Abstract
The biological significance of recently described modifiable residues in the globular core of the bovine nucleosome remains elusive. We have mapped these modification sites onto the Saccharomyces cerevisiae histones and used a genetic approach to probe their potential roles both in heterochromatic regions of the genome and in the DNA repair response. By mutating these residues to mimic their modified and unmodified states, we have generated a total of 39 alleles affecting 14 residues in histones H3 and H4. Remarkably, despite the apparent evolutionary pressure to conserve these near-invariant histone amino acid sequences, the vast majority of mutant alleles are viable. However, a subset of these variant proteins elicit an effect on transcriptional silencing both at the ribosomal DNA locus and at telomeres, suggesting that posttranslational modification(s) at these sites regulates formation and/or maintenance of heterochromatin. Furthermore, we provide direct mass spectrometry evidence for the existence of histone H3 K56 acetylation in yeast. We also show that substitutions at histone H4 K91, K59, S47, and R92 and histone H3 K56 and K115 lead to hypersensitivity to DNA-damaging agents, linking the significance of the chemical identity of these modifiable residues to DNA metabolism. Finally, we allude to the possible molecular mechanisms underlying the effects of these modifications.
The fundamental unit of compaction of eukaryotic DNA is the nucleosome, consisting of two molecules each of the four core histones, H2A, H2B, H3, and H4. Not surprisingly, all four core histone genes are essential. Histones H3 and H4 flank the dyad axis of the structure, bind to the terminal segments of the DNA that enter and leave the nucleosome, and are the most highly conserved histones, suggesting that they play a more prominent regulatory role in chromatin formation. Furthermore, nucleosomes assembled in vitro from histone H3 and H4 tetramers retain the ability to impede transcription (2). As chromatin creates a degree of inaccessibility to the genetic information, proteins involved in DNA-templated reactions must overcome this. Perturbation to chromatin architecture is accomplished by the action of ATP-dependent chromatin remodeling factors, which reposition nucleosomes relative to the underlying DNA (1), and the histone modifying enzymes, which chemically alter histone proteins that comprise the nucleosomes (25, 41). Evidence supports the concerted action of these proteins in dictating transitions between transcriptionally active euchromatin and silent heterochromatin (4, 35).
There is a large repertoire of histone modifications, including lysine (K) and arginine (R) methylation, serine (S) and threonine (T) phosphorylation, and lysine acetylation, ubiquitylation, and sumoylation. Particular examples of these modifications underlie dynamic alterations in histone structure, mostly in the N-terminal “tail” domains of the histones, and are known to act epigenetically to regulate gene expression (19). The histone code hypothesis proposes that numerous effector proteins, which regulate chromatin-based processes, recognize particular patterns of posttranslational modifications in the histone tails (20, 48). Indeed, bromodomain (7, 17, 23) and chromodomain (13, 22, 29) proteins that recognize acetylated and methylated histone residues, respectively, have been identified and are implicated in the regulation of transcription.
Most publications to date have focused on modifications within the amino-terminal tails of the histones. These highly basic stretches of amino acids protrude from the nucleosome core, are flexible and unstructured, and are believed to act as a platform accommodating the binding of chromatin-associated proteins to the modified residues. Although these modifications are quite biologically significant, deletion of the tail domains from the genes encoding individual core histones supports viability (24) and, in fact, “tailless” H3, H2A, and H2B proteins retain the ability to organize into nucleosomes (11), suggesting that the essential function of histone proteins resides in their globular core domain. Indeed, deletions of certain regions of the core domain of histone H4 are lethal (24). Residues that lie in the globular core of the nucleosome have also been shown to be a substrate for modifying enzymes. Histone H3 K79, for example, is methylated by the Dot1 protein in Saccharomyces cerevisiae or the homologous protein in humans, Dot1L (27, 49). Methylation of H3 K79 appears to be cell cycle regulated (14), and its methylation status is important for defining transcriptionally active regions of the genome (36). However, core modifications are not limited to this residue. A report by Zhang et al. (54) provided evidence for the presence of an extensive network of covalent modifications on the globular histone fold domains of Bos taurus histones. They proved that numerous histone core residues have the potential to be acetylated, methylated, and phosphorylated, similar to that of the tail residues. Mapping these residues to the three-dimensional structure of the nucleosome core particle (28) illustrates that these modifiable residues form a pattern of chemical groups along the lateral surface of the nucleosome in addition to mapping to its solvent-exposed surface (4). It should be noted that the Zhang et al. study (54) did not use purified nucleosomes as the starting material, and so it is not necessarily the case that all of these modifications occur on nucleosomes per se; some may occur only on free histones. Importantly, the biological significance of most of these modifications has not been investigated.
In our analysis, we systematically generated a library of histone core domain mutations in the yeast S. cerevisiae at residues corresponding to those identified by Zhang et al. The mutations were designed to structurally mimic either constitutively modified or unmodified versions of potential modification sites. Using this genetic tool, we then explored the requirement of these core modifications in chromatin structure by analyzing the effects of the altered histones on heterochromatin structure and function at transcriptionally silent regions of the genome. We also explored the possibility that modifiable residues within the histone core play roles in the DNA damage response pathway, as has been documented for amino-terminal tail modifications (3, 33, 42). Here, we provide direct physical mass spectrophotometric evidence that one of the residues tested, histone H3 K56, can be acetylated in S. cerevisiae. This result corroborates the genetic data obtained and is in accord with recent results from at least two other groups (30, 51). Our findings are the first to illustrate in vivo that several modifiable residues within histone H3 and H4 cores are vital for heterochromatin integrity. We also demonstrate that several of these residues are required to elicit a normal cellular response to DNA damage, which we propose is likely to be controlled by their modification status.
MATERIALS AND METHODS
Strains and media.
Standard laboratory methods and techniques for budding yeast manipulations were used. The previously described yeast strain JPY12 (38) was employed for all investigations. All media were supplemented with 0.64 mM adenine, except for those monitoring silencing of ADE2. Lead plates were prepared as described elsewhere (5). Synthetic complete (SC) medium was used to prepare plates containing DNA-damaging agents.
Plasmids.
Plasmids pDM18 (43) and pJP11 (38), both carrying HHT2 (encoding histone H3) and HHF2 (encoding histone H4), were utilized in this study. A derivative of pDM18, pEMH7, was constructed in which the 3′ untranslated region of histone H4 was replaced by the ADH1 3′ untranslated region by inserting a 230-bp fragment amplified using primers JB8727 and JB8728 at EcoRI and KpnI restriction sites. Each gene was mutagenized using Stratagene's QuikChange protocol. Initially, pEMH7 was amplified with a distinct set of primers specific for the desired mutation, followed by Dpn1 digestion to eliminate unmutagenized parental strands. All plasmids were sequenced with oligonucleotides JB6505 and JB6504 subsequent to recovery from bacteria to verify the presence of the mutation. A random selection of 10 positively identified mutants was subcloned back into pEMH7 to verify that their associated phenotypes were not dependent upon erroneous mutations incorporated into the backbone of the plasmid during the PCR.
