supplemental material). In contrast, the H2A-H2B side chains directly involved in DNA contacts are only poorly conserved in p14-p16 (Fig. 1A).
We examined the DNA binding properties of the p14-p16 dimer by EMSA and pulldown assays (Fig. 6B; see Fig. S4 in the supplemental material; also data not shown). The p14-p16 heterodimer was unable to bind to DNA shorter than 20 bp (data not shown), and binding to a 35-bp DNA fragment was barely detectable. Binding to DNA fragments of 72 bp and 248 bp was measurable. EMSA with the 72-bp fragment revealed a distinct band that may correspond to a single p14-p16 bound to DNA, but additional heterogeneity of fragment mobility suggested variable positioning and stoichiometry of complexes at a higher protein input (see Fig. S4A and B in the supplemental material). We determined a formal KD of binding (assuming that all heterodimers are functional) of 2.3 μM (see Fig. S4B and C in the supplemental material). Low affinity for DNA has also been observed for the human homologues (40).
N-terminal truncation of p14 or p16 had no effect on DNA binding (Fig. 6B, lanes 8 to 12 and lanes 20 to 24, respectively), whereas the C-terminal truncations showed opposite effects. Deletion of the p14 C terminus reduced binding of the heterodimer to DNA by an order of magnitude (Fig. 6B, lanes 14 to 18). In contrast, deletion of the p16 C terminus enhanced DNA binding drastically compared to the wt (Fig. 6B, lanes 26 to 30), with an apparent KD of 57 nM for p14-p16ΔC binding to the 72-bp fragment (see Fig. S4B and D in the supplemental material). This deletion removes a highly negatively charged tail fragment consisting of 23 glutamates, aspartates, and serines, thereby causing a shift of the theoretical pI of p16 from 4.47 to 9.30. Apparently, this anionic structure prevented tighter binding of the p14-p16 heterodimer to DNA, a feature that has not been observed for the human counterpart (29). Interestingly, a p14ΔC-p16ΔC double deletion mutant showed increased DNA binding, similar to the p14-p16ΔC deletion mutant (data not shown). The effect of the deletion of the anionic p16 tail is thus dominant over the effect of the p14 tail deletion.
Dynamic, but not tight, DNA binding of CHRAC14-CHRAC16 facilitates nucleosome sliding.
We tested whether the different DNA binding affinities of the mutant p14-p16 heterodimers affected ACF-induced nucleosome sliding (Fig. 7A). The N-terminal deletions of both p14 and p16 did not affect nucleosome sliding activity of ACF significantly (Fig. 7A) under these conditions. By contrast, the heterodimer with the C-terminal deletion of p14 was significantly less able to stimulate nucleosome sliding (Fig. 7A, lanes 13 to 18, and B, lanes 13 to 18). Since this variant heterodimer binds DNA much more poorly than the wild type, the result suggests that DNA-binding of p14-p16 is required to facilitate nucleosome sliding.
FIG. 7.
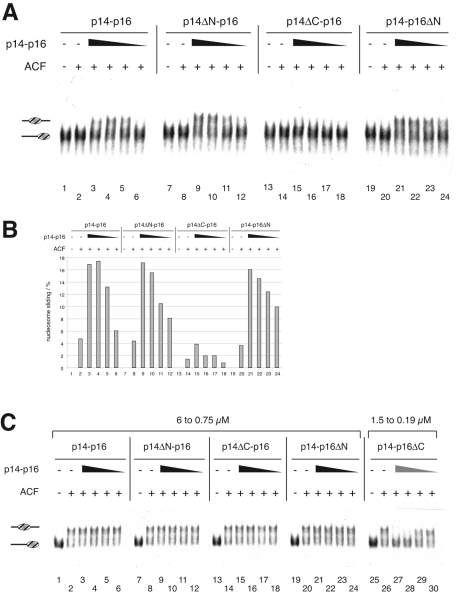
Effect of p14-p16 tails on nucleosome sliding. (A) Deletion of the p14 C terminus leads to loss of sliding enhancement. Sliding reaction mixtures contained 6 nM concentrations of end-positioned nucleosome, 37.5 pM ACF, and 2, 1, 0.5, and 0.25 μM concentrations of intact p14-p16 (lanes 3 to 6), p14ΔN-p16 (lanes 9 to 12), p14ΔC-p16 (lanes 15 to 18), or p14-p16ΔN (lanes 21 to 24). (B) Quantification of nucleosome sliding. The percentage of nucleosomes that had been moved from the end to the center position was determined using a phosphorimager (Fuji). The graph shows the average sliding results from two independent experiments. (C) Deletion of the p16 C terminus inhibits ACF-catalyzed nucleosome sliding. Sliding reactions contained 6 nM concentrations of end-positioned nucleosome, 0.3 nM ACF, and different p14-p16 derivatives (6, 3, 1.5, and 0.75 μM), with the exception of p14-p16ΔC, for which concentrations of 1.5, 0.75, 0.38, and 0.19 μM were used (lanes 26 to 30). +, present; −, absent. Labeling is as described for Fig. 3.
