Abstract
Thyroid transcription factor gene 1 (TTF-1) is a homeobox-containing gene involved in thyroid organogenesis. During early thyroid development, the homeobox gene Nkx-2.5 is expressed in thyroid precursor cells coincident with the appearance of TTF-1. The aim of this study was to investigate the molecular mechanisms underlying thyroid-specific gene expression. We show that the Nkx-2.5 C terminus interacts with the TTF-1 homeodomain and, moreover, that the expression of a dominant-negative Nkx-2.5 isoform (N188K) in thyroid cells reduces TTF-1-driven transcription by titrating TTF-1 away from its target DNA. This process reduced the expression of several thyroid-specific genes, including pendrin and thyroglobulin. Similarly, down-regulation of TTF-1 by RNA interference reduced the expression of both genes, whose promoters are sensitive to and directly associate with TTF-1 in the chromatin context. In conclusion, we demonstrate that pendrin and thyroglobulin are downstream targets in vivo of TTF-1, whose action is a prime factor in controlling thyroid differentiation in vivo.
Differentiated thyroid follicular cells are characterized by the ability to synthesize thyroid hormones and by the expression of a specific set of genes, which includes thyroglobulin (Tg), thyroperoxidase (TPO), thyroid-stimulating hormone receptor (TSH-R), and the specific iodine symporter Na+/I− (NIS). Thyroglobulin, the major secretory product of the thyroid, is the protein precursor for thyroid hormones. It is a 330-kDa glycoprotein that undergoes homodimerization in the endoplasmic reticulum and other posttranslational modifications (i.e., phosphorylation, N- and O-glycosylation, sulfation, and iodination) (13). Thyroglobulin mRNA production is regulated by TSH, as well as by insulin and insulin-like growth factor I (45).
Pendrin is an anion transporter, encoded by the PDS gene. It is expressed in the thyroid gland (4), kidney (43), mammary gland (40), uterus (51), testes (31), and placenta (3). Mutations in the human PDS gene cause the Pendred syndrome (48, 49), which is a relatively common autosomal-recessive disorder characterized by deafness and goiter (37). Deafness is congenital and generally severe, although some late-onset cases can result from light head injury. The cause of goiter, which generally develops around puberty (42), appears to be impaired iodide fixation in the follicular lumen due to a reduced rate of iodide transport across the apical membrane of thyroid gland epithelial cells (39). Pendrin expression has been found in the inner ear as well as in the thyroid gland, which reflects the clinical signs of deafness and goiter (16, 17).
The thyroid-specific expression of various thyroidal genes requires a subset of transcription factors among which thyroid transcription factor 1 (TTF-1; also referred to as Nkx-2.1 and T/EBP) (7). TTF-1 is a 42-kDa homeodomain-containing protein expressed also in the lung and part of the forebrain (10). It belongs to the NK2 class of homeobox proteins that have a tyrosine residue at amino acid 54 of the homeodomain and a conserved 23-amino-acid NK2-specific domain (5). Gene-targeted deletion of the mouse TTF-1 gene is embryonically lethal. These animals have defective thyroid and lung development and brain malformation (29). TTF-1 activates transcription of cotransfected Tg and TPO promoters in nonthyroid cells (7), which suggests that TTF-1 is important for the transcriptional activation of thyroid-specific genes. Interestingly, the presence of the TTF-1 protein does not invariably correlate with active transcription of the Tg and TPO genes in thyroid development (8). From embryonic day 8.5 (E8.5) on, TTF-1 is present in the thyroid bud together with the other thyroid transcription factors TTF-2 and Pax-8, whereas Tg and TPO do not appear before E13. Since TTF-1-null mice lack a thyroid gland, many aspects of the role of TTF-1 in thyroid remain obscure. Similarly, nothing is known about the relationship between TTF-1 and pendrin expression or about the mechanisms underlying pendrin expression in the thyroid gland.
TTF-1 contains three functional domains: an N-terminal transactivation domain, a DNA-binding domain (HD), and a C-terminal transactivation domain (9). There is compelling evidence that TTF-1 functions by forming complexes with other transcription factors on the regulatory regions of target genes. TTF-1 interacts with retinoic acid receptors and associated cofactors (54), nuclear factor 1, members of the AP-1 family, and BR22 (55, 56). It has been suggested that TTF-1 plays a central role in the stabilization of these complexes.