Silencing assays.
JPY12 (38) was transformed with pEMH7 harboring the mutation of interest. Transformants were streaked on yeast extract-peptone-dextrose to allow loss of the wild-type pJP11 (LYS2 HHT1 HHF1) plasmid and replica plated separately to SC-Trp, SC-Lys, and SC-His plates. His+, Trp+, and Lys− colonies were selected and studied further, except in those rare cases in which the mutations required the presence of wild-type histones for viability and were therefore Lys+. Silencing of MET15 at the ribosomal DNA locus was assayed on lead plates as described elsewhere (5). For photography, colonies were incubated at 30°C for 7 days. The strength of ribosomal DNA silencing of the mURA3 marker was monitored by plating fivefold serial dilutions with a starting optical density at 600 nm (OD600) of 0.5 on SC-Trp-His plus 5-fluoroorotic acid (5-FOA) plates. Plates were incubated at 30°C for 4 days and photographed. The silencing of telomeric ADE2 reporter (44) was assayed on SC-Trp plates. Cells were spotted on plates and incubated at 30°C for 3 days and subsequently at room temperature for a further 3 days to allow for color development. All data are representative of at least three independent phenotypic assays undertaken on at least three independent isolates of each mutation in JPY12.
DNA damage sensitivity assays.
Serial dilutions prepared as described above were plated on SC medium for a growth control and on SC medium containing either hydroxyurea (HU), methyl methane sulfonate (MMS), or camptothecin (CPT) at the indicated concentrations. Concentrated stock solutions of each drug were prepared in dimethyl sulfoxide so as to have a final concentration of 1% dimethyl sulfoxide in each plate. Plates were incubated for 3 days at 30°C and photographed, except those that contained HU, which were incubated for a week.
Southern blot analysis.
Genomic DNA was prepared for each strain harboring individual mutations on pEMH7. Genomic DNA was digested for 2 h with KpnI and run on a 0.8% agarose gel. Following transfer of DNA to nylon membrane, the membrane was probed with two 500-bp PCR products internally labeled with [α-32P]dATP. The pEMH7-specific probe was generated using primers JB8455 and JB8456, and the probe specific to ACT1 was created from a yeast genomic DNA templated PCR using primers JB6761 and JB6762. Hybridization of the probes was detected using a phosphorimager screen and quantified using ImageQuant software.
Real-time PCR.
Strains were grown in rich medium supplemented with 0.64 mM adenine and harvested at log phase. RNA was prepared from these cells and treated with DNase, and 450 ng of this RNA was reverse transcribed using Superscript III (Invitrogen). Following RNase H digestion, cDNA equivalent to 25 ng of input RNA was added to a real-time PCR, using the Applied Biosystems SYBR green RT-PCR system. Reactions were run in a 96-well plate using the Applied Biosystems Prism 7900HT fast real-time PCR system. The Ct values for ADE2 expression were compared with that of an internal ACT1 control. The primers used for ACT1 amplification were JB9639 and JB9640, and those for ADE2 amplification were JB9638 and JB9641.
Mass spectrometry analysis (quadrupole time of flight).
For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, excised gel slices from sodium dodecyl sulfate-polyacrylamide gel electrophoresis were digested with trypsin as follows. The gel bands were washed twice in 0.1 M NH4HCO3 (Fluka, Buch, Switzerland) and twice in 50% acetonitrile and subsequently cut into 2- by 2-mm pieces. The gel pieces were shrunk using 100% acetonitrile and then allowed to swell in 10 ng/μl trypsin-0.1 M NH4HCO3 (Promega U.S., Madison, WI) on ice for 20 min. After the rehydration, excess trypsin solution was removed and samples were incubated overnight at 37°C. The digestion was stopped by adding 10 μl of glacial acetic acid, and the supernatant containing the tryptic peptides was harvested. An extraction step was carried out to recover the peptides from the gel slices by adding 50% acetonitrile and incubating at room temperature for 30 min. The supernatant was harvested again and pooled. The pooled peptide extracts were dried down to approximately 5 μl and subjected to LC-MS/MS analysis as follows. Samples were injected onto a 10-cm C18 column (inner diameter, 75 μm) packed with Vydac MS218 5-μm beads (Vydac, Columbia, MD) and eluted using a gradient increasing from 7% solvent B-93% solvent A (solvent A, 0.4% acetic acid, 0.005% heptafluorobutyric acid; solvent B, 90% acetonitrile, 0.4% acetic acid, 0.005% heptafluorobutyric acid) to 45% solvent B-55% solvent A in 30 min (1100 CapLC; Agilent, Palo Alto, CA). Eluting peptides were analyzed using a quadrupole time-of-flight mass spectrometer (QSTAR Pulsar; Sciex, Toronto, Canada). The LC system was connected to the mass spectrometer using a nanoelectrospray source from Proxeon (Odense, Denmark). MS/MS data were searched against a yeast-only database (NCBInr) using Mascot (Matrix Sciences Ltd., London, England). Two missed cleavage sites and the relevant variable modification were allowed, and the mass accuracy was set to 0.2 Da for both precursor and fragment ions.
Mass spectrometry analysis (MALDI).
The mixture of histone proteins was digested with trypsin (20:1 [wt/wt]) in 25 mM ammonia bicarbonate at 37°C for 18 h. Digested peptide mixtures were separated by reversed-phase high-performance LC on a C18 column with an acetonitrile-water-0.1% trifluoroacetic acid solvent system. The collected fractions were dried completely and resuspended in 3 μl of water. The derivatization reaction was carried out by mixing 3 μl of 4-sulfophenyl isothiocyanate (10 μg/μl in 20 mM NaHCO3, pH 9.5) with 1 μl of the fraction at 55°C for 30 min. The reaction was terminated by adding 1 μl of 5% trifluoroacetic acid, and the sample was cleaned by using a micropipette tip (C18 ZipTip; Millipore, Bedford, MA). For mass spectrometer measurements, 0.5 μl of the sample was spotted on a matrix-assisted laser desorption ionization (MALDI) target followed by the addition of 0.5 μl of α-cyano-4-hydroxycinnamic acid matrix and was allowed to dry at room temperature. All MS and MS/MS spectra were acquired in the positive ion mode using a Kratos Analytical (Manchester, United Kingdom) AXIMA-CFR MALDI-TOF mass spectrometer equipped with a pulsed extraction source, a 337-nm pulsed nitrogen laser, and a curved-field reflectron. The acceleration voltage was 20 kV. The MS/MS spectra were interpreted manually or by computer-assisted database searches using the Mascot program (www.matrixscience.com) against the NCBI nr database.
Nucleosome views.
Representations of nucleosomes were generated using PyMOL (8).