Evidently, p14-p16 need to contact both ACF1 and DNA to improve the sliding activity of ACF. How would the improved DNA binding of p14-p16ΔC affect the potential of the heterodimers to stimulate nucleosome sliding? We were unable to assay p14-p16ΔC under standard assay conditions, since its tight binding to the nucleosome interfered with the sliding analysis (not shown). Lowered concentrations of p14-p16ΔC did not interfere with the assay but also did not stimulate nucleosome sliding. To test for a potential negative effect of this mutant we increased the amount of ACF in the sliding reactions to levels that catalyzed complete nucleosome mobilization in the absence of p14-p16 (Fig. 7C). Under those conditions, the p14-p16ΔC deletion mutant inhibited the sliding reaction (lanes 25 to 30), whereas none of the other deletion mutants affected ACF activity, even at four-times-higher concentrations (lanes 7 to 24). Interestingly, the presence of p14-p16ΔC did not affect the ATPase activity of ACF (see Fig. S5 in the supplemental material), indicating that p14-p16 affect the efficiency of the ATPase to translocate the DNA relative to the octamer surface. In summary, our data suggest that dynamic, but not tight, nucleosome binding of p14-p16 stimulates ACF-induced nucleosome sliding.
DISCUSSION
Dynamic DNA interactions of CHRAC14-CHRAC16 facilitate nucleosome sliding.
We recently observed an enhancement of ACF-dependent nucleosome sliding by HMGB1, an abundant structure-specific DNA binding protein (6). Because the activating function of HMGB1 did not correlate with the strength of DNA binding but depended on a dynamic (i.e., weak) interaction with DNA, we suggested that HMGB1 might act as a DNA chaperone that promotes the distortion of DNA at its entry into the nucleosome. Manipulating DNA-histone interactions at this strategic site is likely to be a rate-limiting step in the current prevailing models of nucleosome mobilization (31, 46). Our analysis of the properties of the p14-p16 heterodimer revealed a number of similarities to HMGB1. Both entities enhance the catalysis of nucleosome sliding at limiting ACF concentrations. Both bind DNA weakly and nonspecifically. In either case, the dynamics of DNA interaction is assured by the presence of highly acidic C termini (on HMGB1 and on p16). Deletion of these charged C termini leads to a dramatic increase of DNA binding by the remaining histone folds and HMG domains, respectively. Under those conditions, ACF-dependent nucleosome sliding is not stimulated but repressed, indicating that tight binding leads to a locking of nucleosomal positions, as has been observed upon interaction of linker histone with nucleosomes (24, 25, 39).
The striking analogies between the properties of HMGB1 and the p14-p16 heterodimer lead us to speculate that the small histone fold subunits of CHRAC may serve as a built-in DNA chaperone that aids the disruption of DNA-histone interactions during the remodeling process by transiently providing a DNA binding surface. The histone fold heterodimer of p14-p16 resembles the geometry of H2A-H2B but lacks the tight grip of their interacting side chains. p14-p16 thus provide a surface that may lend itself for transient deposition of a segment of DNA stripped off the histone octamer surface. Furthermore, the acidic C-terminal tail of CHRAC16 might be in place to serve as a transient acceptor for a positively charged histone surface, such as the N terminus of histone H3 that reaches out into the linker DNA.