Another component of the NK2 family is Nkx-2.5, which is a homeodomain-containing protein, originally identified as a potential vertebrate homologue of the Drosophila gene tinman (30). Nkx-2.5 plays a crucial role in heart morphogenesis (5). Its transcripts are found in the heart (26), in heart progenitor cells (33), in mammary gland during lactation (11), and in the thyroid primordium during development (26). NKX-2.5 transcripts were identified in the pharyngeal endoderm at stage E8.5 to E9.5 in a subset of cells that start to differentiate and migrate antero-ventrally to constitute the thyroid gland (5). At later stages, pharyngeal expression of NKX-2.5 is limited to the area corresponding to the thyroid primordium (5) and is undetectable thereafter. Several mutations in the human NKX-2.5 gene, including the N188K mutation (2), have been found in patients with congenital heart disease (23, 24, 27, 47, 57).
Transmission is autosomal dominant, with incomplete penetrance (2, 52, 53). Because Nkx-2.5 acts as a homodimer through its homeodomain (28), dimerization is impaired by some disease-causing mutants that are dominant negative over the wild-type counterpart (57). We previously demonstrated that Nkx-2.5 induces the expression of the human type 2 deiodinase (12), a selenoenzyme critical in thyroid hormone metabolism, expressed in both human thyroid and heart (44). However, the role of Nkx-2.5 in thyroid development remains poorly understood.
The aims of our study were to examine the mechanisms underlying TTF-1-driven transcription and the effect of reduced TTF-1 activity on thyroid cells in vivo. Here we demonstrate that Nkx-2.5 interacts with TTF-1 and that a dominant-negative isoform (N188K) blocks TTF-1 transcriptional activity by titrating it away from its target DNA. We also demonstrate that TTF-1 is an upstream regulator of the pendrin and thyroglobulin genes in thyroid cells, thereby controlling thyroid differentiation in vivo.
MATERIALS AND METHODS
Plasmids and expression constructs.
Plasmids encoding TTF-1, Nkx-2.5, and N188K are described elsewhere (12). To obtain the C-terminal-deleted Nkx-14 construct, we ran a PCR using wild-type Nkx-2.5 as template, and oligonucleotides Nk1s and Nk2r (Table 1). The resulting fragment was subcloned into the pFLAG-CMV2 plasmid (Sigma Chemical Co., St. Louis, MO). The generated plasmid was sequenced in a double orientation for sequencing control. For RNA interference (RNAi) experiments in Fischer rat thyroid line 5 (FRTL-5) cells, chemically synthesized oligonucleotides encoding rat TTF-1-specific double-stranded RNA including the loop and polymerase III (Pol III) termination sequence were converted to double-stranded DNA and subcloned downstream to an RNA Pol III promoter into a modified pcDNA3.1 plasmid (kindly provided by A. Leonardi). The generated plasmids, designated Si1 and Si2, were sequenced in a double orientation for sequencing control. A scrambled sequence that did not show any identity with genes in the GenBank database was used as a negative control for the control transfected cells (Ctr). The chloramphenicol acetyltransferase (CAT) reporter plasmids C5-CAT (36) and Tg-CAT (50) and deletion mutant plasmids Δ1, Δ14, Δ35, and ΔG7 have been described elsewhere (14).
TABLE 1.