RESULTS
Mapping modifiable histone residues on the S. cerevisiae nucleosome structure.
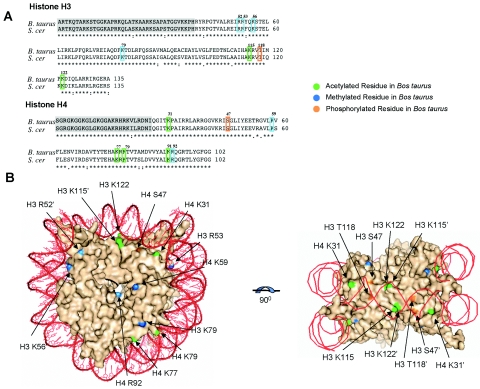
The positions of modified residues in the globular core of the bovine histones were mapped onto the amino acid sequence of S. cerevisiae histone H3 and H4 proteins. As depicted in Fig. 1A, all of the modifiable residues identified from bovine histones have unambiguous counterparts in the yeast sequence. In fact, residues H4 S47, H3 K79, and T118 were invariant over a wide representation of eukaryotic species, and residues H3 R52, K56, K115, K122, H4 K31, K59, K79 K91, and R92 showed amino acid variation only among deeply rooted eukaryotes, such as Giardia lamblia (see Fig. S1 in the supplemental material). The preservation of the exact positioning of these modifiable residues in the yeast nucleosome was verified by comparing the structural context of each amino acid in the Xenopus laevis (28) and S. cerevisiae (50) nucleosome crystal structures, the former of which shares more sequence identity with the bovine histones. The location of each modifiable residue is highlighted on the yeast nucleosome crystal structure in Fig. 1B.
FIG. 1.
(A) Sequence alignment of histones H3 and H4 from Saccharomyces cerevisiae and Bos taurus, generated using CLUSTALW. Shaded residues represent the N-terminal tails of the histone proteins. Modified residues identified in bovine histones are labeled and highlighted in both sequences according to the type of modification. (B) Mapping of modifiable residues on the surface of the yeast nucleosome crystal structure (50). A surface representation of the nucleosome, without histone tails, is shown and is viewed down the DNA superhelical axis. Potentially acetylated residues are green, methylated residues are blue, and phosphorylated residues are orange. The DNA helix is represented in bright green. (C) Rotation of view in panel B 90° around the horizontal axis. This is defined as the lateral surface of the nucleosome.
Generation of S. cerevisiae histone mutations.
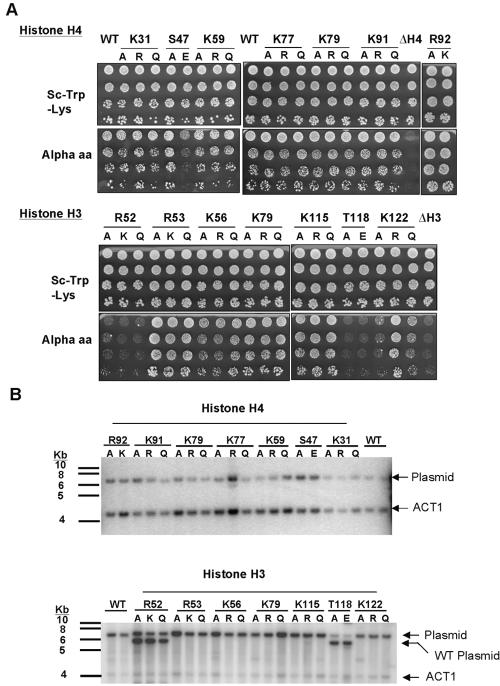
The mutagenic strategy undertaken generated alleles at each potentially modifiable residue so as to mimic both modified and unmodified versions of the amino acid. Acetylated lysines, for example, were mutated to both arginine and glutamine, which are the natural amino acids that most closely mimic deacetylated and acetylated lysine, respectively. All lysine residues were treated in this way, even those known only to be methylated in bovine, because there are instances of lysine residues that can be either acetylated or methylated. Additionally, all modified residues tested were systematically replaced with alanine. This generated a library of 39 histone alleles at 14 core residues in both histone H3 and histone H4 (Table 1). We anticipated that many of these substitutions would be lethal, given the unusually high conservation of histone sequences from yeast to mammals. To probe this, a previously developed plasmid shuffle strategy was used (38). In this strategy, histone function permits growth on medium containing α-amino-adipate, which selects against the wild-type histone gene present on the LYS2 “shuffle” plasmid pJP11 (38). As shown in Fig. 2A, all substitutions in histone H4, and the majority of altered histone H3 proteins, supported viability. Notably, only changes at two histone H3 residues, R52 and T118, were lethal. A few mutant strains had a slow growth phenotype (see Table S1 in the supplemental material). These include those strains harboring histone H4 S47E, histone H3 K122A, and histone H3 K122Q mutations. To determine to what extent strains containing viable substitutions compensated for the presence of the aberrant histones by increasing the copy number of these histone genes, Southern blotting analysis was employed. Data showed that the ratio between the abundance of the plasmid and a single-copy gene, ACT1, was not significantly altered in the mutated strains compared with wild type, indicating that, surprisingly, the majority of these substitutions of invariant amino acids were very well tolerated in yeast cells (Fig. 2B).
TABLE 1.
Library of histone alleles engineered through site-directed mutagenesis
| Histone | Residue | Modification | Substitutions |
|---|---|---|---|
| Histone H4 | Lys 31 | Acetylation | K31A, K31R, K31Q |
| Ser 47 | Phosphorylation | S47A, S47E | |
| Lys 59 | Methylation | K59A, K59R, K59Q | |
| Lys 77 | Acetylation | K77A, K77R, K77Q | |
| Lys 79 | Acetylation/methylation | K79A, K79R, K79Q | |
| Lys 91 | Acetylation | K91A, K91R, K91Q | |
| Arg 92 | Methylation | R92A, R92K | |
| Histone H3 | Arg 52 | Methylation | R52A, R52K, R52Q |
| Arg 53 | Methylation | R53A, R53K, R53Q | |
| Lys 56 | Acetylation/methylation | K56A, K56R, K56Q | |
| Lys 79 | Methylation | K79A, K79R, K79Q | |
| Lys 115 | Acetylation | K115A, K115R, K115Q | |
| Thr 118 | Phosphorylation | T118A, T118E | |
| Lys 122 | Acetylation | K122A, K122R, K122Q |
FIG. 2.
(A) Plasmid shuffle experiment of mutant histones to determine their viability. Strains expressing aberrant histones in the presence of wild-type proteins were plated with an initial OD600 of 0.5 and serially diluted fivefold on both SC-Trp-Lys and on plates containing α-amino-adipate, which selects against the wild-type histone gene present on the LYS2 “shuffle” plasmid. As a negative control, the JPY12 yeast strain was transformed with a plasmid from which either histone H3 or H4 was completely deleted. (B) Southern blot analysis to determine whether cells expressing the mutated histones were compensating for their presence by increasing the copy number of the plasmid harboring the altered histone. Genomic DNA was extracted from each strain and probed for ACT1, a single-copy gene, and the plasmid backbone in a Southern blot analysis. The ratio of these band intensities was then compared with that of cells containing wild-type histones.