It has been suggested that some nucleosome remodeling enzymes use H2A-H2B heterodimer exchange to facilitate remodeling (18). In this respect, the presence of histone-fold subunits in CHRAC with an overall structure and charge distribution similar to histones H2A-H2B is worth noting. Replacement of H2A-H2B with p14-p16 would lead to significant nucleosome destabilization. In the nucleosome, the region following helix αC in histone H2A forms a two-stranded β-sheet with the C-terminal end of a neighboring H4 histone, which stabilizes the octamer structure (33). In p16, this region corresponds to helix αC, which packs against helices α2 and α3. In a hypothetical model where the p14-p16 heterodimer would replace H2A-H2B in the nucleosome, the p16 helix αC would prevent a similar interaction with the histone H4 C terminus and might considerably destabilize the nucleosome. However, the fact that we never observed a destabilization of nucleosomes during CHRAC-induced remodeling argues against such a scenario (32, 47). In addition, the observed differences in the core structures argue against a possible exchange during remodeling.
CHRAC is an evolutionarily conserved machinery. Recently, Kukimoto and colleagues reported on the stimulatory role of the human homologues of p14 and p16 on human ACF, but their study did not provide a mechanistic explanation (29). The human p14-p16 homologues, hCHRAC17 and hCHRAC15, also contain acidic glutamate- and aspartate-rich C termini of different length. However, in unresolved contrast to our findings, Kukimoto and colleagues reported a reduced DNA binding upon deletion of the negatively charged tail domains (29).
Physiological function of p14-p16.
The question of whether ACF and CHRAC exist as independent entities in living cells is still unanswered. The lack of suitable mutations in metazoan cells or reagents to localize the histone fold subunits in nuclei with confidence have hindered the exploration of their physiologic functions. Under these circumstances, the existence of homologous proteins in yeast provides valuable information. Recently, the Saccharomyces cerevisiae histone fold proteins Dls1p and Dpb4p were shown to associate with the Isw2 remodeling complex to form an entity reminiscent of CHRAC (26, 35). The similarity between the yeast Isw2 and the metazoan ACF complexes was previously not appreciated due to the very limited similarity between ACF1 and Itc1p, the largest subunit of the Isw2 complex. Strikingly, the two proteins only show similarity in their very N terminus with a recognizable WAC motif (35), which we and others (29) showed to be involved in the interaction with the histone fold subunits. The precise role of Dls1p and Dpb4p is still unclear, since yeast CHRAC appears to counteract telomeric silencing (26) but, on the other hand, is involved in the repression of some target genes (35). The situation is complicated by the fact that Dpb4p is also a subunit of a DNA polymerase ɛ complex, and mutant phenotypes may therefore reflect composite functions. Isw2-dependent repression of transcription and nucleosome repositioning is variably effected by mutation of the DLS1 gene at different gene loci (35), suggesting the possibility that two complexes related to ACF and CHRAC also exist in yeast, differing only by the presence of the histone fold subunits. Resolution of these issues will be facilitated by localization of all CHRAC components in living cells.
Supplementary Material
Acknowledgments
We thank J. A. Marquez and M. Röwer for access and support with the Cartesian crystallization robot. We also thank R. Ravelli and W. Shepard for access and support with ESRF beam lines ID14-4 and ID29; J. Brzeski, D. Fyodorov, and J. Kadonaga for ACF1 and p14-p16 expression constructs; and N. Mücke and J. Langowski for sharing unpublished analytical ultracentrifugation data.
This work was supported by Deutsche Forschungsgemeinschaft through TR5 and SFB594. C.F.-T. and T.G. acknowledge support by a long-term EMBO postdoctoral fellowship and an EMBL predoctoral fellowship, respectively.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Badenhorst, P., M. Voas, I. Rebay, and C. Wu. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16:3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxevanis, A. D., G. Arents, E. N. Moudrianakis, and D. Landsman. 1995. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 23:2685-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, P. B., and W. Hörz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 4.Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus, A. C. Lavigne, I. Davidson, and D. Moras. 1998. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94:239-249. [DOI] [PubMed] [Google Scholar]
- 5.Bochar, D. A., J. Savard, W. Wang, D. W. Lafleur, P. Moore, J. Cote, and R. Shiekhattar. 2000. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonaldi, T., G. Längst, R. Strohner, P. B. Becker, and M. E. Bianchi. 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozhenok, L., P. A. Wade, and P. Varga-Weisz. 2002. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54(Pt 5):905-921. [DOI] [PubMed] [Google Scholar]
- 9.Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24:958-961. [Google Scholar]