Oligonucleotides used to generate plasmids and for EMSA
| Oligonucleotide | Sequence (5′-3′) | Orientation |
|---|---|---|
| rPDN1s | GTCATAGGGAGACCATAAGACAG | Sense |
| rPDN2r | CGCGACGAGGGAAGACACTCAGA | Antisense |
| rpPDNmutS | GATCAAACTCGTGTGCCTTTTA | Sense |
| rpPDNmutR | TAAAAGGCACACGAGTTTGATC | Antisense |
| pGL31S | GGTACCGAGCTCTTACGCGTGCTAG | Sense |
| rpPDNmR | AAGGGGAAGCCCTGGGTCCTG | Antisense |
| rPDS1S | GTCATAGGGAGACCATAAGACAG | Sense |
| rPDS1R | AGAACTCAACCATCACTCTGTCA | Antisense |
| rPDS3S | GATCCTGTGGTTTTGGTGCTGC | Sense |
| rPDS3R | GTGGCATCTCTGGGTTGGTCC | Antisense |
| rTG3S | CCCATGTCCTGGAGTGGTCAC | Sense |
| rTG3R | GCTGCTACCAGGCAGACTGAG | Antisense |
| rpPDNrac1 | CCTTGACTCGGTATTTTGGGA | Antisense |
| rPDN+57R | TACACCGGCCGCGACACCGCATA | Antisense |
| rPENr | TCCACGCACACTCTGGGCTGTAAC | Antisense |
| 3E-TKLucU | CTCAAACTCAAGTGCCTTTTCAAACTCAAGTGCCTTTTCAAACTCAAGTGCCTTTC | Sense |
| 3E-TKLucL | TCGAGAAAGGCACTTGAGTTTGAAAAGGCACTTGAGTTTGAAAAGGCACTTGAGTTTGAGAGCT | Antisense |
To isolate the rat pendrin 5′ untranslated region (UTR), a genomic rat DNA was used as the template in a PCR with oligonucleotides rPDN1s and rPDN2r (Table 1). The 3,074-nucleotide PCR product, which corresponds to the rat pendrin cDNA promoter and 5′ UTR, was subcloned into pGL3-basic vector (Promega, Madison, WI) to generate the rPdsLuc3.0 plasmid.
The rPdsLuc2.2 construct was generated by digesting the rPdsLuc3.0 plasmid with XhoI and religating the backbone vector. The rPdsLuc3.0 construct containing a transition from AA to GT in the E site in the context of the rPdnLuc3.0 construct was generated using recombinant PCR with two sets of oligonucleotides (the pair rpPDNmutS and rpPDNmutR and the pair pGL31S and rpPDNmR). Briefly, the two PCR products (pGL31S/rpPDNmutR and rpPDNmutS/rpPDNmR) were combined by PCR using pGL31S and rpPDNmR as outside oligonucleotides, and the final PCR product was reinserted into rPdsLuc3.0 digested with BstXI and BglII (Table 1).
To generate plasmids with a repeated pendrin putative “TTF-1 site” (3E-TKLuc or 3mE-TKLuc), oligonucleotides containing three tandem repeats of the TTF-1 binding sequence (5′-CTCAAACTCAAGTGCCTTTTCAAA-3′; sites underlined and bold are mutated in the E site [mE]) were chemically synthesized, annealed to the antisense oligonucleotides, and subsequently cloned into pTKLuc digested with SacI/XhoI (Table 1).
DNA transfection and CAT and Luc expression assays.
For CAT and luciferase (Luc) assays, we transfected HeLa and FRTL-5 cells using the calcium phosphate and FuGene (Roche Applied Science, Mannheim, Germany) precipitation method, respectively. For each 60-mm dish, 3 μg of CAT reporter vector were cotransfected with 0.2 μg of TTF-1-expressing vector and with 0.3 μg of Rous sarcoma virus-Luc vector as an internal control. For the uninduced control, the pFLAG-CMV-2 empty vector (where CMV is cytomegalovirus) was used in the same quantity as the Nkx-2.5 vector. CAT activities were measured 48 h after transfection, and differences in transfection efficiency were corrected relative to the level of luciferase activity. Each construct was analyzed in duplicate in at least three separate transfections. Luciferase assays were performed using a dual-luciferase reporter assay system (Promega, Madison, WI) 48 h after the transfection. Data (Luc/Renilla) are reported as means ± standard deviation (SD).
FRTL-5 cells stably expressing N188K and TTF-1-silenced clones.
FRTL-5 cells stably expressing NkxN188K were obtained by transfecting N188K plasmid (7 μg) into FRTL-5 cells using FuGene transfection reagent (Roche Applied Science, Mannheim, Germany). Two days after transfection, the cultures were grown in the presence of G418 (200 ng/ml). Forty clones were selected and screened by Western blot analysis using anti-Flag antibody (M2; Sigma, Chemical Co.). To obtain TTF-1-silenced FRTL-5 stable clones, we cotransfected Si2 (7.5 μg) and SiCtr plasmids with pPURO resistance vector (0.5 μg) into FRTL-5 cells. Two days after transfection, the cultures were grown in the presence of puromycin (1 μg/ml). Puromycin-resistant clones were isolated after 2 weeks of continuous selection. Thirty clones were selected and screened for the levels of endogenous TTF-1 by Western blot analysis.
Immunoprecipitations and Western blot analysis.