High-throughput phenotypic analysis of histone mutations.
All of the engineered histone mutations were examined for silencing properties in the ribosomal DNA and in telomeres and also for sensitivity to three distinct DNA-damaging treatments, as described in Materials and Methods. The majority of the histone mutants showed one or more phenotypes in these assays. For instance, we found that telomeric silencing was negatively affected by 23 of the 39 alleles, 50% of the mutants induced an lrs (loss-of-ribosomal DNA silencing) phenotype, and one-third were more sensitive to HU. These data are summarized in Table 3, below, and in Table S1 in the supplemental material. In the following sections we examine specific cases where evidence was obtained which is consistent with control of heterochromatin or DNA damage sensitivity by the covalent modification state.
TABLE 3.
Summary of phenotypes observed for indicated histone allelesa
| Substitution | Growth in absence of wild type | ribosomal DNA silencing | Telomeric silencing | Growth on HU |
|---|---|---|---|---|
| WT H3/H4 | +++ | +++ | +++ | +++ |
| H4 K31A | +++ | ++++ | ++++ | +++ |
| H4 K31R | +++ | +++++ | ++++ | +++ |
| H4 K31Q | +++ | ++++ | ++++ | +++ |
| H4 K59A | ++ | ++ | + | ++ |
| H4 K59R | ++ | +++ | + | +++ |
| H4 K59Q | +++ | ++ | + | ++ |
| H4 K77A | +++ | +++++ | + | +++ |
| H4 K77R | +++ | +++ | +++++ | +++ |
| H4 K77Q | +++ | +++++ | +++ | +++ |
| H4 K79A | +++ | + | + | +++ |
| H4 K79R | +++ | +++ | +++ | +++ |
| H4 K79Q | +++ | + | + | +++ |
| H3 K56A | +++ | + | +++ | + |
| H3 K56R | +++ | +++ | +++ | + |
| H3 K56Q | +++ | + | ++ | ++ |
The mutants were analyzed and their phenotypes were scored arbitrarily with respect to wild type. For a given process, wild type was denoted +++, and those mutants with a negative effect were scored as ++ or +, depending on the severity of the mutation. Alleles that induced an increase in silencing are indicated with ++++ or +++++, the latter of which corresponds to almost-complete repression of the reporter gene.
Modifiable nucleosome core residues are important for transcriptional silencing at telomeres and ribosomal DNA.
Using a genetic approach, we sought indirect evidence for the existence of nucleosome core modifications in S. cerevisiae and to infer the potential modification status of each residue at three transcriptionally silent regions, a strategy previously employed to study tail modifications (21, 31). The intrinsic logic is that mutations that represent different modified states of a residue will display contrasting phenotypes, enabling inference of the potential modification state. To assess the effect of altered histones on heterochromatin, we studied transcriptional silencing, a genetically tractable yeast system that allowed us to monitor the existence of various states of heterochromatin.
Histone H4.
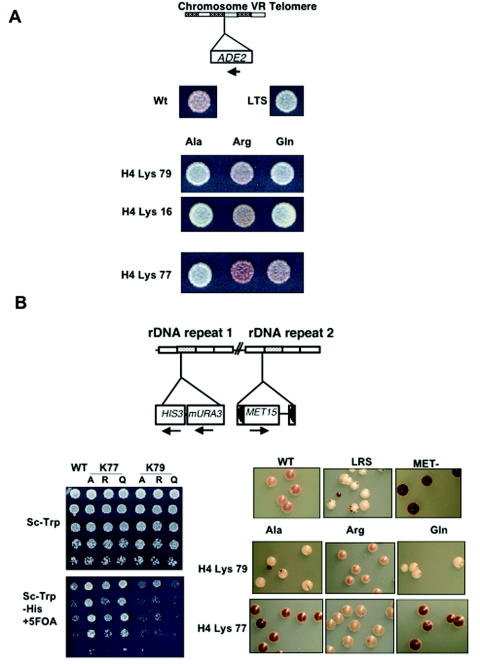
Peptide mass fingerprinting of bovine histone H4 indicated that K77 and K79 are acetylated in vivo in that species (54). To study the effects that mutation of these residues has on telomeric silencing in yeast, we employed strains that utilized an ADE2 reporter at the telomeres of chromosome V. Substitutions at H4 K79 displayed a pattern of phenotypes for telomeric silencing similar to that of a well-characterized acetylated residue, histone H4 K16. We found that the K79Q substitution leads to a loss of telomeric silencing, whereas K79R has no effect on silencing, suggesting that the positive charge is required for telomere silencing. These results are consistent with K79 being maintained in a deacetylated state at silent telomeres (Fig. 3A). The substitutions at histone H4 K77, however, gave the opposite result: K77Q appears normal for telomeric silencing, whereas K77R causes silencing to increase. These results would be consistent with the acetylation of K79 at silent telomeres (Fig. 3A). Consistent with the notion that modification of these residues is involved in the regulation of telomeric silencing, both H4 K77A and H4 K79A aberrant histone proteins cause telomeric silencing to be lost. Quantitative real-time PCR was undertaken to verify that the color changes reported in the silencing assay accurately represent differences in levels of ADE2 transcript. Values obtained did indeed corroborate the genetic data, as we saw that the trend in ADE2 mRNA levels for the mutants mirrored their effects on silencing (Table 2).
FIG. 3.
(A) Telomeric silencing assay of histone H4 K16, K77, and K79 substitutions. Strains containing an ADE2 reporter inserted into the telomeric regions of chromosome V were transformed with plasmids containing the substituted histones and plated onto SC-Trp plates. After incubation at 30°C for 2 days, plates were put at 4°C for a further 7 days to facilitate the development of the red color. WT and loss-of-telomeric-silencing (LTS) strains are shown for comparisons. (B) ribosomal DNA (denoted rDNA in the figure) silencing assay of histone H4 K77 and K79 substitutions utilizing a strain containing mURA3 and a MET15 marker inserted into ribosomal DNA repeats. Cells were plated with an initial OD600 of 0.5 and serially diluted fivefold on SC-Trp for a growth control and SC-Trp-His plus 5-FOA to analyze silencing of the mURA3 reporter. Additionally, colonies were streaked onto complete medium containing PbNO3 and incubated at 30°C for 5 days. Brown color formation was analyzed after 1 week at 4°C. WT, loss-of-ribosomal DNA-silencing (LRS), and met15 null strains are shown as controls.
TABLE 2.