- 10.CCP4. 1994. Collaborative computational project number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50:760-776. [DOI] [PubMed] [Google Scholar]
- 11.Corona, D. F., A. Eberharter, A. Budde, R. Deuring, S. Ferrari, P. Varga-Weisz, M. Wilm, J. Tamkun, and P. B. Becker. 2000. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC). EMBO J. 19:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corona, D. F., and J. W. Tamkun. 2004. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta 1677:113-119. [DOI] [PubMed] [Google Scholar]
- 13.Corona, D. F. V., G. Längst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 14.Eberharter, A., and P. B. Becker. 2004. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 117:3707-3711. [DOI] [PubMed] [Google Scholar]
- 15.Eberharter, A., S. Ferrari, G. Längst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 20:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberharter, A., G. Längst, and P. B. Becker. 2004. A nucleosome sliding assay for chromatin remodeling factors. Methods Enzymol. 377:344-353. [DOI] [PubMed] [Google Scholar]
- 17.Eberharter, A., I. Vetter, R. Ferreira, and P. B. Becker. 2004. Acf1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 23:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165-173. [DOI] [PubMed] [Google Scholar]
- 19.Fyodorov, D. V., and J. T. Kadonaga. 2002. Binding of Acf1 to DNA involves a WAC motif and is important for ACF-mediated chromatin assembly. Mol. Cell. Biol. 22:6344-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff, Y. G., S. Werten, C. Romier, L. Carre, O. Poch, D. Moras, and I. Davidson. 2000. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goppelt, A., G. Stelzer, F. Lottspeich, and M. Meisterernst. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15:3105-3116. [PMC free article] [PubMed] [Google Scholar]
- 22.Grüne, T., J. Brzeski, A. Eberharter, C. R. Clapier, D. F. Corona, P. B. Becker, and C. W. Müller. 2003. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12:449-460. [DOI] [PubMed] [Google Scholar]
- 23.Hagemeier, C., A. Cook, and T. Kouzarides. 1993. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 21:4998-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, D. A., and A. N. Imbalzano. 2000. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39:11649-11656. [DOI] [PubMed] [Google Scholar]
- 25.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Iida, T., and H. Araki. 2004. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 28.Jones, T., J. Zhou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 29.Kukimoto, I., S. Elderkin, M. Grimaldi, T. Oelgeschlager, and P. D. Varga-Weisz. 2004. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol. Cell 13:265-277. [DOI] [PubMed] [Google Scholar]
- 30.Längst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 31.Längst, G., and P. B. Becker. 2004. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta 1677:58-63. [DOI] [PubMed] [Google Scholar]
- 32.Längst, G., E. J. Bonte, D. F. V. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 33.Luger, K., A. W. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 34.Lutzmann, M., R. Kunze, A. Buerer, U. Aebi, and E. Hurt. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McConnell, A. D., M. E. Gelbart, and T. Tsukiyama. 2004. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol. Cell. Biol. 24:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellor, J., and A. Morillon. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677:100-112. [DOI] [PubMed] [Google Scholar]
- 37.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls, A., R. Bharadwaj, and B. Honig. 1993. GRASP-graphical representation and analysis of surface properties. Biophys. J. 64:A166. [Google Scholar]
- 39.Pennings, S., G. Meersseman, and E. M. Bradbury. 1994. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA 91:10275-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poot, R. A., G. Dellaire, B. B. Hulsmann, M. A. Grimaldi, D. F. Corona, P. B. Becker, W. A. Bickmore, and P. D. Varga-Weisz. 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19:3377-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romier, C., F. Cocchiarella, R. Mantovani, and D. Moras. 2003. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278:1336-1345. [DOI] [PubMed] [Google Scholar]
- 42.Santoro, R., J. Li, and I. Grummt. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32:393-396. [DOI] [PubMed] [Google Scholar]
- 43.Strohner, R., A. Nemeth, K. P. Nightingale, I. Grummt, P. B. Becker, and G. Längst. 2004. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 24:1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Travers, A. A. 2003. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 4:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. (Erratum, 389: 1003.) [DOI] [PubMed] [Google Scholar]
- 48.Xiao, H., R. Sandaltzopoulos, H. Wang, A. Hamiche, R. Ranallo, K. Lee, D. Fu, and C. Wu. 2001. Dual functions of the largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8:531-543. [DOI] [PubMed] [Google Scholar]
- 49.Zemzoumi, K., M. Frontini, M. Bellorini, and R. Mantovani. 1999. NF-Y histone fold alpha1 helices help impart CCAAT specificity. J. Mol. Biol. 286:327-337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.