Total cell lysates of transiently transfected BOSC-23 cells were prepared in Busslinger lysis buffer (20 mM Tris, pH 7.9, 120 mM KCl, 5 mM MgCl2, 0.2% Nonidet-P40, 0.2 mM EDTA, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM sodium vanadate, 50 mM NaF). Total cell extracts (500 μg) were immunoprecipitated with anti-Flag monoclonal antibody conjugated to agarose beads or a corresponding anti-c-Myc as a negative control (Red anti-Flag M2 and anti-c-Myc affinity gel; Sigma Chemical Co). Two hours later, immunocomplexes were washed five times with lysis buffer, resuspended in sample buffer without β-mercaptoethanol, and heated at 65°C for 5 min. The supernatant was transferred to fresh tubes containing 5% β-mercaptoethanol. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto an Immobilon P (Millipore, Bedford, MA) membrane for 12 h at a constant current of 150 mA. Nkx-2.5 and TTF-1 were immunodetected with a monoclonal anti-Flag antibody (M2) and a rabbit polyclonal antibody against TTF-1 (14), respectively. Tg was immunodetected with a rabbit polyclonal antibody kindly provided by B. Di Jeso.
To obtain nuclear extracts from HeLa cells transiently cotransfected with Lipofectamine, (Invitrogen, Gaithersburg, MD), 5 μg of TTF-1, N188K, or CMV-Flag vector were mixed with 1.6 ml of OptiMem and 40 μl of Lipofectamine for 45 min and then added to the cells in 6 ml of total OptiMem. After 48 h, cells were washed and harvested by scraping in 2 ml of 1× phosphate-buffered saline, pH 7.4. After centrifugation at 500 × g, pellets were frozen at −80°C until required for the preparation of nuclear extracts. BOSC-23 (clone CRL-11270; American Type Culture Collection) cells were cotransiently cotransfected with Lipofectamine for immunoprecipitation assays. Four micrograms of total DNA was used for each 60-mm dish. Cells were collected 48 h after transfection.
mRNA preparation and Northern blot analysis.
Total RNA was extracted from cells using Trizol (Life Technologies Ltd., Paisley, Scotland), according to the manufacturer's directions. Poly(A)+ was isolated from 800 μg of total RNA for each sample using the Oligotex mRNA Midi kit (QIAGEN, Chatsworth, CA). After isolation, total RNA or poly(A)+ was spectrophotometrically quantified at 260 nm. Twenty micrograms of total RNA or 8 μg of poly(A)+ was resolved by electrophoresis through a formaldehyde gel and subjected to Northern blotting using Hybond N (Amersham-Pharmacia, Piscataway, NJ) according to the manufacturer's indications. For Tg and Pds detection, Northern blots were hybridized with cDNA fragments corresponding to rat Tg and Pds, respectively, and radiolabeled using a Prime-Lt II random primer labeling lit (Stratagene, La Jolla, CA). The signal was revealed by autoradiography using X-Omat film (Kodak).
Real-time PCR.
We used real-time PCR to detect variations in Nkx-2.5, Tg, NIS, and β-actin gene copies as described elsewhere (11). The cDNAs were amplified by PCR in an ABI Prism 7700 sequence detector (PE Biosystems, Foster City, CA) using the fluorescent double-stranded DNA-binding dye SYBR Green (Applied Biosystem, Warrington, United Kingdom). Specific primers for the Nkx-2.5, Tg, NIS, and β-actin genes (Table 2) were designed to work under the same cycling conditions (95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min) and generated products of comparable sizes (around 150 to 200 bp for each amplification). Primer combinations were positioned to span an exon-exon junction to avoid genomic DNA interference. All samples were run in triplicate.
TABLE 2.
Comparative analysis by real-time PCR of a subset of thyroid-specific genes
| Gene name | Accession no. | Relative inductiona |
|---|---|---|
| Tg | NM_030988 | −21.6 |
| Txnrd1 | NM_031614 | −5.3 |
| Pds | NM_019214.1 | −3.8 |
| TSH-R | NM_012888 | −2.15 |
| Nis | NM_052983 | −1.5 |
| TTF-1 | D38035 | −1.3 |
| Pax-8 | X94246 | −1.2 |
| TTF-2 | Y11321 | −1.07 |
| Tpo | NM_019353 | 1.17 |
| Tubulin | NM_022298 |
Relative induction (n-fold) is calculated in terms of Cl.1 versus pcDNA and represents the mean of three separate real-time PCRs run in duplicate.