Real-time PCR analysis of effects of histone alleles on expression of telomeric ADE2a
| Substitution | ΔΔCt (+/−SD) | Relative expression of subtelomeric ADE2 |
|---|---|---|
| WT H4 | 0 | 1 |
| LTS allele | −1.36 (0.11) | 2.6 |
| H4 K16A | −1.32 (0.16) | 2.5 |
| H4 K16R | 0.34 (0.15) | 0.8 |
| H4 K16Q | −1.85 (0.13) | 3.6 |
| H4 K77A | −1.31 (0.09) | 2.5 |
| H4 K77R | 1.14 (0.18) | 0.4 |
| H4 K77Q | −0.50 (0.02) | 1.4 |
| H4 K79A | −1.0 (0.37) | 2 |
| H4 K79R | −0.68 (0.01) | 1.6 |
| H4 K79Q | −1.07 (0.41) | 2.1 |
RNA was prepared from the indicated substitution strains and reverse transcribed into cDNA. Quantitative PCR was undertaken to amplify both ADE2 and ACT1 cDNA in the presence of SYBR green. ΔΔCt was calculated using the formula (Ct ADE2 − Ct ACT1)mutant − (Ct ADE2 − Ct ACT1)wild type. Values were also compared with those of an RT-negative control for each sample. Results are representative of RNA that was prepared from two individual colonies of each sample, both of which were assayed in quadruplicate.
The ribosomal DNA locus on chromosome XII is also maintained within a transcriptionally repressed chromatin structure (15, 45). A previously described strain was utilized that contains both MET15 and mURA3 insertions in the ribosomal DNA locus (38). Silencing could therefore be analyzed using two independent reporter systems. Consistent with the telomeric silencing results, K79R maintained wild-type levels of ribosomal DNA silencing, whereas K79Q induced an lrs phenotype as indicated by its white coloring on lead plates and its increased sensitivity to 5-FOA (Fig. 3B).
The support for acetylation of residue H4 K77 in the ribosomal DNA is less apparent. The K77R substitution has no effect on ribosomal DNA silencing, consistent with it being deacetylated at the ribosomal DNA locus. However, neither the K77Q nor the K77A mutation leads to the lrs phenotype but, rather, confers an irs (increased ribosomal DNA silencing) phenotype (Fig. 3B), suggesting a more repressed chromatin structure. This suggests that either (i) the positive charge of lysine 77 limits the amount of ribosomal DNA silencing, or (ii) perhaps K77 modification does not play a role in silencing at this locus.
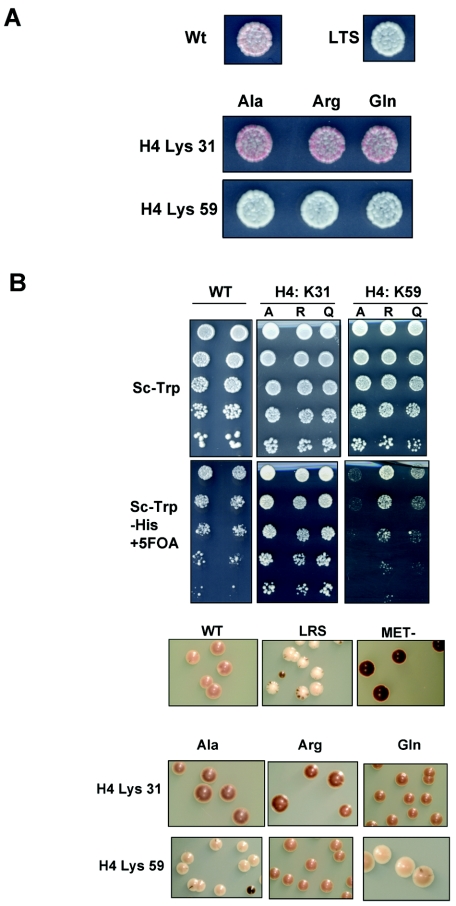
Additionally, our genetic analysis of K59 in histone H4 supports a role for its modification at ribosomal DNA. An earlier study suggested that ribosomal DNA silencing was not affected by mutations in this residue (54), but we saw clear silencing defects in the H4 K59A and K59Q mutants (Fig. 4B). However, as K59 has been shown to be a substrate for methylation in B. taurus, we are unable to draw conclusions from the K59Q mutation. Nonetheless, we can conclude from this analysis that the positive charge on an unmethylated K59 is necessary for effective ribosomal DNA silencing. The discrepancy with the earlier study is likely due to the fact that the strength of ribosomal DNA silencing varies between strain backgrounds, and our silencing studies are done in a strain background in which differences are easily detected (38, 45).
FIG. 4.
Transcriptional silencing assays for reporter strains expressing substitutions at histone H4 K59 and K31. (A) Cells were plated and analyzed as for Fig. 3. (B) ribosomal DNA silencing was monitored by plating and analysis as for Fig. 3.
Histone H3.
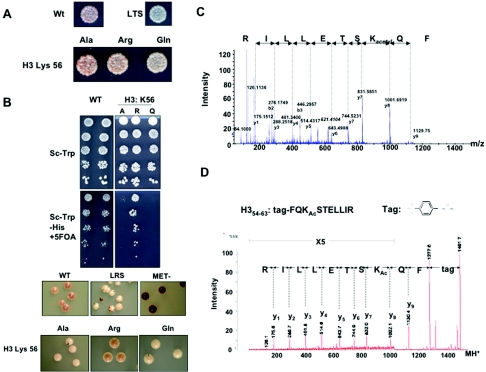
In an earlier study, the peptide fragment containing residues 52 to 62 of histone H3 showed an increase of mass of 14 Da by mass spectrometry that was consistent with methylation of a residue contained within this sequence (54). The possible candidates for this modification were R52, R53, and K56. We analyzed mutations at all three residues and noted that the genetics were consistent with K56 acetylation leading to a loss of silencing in the ribosomal DNA and to a lesser extent in the telomeres (Fig. 5). The K56Q mutant led to mild derepression of the reporter gene at telomeres, whereas the aberrant histone K56R retained wild-type levels of silencing (Fig. 5A). Substitution of K56 with glutamine additionally interfered with heterochromatin formation at ribosomal DNA and led to an lrs phenotype, whereas replacement of lysine with an arginine residue had no effect (Fig. 5B). Assuming that the presence of arginine mimics a persistent deacetylated state, these data suggest that the lack of modification at K56 sites is required for efficient silencing at both ribosomal DNA and telomeres. Consistent with this hypothesis, K56A caused a loss of ribosomal DNA silencing. Although the initial data generated from B. taurus suggested that K56 was monomethylated, the genetic analysis supported its acetylation in S. cerevisiae. We therefore undertook mass spectrometry analysis to reconcile these conflicting data and determine the exact state of K56 in yeast. We showed by both MALDI post-source decay (Fig. 5D) and quadrupole time-of-flight mass spectrometry analysis (Fig. 5C) that K56 is acetylated in bulk yeast histones (Fig. 4C). (K56 acetylation was first reported to us by Hiroshi Masumoto and Alain Verreault at London Research Institute, who have obtained extensive independent evidence for this modification [personal communication].) Additionally, during the preparation of the manuscript H3 K56 acetylation was reported by another independent group (51).