Template concentration was calculated from the cycle number when the amount of PCR product passed a threshold established in the exponential phase of the PCR. The relative amounts of gene expression were calculated with β-actin expression as an internal standard (calibrator). The results, expressed as the relative difference N (for N-fold) in target gene expression, was determined with the formula: Ntarget = 2Ct(sample) − Ct(calibrator), where Ct is cycle threshold.
Semiquantitative EMSA.
We evaluated the binding of TTF-1 to DNA in the presence of Nkx-N188K by electrophoretic mobility shift assay (EMSA) using double-stranded end-labeled oligonucleotides D and C (12). Antisense oligonucleotides were labeled with a T4 polynucleotide kinase (New England Biolabs, Boston, MA) and radioactive [γ-32P]ATP. Double-stranded oligonucleotides were purified by passing them through nick columns containing Sephadex G-50 DNA grade resin (Amersham-Pharmacia).
Nuclear extracts were prepared as previously described (12) from transfected HeLa or FRTL-5 cells. EMSAs were performed with 30-μl reaction mixtures at room temperature at a final concentration of 20 mM Tris-HCl, pH 7.5, 75 mM KCl, 1 mM dithiothreitol, 10% glycerol, and 1 mg/ml poly(dI-dC). Binding reaction mixtures containing 1 μg of nuclear extracts and 10 fmol of the probe were equilibrated for 30 min at room temperature and separated by electrophoresis through a 6% polyacrylamide gel containing 0.5× Tris-borate-EDTA. Binding specificity was assessed by competition with a 100-fold molar excess (or as indicated) of cold competitors incubated with proteins 15 min before the probe was added.
Identification of the transcription start site.
The transcription start site (TSS) of the rat Pds gene was determined with the 5′ RACE system, version 2.0 (Invitrogen Laboratories, Inc.), for rapid amplification of cDNA ends (RACE), using total RNA from FRTL-5 cells and oligonucleotides specific for the rpPDNrac1 gene for first-strand cDNA synthesis. A homopolymeric tail was added to the 3′ end of the synthesized cDNA, and tailed cDNA was amplified by PCR (first PCR) using nested specific oligonucleotides (rPDN+57R) and the anchor primer provided with the kit (Table 1). The resulting DNA fragments were reamplified (second PCR) using an internal oligonucleotide (rPENr and the anchor primer provided with the kit; RACE products) producing three different DNA fragments corresponding to the indicated three alternative TSS (Fig. 5B). RACE products were subcloned into TOPO-TA vector (Invitrogen, Laboratories, Inc.) before sequencing. Of the 13 clones analyzed, three corresponded to the TSS −526 bp from ATG, five corresponded to the TSS −504 bp from ATG, and three corresponded to the TSS −486 bp from ATG. Given the relative frequencies of the sequenced PCR products, none of them could be identified as the major TSS. Reverse-transcription PCR (RT-PCR) analysis of 5 μg of reverse-transcribed total mRNA from FRTL-5 cells gave results consistent with the sites identified.
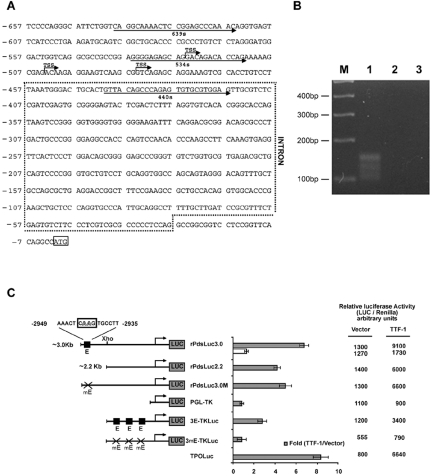
FIG. 5.
The Pds gene is a novel target of TTF-1. (A) Sequence of the −657 bp of the rat PDS 5′ UTR. Intron sequences (430 bp) as deduced by RT-PCR and RACE are boxed. TSSs as deduced by 5′ RACE are indicated by arrows. (B) RACE products (35 cycles), from reverse-transcribed total mRNA (1 μg) from FRTL-5 cells, were subjected to PCR with nested oligonucleotides as described (see Material and Methods), revealed by ethidium bromide staining, and photographed under UV light. The figure refers to one PCR, which was run twice with the same material with comparable results. M, 100-bp DNA ladder. Lanes 1 to 3 show the results of PCRs with nested oligonucleotides. Lane 1, RACE products obtained by using the DNAs from first PCR as template; lane 2, same as lane 1, but no DNA from first PCR used as template; lane 3, no cDNA. DNA products purified from lane 1 were subcloned into TOPO-TA vector, and 15 colonies were sequenced in double orientations. (C) Activities of various PDS promoter constructs. Luciferase reporter constructs (3 μg) containing various lengths of PDS promoter were cotransfected in HeLa cells with a plasmid encoding TTF-1 (0.2 μg/60-mm-diameter dish) or CMV-Flag together with 0.3 μg of Rous sarcoma virus-Renilla as an internal control. The relative activities of each Luc-reporter plasmid with cotransfected TTF-1 or CMV-Flag expressed as arbitrary units are also indicated. Deletion of the promoter region between 3.0 and 2.2 kb significantly decreased the relative activation (n-fold) of the promoter activity by TTF-1. Mutation of site E (CAAG to CGTG) in the context of the rPDSLuc3.0 construct was obtained by site-directed mutagenesis (rPDSLuc3.0 M) and analyzed in similar experiments. A plasmid with three-E tandem motifs upstream to the thymidine-kinase minimal promoter (3E-TKLuc) or its mutated counterpart (3mE-TKLuc) was tested in similar experiments. White bars indicate the increase (n-fold) in induction when 0.2 μg of TTF-1 RNAi plasmid was cotransfected (together with TTF-1) as a control of specificity. The relative activity of each luciferase reporter plasmid with cotransfected TTF-1 or CMV-Flag, expressed in arbitrary units, is also indicated. The data shown are means ± SD of the luciferase/Renilla ratios from four experiments performed in duplicate.
ChIP.
Briefly, approximately 8 × 106 cells were fixed with 1% formaldehyde in growth medium at 37°C for 30 min. Glycine was added to a final concentration of 0.125 M to stop cross-linking. Sonicated, cross-linked chromatin was centrifuged at 12,000 rpm for 30 min to remove insoluble material. The soluble chromatin was diluted 10-fold in dilution buffer and used directly for chromatin immunoprecipitation (ChIP) assays. Five percent of total DNA was used as a positive control for PCR assays (denoted “input DNA”). Six optical density (A260) units of chromatin from FRTL-5 or clone Cl.1 were mixed with 10 μl (0.3 μg/μl) of anti-TTF-1 (32) and incubated at 4°C for 3 h before precipitation with protein A/G Sepharose (Sigma Chemical Co., St. Louis, MO). After five rounds of washing, bound DNA-protein complexes were eluted by incubation with 1% sodium dodecyl sulfate-0.1 M NaHCO3 elution buffer. Formaldehyde cross-links were reversed by incubation in 200 mM NaCl at 65°C. Samples were extracted twice with phenol-chloroform and precipitated with ethanol. DNA fragments were recovered by centrifugation, resuspended in 50 μl of H2O, and used for real-time PCRs with oligonucleotide pairs rPDS1S ([minus]3,025 bp from ATG) and rPDS1R, rPDS3S (−14,545 bp from ATG) and rPDS3R, and rTG3S (−227 bp from ATG) and rTG3R (Table 1).
RESULTS
A dominant-negative Nkx-2.5 isoform (N188K) inhibits TTF-1 transcriptional activity.