FIG. 5.
Acetylation of H3 K56 in S. cerevisiae. (A) Silencing reporter strains expressing alanine, glutamine, and arginine substitutions at histone H3 K56 were plated and analyzed as for Fig. 3. (B) ribosomal DNA silencing strains were plated and analyzed as for Fig. 3. WT, loss-of-ribosomal DNA-silencing (LRS), and met15 null strains are shown as controls. (C) MS/MS spectrum of the doubly charged lysine acetylated peptide ion, FQKacetylSTELLIR. The complete sequence including the acetylated lysine residue was deduced from the y-ion series as shown. Diagnostic fragment ions (53) for an acetylated lysine residue are observed at m/z 84.1 and m/z 126.1. (D) The MALDI postsource decay spectrum of a histone H3 peptide isolated from a trypsin-digested total yeast histone preparation. The peptide was N-terminally sulfonated prior to mass spectrometric analysis.
A second possible site of modification within the core of histone H3 is K115 acetylation. In support of this, the substitutions of this residue show the predicted pattern of transcriptional silencing phenotypes for a deacetylated residue both at telomeres and at ribosomal DNA (see Table S1 in the supplemental material). Similarly, it appears that the previously observed acetylation of histone H3 K122 might affect ribosomal DNA silencing. Mutations that incorporate either a glutamine or an arginine residue at these sites result in lrs and irs phenotypes, respectively, which suggests that the unmodified version of K122 is important for ribosomal DNA silencing but also indicates that a permanently deacetylated state is undesirable. Therefore, our genetic results could be interpreted as follows: tight regulation of the modified state of K122 is required at ribosomal DNA (see Table S1 in the supplemental material).
Predicted modifiable core histone residues that play a modification-independent role in heterochromatin formation.
Certain predicted modifiable residues, however, did not show phenotypic patterns consistent with modification state (Table 3). For example, all substitutions made at histone H4 K31 and H4 S47 resulted in increased telomeric and ribosomal DNA silencing, suggesting that the mutations generate a form of chromatin that is presumably less accessible to the transcriptional machinery. Conversely, mutations at H4 K59 and R92 and H3 R52, T118, and K122 facilitated the complete expression of the ADE2 reporter at telomeres, regardless of the replacement. This suggests that telomeric heterochromatin depends upon the identity of the particular amino acid side chain at each of these sites (see Fig. 4A for representative data). However, we cannot assume that these amino acids are not sites of modification in S. cerevisiae, as indeed we have previously discussed that the mutation of residues H4 K59 and H3 K122 produced phenotypes at rRNA that supported their acetylation. Perhaps the modifications of H4 K31, S47, and R92 and H3 R52 and T118 might be critical for other unexplored cellular processes, or perhaps the substitutions do not faithfully mimic the effects of the modifications in these instances.
The DNA damage response is impaired by histone H3 and H4 core mutations.
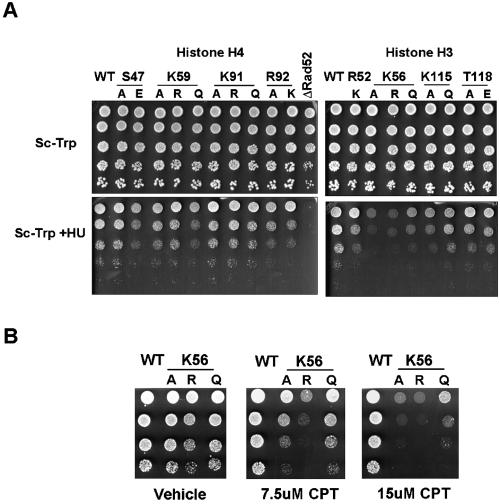
To determine whether mechanisms underlying DNA-templated cellular processes utilize modification of the nucleosome core, we initially explored the sensitivity of each of the 39 histone alleles to various DNA-damaging reagents, including MMS, HU, and CPT. None of the single amino acid substitutions conferred resistance or hypersensitivity to MMS. However, as depicted in Fig. 6, certain mutant histones led to an impaired response to HU treatment. Histone H4 variant proteins S47E, K59A, K59Q, R92A, and R92K displayed growth defects on medium containing 200 mM HU in comparison to wild-type H4 strains. Residues S47, K59, and K91 therefore showed contrasting phenotypes for the different substitutions, suggesting that it is the modification at these sites that may control accessibility to DNA damaging agents or the efficacy of DNA repair pathways. The exact structural attributes of residue H4 R92 on the other hand appear necessary for recovery from DNA damage, as all substitutions made at this residue confer increased HU sensitivity. Histone H3 K115A, K115Q, T118A, T118E, K56A, and K56Q mutants also were hypersensitive to HU, suggesting that the modification of these amino acids is influencing DNA repair. Of the above mutants, only H3 K56A and H3 K56Q were sensitive to CPT (Fig. 6B). The differential sensitivity to these two drugs, both of which are proposed ultimately to produce double-stand breaks, may be explained by their different modes of action. HU inhibits ribonucleotide reductase, leading to replication defect and subsequent fork collapse, whereas CPT is a topoisomerase I inhibitor. Presumably, the detailed structure, positioning, or timing of these two types of double-strand breaks is different enough that they end up packaged differently in the different types of mutant nucleosomes.
FIG. 6.
The DNA repair response is affected by substitutions of core nucleosome residues. (A) Strains expressing mutated histones were plated with an initial OD600 of 0.5 and serially diluted fivefold on SC as a growth control and SC plus 200 mM HU to analyze the ability of these cells to recover from DNA damage-induced stress. A rad52 null strain was used as a positive control. (B) Cells harboring substitutions at H3 K56 were examined on plates containing CPT at the indicated concentrations. The initial density of cells corresponded to an OD600 of 0.5, and cells were serially diluted fivefold.
DISCUSSION
Histones have been mutagenized and subjected to a wide variety of genetic screens (47). Numerous investigators have mutagenized individual histone residues to test specific hypotheses for their roles. More general screens have been performed for histone mutants that perturb interactions with chromatin remodelers (12, 18, 26, 40) or that decrease or increase silencing (10, 38) and histone variants important in centromere function (16). In each case, multiple alleles with the desired phenotypes were isolated, although the screens have probably not been done to saturation, suggesting there is much more to learn through such an approach.