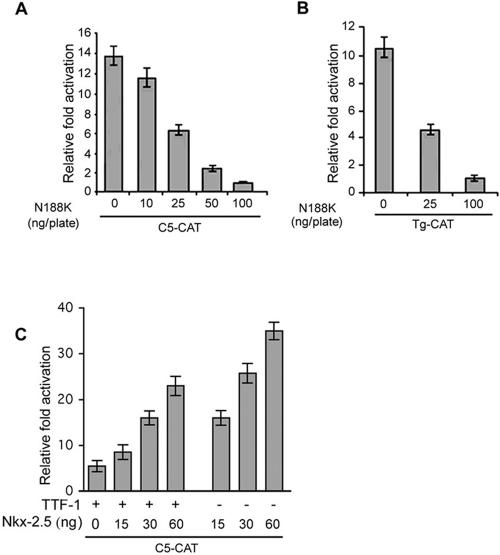
We recently demonstrated that the naturally occurring Nkx-2.5 mutant (N188K), which does not bind DNA or transactivate its target promoters, exerts a dominant-negative effect on the wild-type protein (12). Given the simultaneous expression of Nkx-2.5 and TTF-1 in the thyroid primordium, the virtually identical consensus DNA binding sites, and the 85% identity in the homeodomains of the two proteins, we investigated whether Nkx-2.5 affects TTF-1 transcriptional activity. To this end, we evaluated the effect of cotransfected N188K or wild-type Nkx-2.5 on TTF-1 activation of the artificial C5-CAT promoter. HeLa cells were transiently cotransfected with C5-CAT, TTF-1, and increasing amounts of N188K (Fig. 1A) or wild-type Nkx-2.5 (Fig. 1C). While Nkx-2.5 is a more potent inducer of C5-CAT than wild-type TTF-1 (Fig. 1C), N188K dose-dependently blocked TTF-1 induction, and even at a low concentration (100 ng/dish versus 200 ng/dish of TTF-1) it completely blocked TTF-1-induced C5-CAT promoter activation. TTF-1 transcriptional activity was specifically and completely abrogated in two physiologically relevant promoters that are in vitro targets of TTF-1, i.e., type 2 deiodinase (HD2-CAT) and Tg promoter constructs, in both HeLa and FRTL-5 cells (Fig. 1B, and data not shown). These results indicate that N188K is able to repress the effect exerted by TTF-1 on its downstream target promoters.
FIG. 1.
N188K represses TTF-1-dependent transcription in HeLa cells. (A and B) Cells were transiently cotransfected with C5-CAT or the thyroglobulin promoter (0.3 μg/plate), TTF-1 (0.2 μg/plate), and the indicated increasing amounts of N188K. Cells were harvested and assayed for CAT activity. Values are the mean of at least three experiments ±SD. (C) C5-CAT was cotransfected in HeLa cells with TTF-1 (15 ng) and the indicated amounts of Nkx-2.5 expression plasmids. Relative activation (n-fold) is the ratio between values obtained with and without cotransfection of the transcription factors. CMV-Luc was used as internal control, and CAT values were normalized to Luc activity.
N188K represses TTF-1-mediated transcription in thyroid cells and modifies the expression of a subset of thyroid-specific genes.
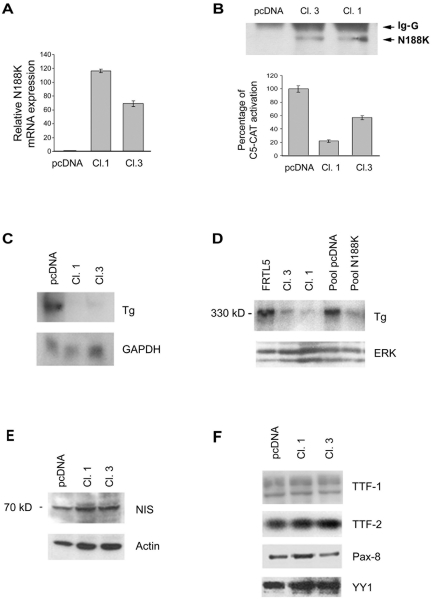
The finding that N188K is a potent repressor of TTF-1 prompted us to use N188K to reduce TTF-1 activity in thyroid cells. To this aim we generated FRTL-5 clones stably expressing Flag-tagged N188K under the control of a constitutively active promoter (CMV). After transfection, we selected cells that grew in antibiotic-selective medium (see Materials and Methods). Among the 40 clones analyzed (FRTL-5/N188K), clones 1 and 3 were found to express different N188K transgene levels at the mRNA (Fig. 2A) and protein level (Fig. 2B, upper panel). First, we measured TTF-1 activity using the TTF-1-responsive promoter construct C5-CAT. C5-CAT promoter activity, reported as 100% in mock-transfected cells (pcDNA clone), was significantly reduced in clone 1 (to 20% of total activity) and, to a lesser extent, in clone 3 (to 55% of total activity) (Fig. 2B, lower panel). As expected, the different promoter activities were not due to a reduction in endogenous TTF-1 protein levels as measured by Western blotting (Fig. 2F). We used these clones to study the effects of TTF-1 abrogation in a fully differentiated thyroid cell. The FRTL-5/N188K cells were viable and their replication rate did not differ from that of parental clones under normal conditions (data not shown).