In this study we have taken a more structural approach, probing systematically the residues known to be modifiable and testing their biological roles. A conclusion that may seem surprising is that the identity of these modifiable residues, which are very highly phylogenetically conserved, can be changed without killing the cell—we infer that drastically different side chains do not compromise the basic function of these histones to form nucleosomes. On the other hand, since these residues are on the nucleosome surface at a solvent interface and can clearly function in one or more chemically modified states, it is perhaps not so surprising that the side chains can be modified. On the other hand, the percentage of these mutants that display obvious phenotypes relating to altered silencing (92%) or sensitivity to DNA-damaging agents (33%) is astonishingly high and suggests possible active roles for the nucleosome surfaces associated with these residues.
At least eight residues within the histone fold domains of bovine histones H3 and H4 are subject to posttranslational modification; however, the functional significance of these modifications is unknown. The genetic data presented here indicate that several of these residues are critical for epigenetic regulation of gene expression in S. cerevisiae. Moreover, different amino acid substitutions confer distinct and indeed opposite phenotypes on gene silencing, strongly suggesting that these core residues are also subject to posttranslational modification in S. cerevisiae. The mechanistic pathways underlying the consequences of these modifications may involve direct effects on nucleosomal structure or packing, or they may implicate the interaction with a nonhistone protein, or both. By considering the observed phenotypes of mutations engineered at these sites with respect to the precise location of the specific residue on the nucleosome structure, we generated testable hypotheses to address this.
Lateral surface residues.
Histone H4 residues K77 and K79 are both located on the lateral surface of the nucleosome within the DNA-protein interface (Fig. 1B). H4 K77 does not directly contact the DNA but is positioned close enough that an interaction could be facilitated through a water molecule (6). H4 K79, however, directly interacts with the DNA. Both residues are located in a region of histone H4 called the L2 loop, which has been shown in the nucleosome crystal structure to interact with the H3 L1 loop, generating a DNA binding surface known as L1L2 (28). The lack of an observable silencing phenotype for the K79R substitution indicates that the presence of a positive charge at this location is sufficient for normal histone H4 functions at heterochromatin. As the K79Q mutation leads to a destabilization of heterochromatin, we can hypothesize that acetylated K79 will interfere with DNA-protein interactions and render the nucleosome more mobile, suggesting that its acetylation increases the accessibility of DNA. This is consistent with the recently proposed “regulated nucleosome mobility” model (4), which suggests that modifications that alter histone-DNA interactions regulate the equilibrium between mobile and relatively stationary nucleosomes. In contrast, the H4 K77 mutations are less easily interpreted by this model. Mutations in residue K77 are consistent with acetylation of this residue being essential for telomeric silencing but inhibitory to silencing at the ribosomal DNA locus. This paradox can be reconciled by the fact that different proteins are required in the establishment of silencing at each of these loci, and perhaps K77 modification is important for this distinction. This observed effect at telomeres is consistent with findings that knockout of class I histone deacetylases, which presumably leads to increased acetylation of histones (and perhaps other proteins), paradoxically enhances silencing in several organisms (9, 37, 46).
Other modifiable residues that lie on the lateral surface of the nucleosome include H3 K115, K122, H4 K31, and S47. Similar to H4 K77, they all have the potential to participate indirectly in DNA binding. K115 and K122 specifically lie in the L2 loop region of H3, which interacts with H4 L1 loop, forming another L1L2 DNA binding surface in the dyad axis of the nucleosome. Both residues were shown to be acetylated in bulk histones from B. taurus. The phenotypes we observed for K115 mutations are consistent with K115 acetylation in the regulation of DNA-protein interactions, as the K115Q substitution causes derepression at both ribosomal DNA and telomeres, whereas the K115R mutation has no effect. Similar observations were made for H3 K122 mutant phenotypes at the ribosomal DNA locus. However, at telomeric regions, the contribution of K122 appears to be dependent on side chain identity, as arginine cannot substitute for its function, and both K122A and K122Q also result in the loss of silencing. Therefore, as for H4 K77, it appears that H3 K122 behaves differently at distinct silent regions of the genome.
The decrease in accessibility of DNA to the transcription machinery observed at telomeres for all substitutions at H4 K31 and S47 is intriguing. In spite of their location directly beneath the DNA on the lateral surface, alterations to their chemical make-up, mimicking a persistently modified state, actually lead to an increase in silencing. The molecular mechanisms of this remain to be elucidated, but hypotheses predicted by these data are that (i) these residues play a role in recognition of a silencing inhibitor in the unmodified state, or (ii) they have a direct effect on nucleosome compaction.
H3 T118I has previously been identified as a SWI/SNF-independent or SIN allele (26). Sin− mutations are thought to suppress mutations in the ATP-dependent nucleosome remodeling complex by causing increased nucleosome mobility. In the crystal structure, H3 T118 directly interacts with DNA and H4 R45, which protrudes into the DNA minor groove, and this latter interaction is presumed to ensure a nonspecific interaction between R45 and DNA. A nucleosome containing H3 T118I has been crystallized (34) and indeed exhibits weakened DNA contacts in this region. Furthermore, H3 T118I suppresses a SUC2 UAS deletion mutation, thereby allowing transcription of this gene solely from its basal promoters (39). This additionally supports its effect on DNA accessibility through increased nucleosome mobility. Phosphorylation of H3 T118 is predicted to similarly weaken histone-DNA interactions; however, this modification is presumably tightly regulated and/or highly localized within the cell, since replacing H3 T118 with glutamic acid (or alanine) is lethal to yeast and both H3 T118E and T118A exert a dominant-negative effect at silent loci. In addition, T118A showed a slight increase in sensitivity to HU. Undoubtedly, further analysis is required to uncover the exact biological significance of phosphorylated T118. It should be noted that both potentially phosphorylated residues, H4 S47 and H3 T118, lie in close proximity on the nucleosome surface, therefore having the potential to generate an acidic patch.
H3 K56 is located near a terminal DNA segment that enters (or leaves) the nucleosome. Its side chain is surface exposed, and it is positioned close to the edge of the lateral surface of the nucleosome. Our mass spectrometry results verify acetylation of this residue in S. cerevisiae, and it is evident from our genetic data that this modification plays a role in heterochromatin formation. K56 mutants also show an increase in sensitivity to numerous DNA damage-causing agents. Given its location on the three-dimensional structure of the nucleosome, we hypothesize that acetylation of K56 might induce detachment of DNA from the nucleosome, increasing DNA accessibility to both transcription machinery and repair proteins. Consistent with this theory, we show that the unmodifiable K56R variant protein, which presumably would not facilitate such unraveling, is the most sensitive to DNA-damaging drugs. As many ATP-dependent nucleosome remodeling complexes act through this mechanism, it will be interesting to probe for interactions between enzymes that modulate K56 modification and such remodeling complexes. Indeed, the recent independent report of H3 K56 acetylation (51) showed that this modification is required for recruitment of Snf5, a SWI/SNF nucleosome remodeler to both the HTA1 and SUC2 genes.
Residues on the nucleosome face.
Certain modifiable residues map to the face of the nucleosome. One such residue is histone H4 K59, clearly shown to be methylated in bovine histones. Replacement of this lysine with alanine, arginine, or glutamine inhibited the silencing mechanisms at telomeres. The exposure of this side chain on the nucleosome surface suggests that it can act as part of a binding surface that regulates internucleosomal interactions or is recognized by a silencing factor (e.g., COMPASS). In the latter case, as no amino acid substitution accurately mimics the methylated state, we speculate that such a factor could interact with methyl lysine. At the ribosomal DNA, however, we see that silencing is maintained in the presence of H4 K59R, consistent with heterochromatin formation at ribosomal DNA relying on the unmodified residue.
Finally, there are two modifiable residues in histone H4, namely K91 and R92, that are not surface exposed but buried in the nucleosome core. They are located close to the interface between the H3/H4 tetramer and H2A/H2B dimer, and K91 participates in a salt bridge with H2B E63. During the preparation of the manuscript, it was revealed by Ye et al. (52) that the acetylation of K91 is important for regulating chromatin assembly. Our data are consistent with theirs in that the unmodifiable K91R variant protein can substitute for the native protein and no phenotypes were observed for this mutation in the assays under examination. K91Q, however, showed slight loss of ribosomal DNA silencing as well as increased sensitivity to hydroxyurea, indicating that perturbing this interaction may cause generalized destabilization of chromatin structure. Either substitution of R92 to alanine or lysine eliminated silencing at telomeres and rendered the cell more sensitive to DNA damage, indicating an important role for this residue in the nucleosome core.
The genetic analysis presented here provides insights into the role of previously uncharacterized nucleosome core modifications. Our systematic mutational analysis has permitted us to probe for the influence of these modifications on silencing as well as on the DNA repair response pathway. While our observations are consistent with the modification of several of these residues in S. cerevisiae, we acknowledge that the results could reflect the major charge changes introduced at these locations. However, earlier insights into tail modifications gained using this strategy (31) and the recent independent identification of both H3 K56 and H3 K91 acetylation in S. cerevisiae lend credence to our claim and suggest that direct physical evidence supporting the remaining sites of modification may be uncovered in the future. Although at this point we can only speculate as to the detailed mechanisms that implement the functions of these modifications, it is evident that they are biologically significant and warrant further investigation. Several studies have shown that combining mutations in the N-terminal modifiable residues can lead to complex and sometimes synergistic phenotypes (10, 31, 32). Considering the potentially complex network defined by the modifiable residues on the nucleosome core, it would be interesting for future work to include an analysis of different combinations of these core mutations.
Supplementary Material
Acknowledgments
This work was supported by the Technology Center for Networks and Pathways of Lysine Metabolism (NIH RR020839).
We are grateful to Alain Verreault and Hiroshi Masumoto for sharing unpublished data and Pamela Meluh for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 2.Chang, C. H., and D. S. Luse. 1997. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 272:23427-23434. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656-660. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. [DOI] [PubMed] [Google Scholar]
- 5.Cost, G. J., and J. D. Boeke. 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12:939-941. [DOI] [PubMed] [Google Scholar]
- 6.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319:1097-1113. [DOI] [PubMed] [Google Scholar]
- 7.de la Cruz, X., S. Lois, S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Do protein motifs read the histone code? Bioessays 27:164-175. [DOI] [PubMed] [Google Scholar]
- 8.Delano, W. L. 2002. The PyMOL molecular graphics system. Delano Scientific, San Carlos, Calif.
- 9.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384:589-591. [DOI] [PubMed] [Google Scholar]
- 10.Dion, M. F., S. J. Altschuler, L. F. Wu, and O. J. Rando. 2005. From the cover: genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327:85-96. [DOI] [PubMed] [Google Scholar]
- 12.Duina, A. A., and F. Winston. 2004. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell. Biol. 24: 561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eissenberg, J. C. 2001. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene 275:19-29. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 15.Fritze, C. E., K. Verschueren, R. Strich, and R. Easton Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 18.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka, M., and M. M. Smith. 2003. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 13:154-160. [DOI] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, L. M., P. S. Kayne, E. S. Kahn, and M. Grunstein. 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, D. O., I. G. Cowell, and P. B. Singh. 2000. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22:124-137. [DOI] [PubMed] [Google Scholar]
- 23.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 26.Kruger, W., C. L. Peterson, A. Sil, C. Coburn, G. Arents, E. N. Moudrianakis, and I. Herskowitz. 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9:2770-2779. [DOI] [PubMed] [Google Scholar]
- 27.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 28.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 29.Lusser, A., D. L. Urwin, and J. T. Kadonaga. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12:160-166. [DOI] [PubMed] [Google Scholar]
- 30.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294-298. [DOI] [PubMed] [Google Scholar]
- 31.Megee, P. C., B. A. Morgan, B. A. Mittman, and M. M. Smith. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247:841-845. [DOI] [PubMed] [Google Scholar]
- 32.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 33.Moore, J. D., and J. E. Krebs. 2004. Histone modifications and DNA double-strand break repair. Biochem. Cell Biol. 82:446-452. [DOI] [PubMed] [Google Scholar]
- 34.Muthurajan, U. M., Y. Bao, L. J. Forsberg, R. S. Edayathumangalam, P. N. Dyer, C. L. White, and K. Luger. 2004. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23:260-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. [DOI] [PubMed] [Google Scholar]
- 36.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 37.Olsson, T. G., K. Ekwall, R. C. Allshire, P. Sunnerhagen, J. F. Partridge, and W. A. Richardson. 1998. Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res. 26:3247-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, J. H., M. S. Cosgrove, E. Youngman, C. Wolberger, and J. D. Boeke. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32:273-279. [DOI] [PubMed] [Google Scholar]
- 39.Prelich, G., and F. Winston. 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 42.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 43.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 45.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. S., E. Caputo, and J. D. Boeke. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19:3184-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, M. M., and M. S. Santisteban. 1998. Genetic dissection of histone function. Methods 15:269-281. [DOI] [PubMed] [Google Scholar]
- 48.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 49.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745-756. [DOI] [PubMed] [Google Scholar]
- 50.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375-385. [DOI] [PubMed] [Google Scholar]
- 52.Ye, J., X. Ai, E. E. Eugeni, L. Zhang, L. R. Carpenter, M. A. Jelinek, M. A. Freitas, and M. R. Parthun. 2005. Histone H4 lysine 91 acetylation A core domain modification associated with chromatin assembly. Mol. Cell 18: 123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, K., P. M. Yau, B. Chandrasekhar, R. New, R. Kondrat, B. S. Imai, and M. E. Bradbury. 2004. Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics 4:1-10. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, L., E. E. Eugeni, M. R. Parthun, and M. A. Freitas. 2003. Identification of novel histone posttranslational modifications by peptide mass fingerprinting. Chromosoma 112:77-86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.