FIG. 2.
Generation and characterization of FRTL-5 clones stably expressing Flag-tagged N188K. (A) FRTL-5 cells were transfected with Flag-tagged N188K or parental empty vector (pcDNA) as described in Materials and Methods. N188K mRNA levels were measured in two selected clones by real-time PCR. (B) N188K protein levels were measured by immunoprecipitation and Western blot analysis with anti-Flag antibody (upper panel). FRTL-5 clones were transiently transfected with C5-CAT and CMV-Luc as an internal control to assess the effect of N188K on C5-CAT activity (lower panel). The C5-CAT/CMV-Luc ratio in control cells (pcDNA) is arbitrarily reported as 100%. (C) Northern blot analysis with 20 μg of total RNA from mock-transfected FRTL-5 (pcDNA), Cl.1, and Cl.3 cells probed with a cDNA fragment corresponding to rat Tg. (D to F) Western blot analyses of Tg (D), NIS (E), TTF-1 (F), TTF-2 (F), and Pax-8 (F) expression in pcDNA and Cl.1 and Cl.3 cells. The membranes were subsequently stripped and reprobed with antibodies against the indicated antibodies or β-actin and YY-1 for loading control for total or nuclear extracts, respectively. IgG, immunoglobulin G; ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We next examined the effect of the reduced TTF-1 activity induced by N188K on the levels of thyroglobulin, whose promoter is a well-established downstream target of TTF-1 in vitro (6, 25, 34, 50). We measured Tg mRNA levels in FRTL-5/N188K clones 1 and 3 versus mock-transfected cells (pcDNA). Interestingly, Tg mRNA levels were dramatically decreased in both clones (Fig. 2C). Consistently, Tg protein levels were lower in clones 1 and 3 and in the mass population of N188K-transfected cells than in mock-transfected controls or in FRTL-5 parental cells (Fig. 2D). Sodium-iodide symporter (NIS) protein levels, which are considered nonresponsive to TTF-1 in vivo (46), did not decrease in transfected clones (Fig. 2E), thereby acting like transcription factors TTF-2 and PAX-8 (Fig. 2F). These data provide the first evidence that selected “in vivo targets” of TTF-1 such as thyroglobulin are affected in vivo by reduced TTF-1 activity.
N188K titrates endogenous TTF-1 away from its target DNA sequence by physically interacting with it.
To identify the mechanisms whereby N188K interferes with the effect of TTF-1, we used coimmunoprecipitation assays to determine whether Nkx-2.5 and TTF-1 physically interact. Flag-tagged Nkx-2.5- and TTF-1-encoding vectors were transiently cotransfected in BOSC-23 cells. A significant amount of TTF-1 was detected in Nkx-2.5 immunoprecipitates (see Fig. S1A in the supplemental material), thereby indicating that Nkx-2.5 enters into contact with TTF-1. We tested various TTF-1 deletion mutants to map the TTF-1 region(s) involved in the interaction with Nkx-2.5 and found that the TTF-1 homeodomain is required for binding to Nkx-2.5 (see Fig. S1A and B in the supplemental material).
We used a similar approach to map the TTF-1-interacting domain on the Nkx-2.5 protein. Coimmunoprecipitation analyses with an Nkx C-terminal deleted mutant (Nkx-14), the above-described N188K (in which the mutation affects the homeodomain), and wild-type TTF-1 demonstrated that the Nkx-2.5 C-terminal region is required for interaction with TTF-1 (see Fig. S1C and D in the supplemental material).
To examine further the molecular mechanisms by which N188K affects TTF-1, we used EMSA to assess TTF-1 binding to its consensus DNA (D, identified in the hDio2 promoter [21]) with and without N188K. A TTF-1/D complex present in nuclear extracts from TTF-1-transfected HeLa cells disappears in cells cotransfected with TTF-1 and N188K (see Fig. S2A in the supplemental material). In the FRTL-5/N188K clones, competition studies showed that TTF-1 binding to oligonucleotide C, which corresponds to the TTF-1 binding site on the rat thyroglobulin promoter (6), was significantly weaker in clone 1 than in mock-transfected cells (pcDNA) (see Fig. S2C in the supplemental material).
A previous study demonstrated that TTF-1 and Pax-8 make contact through the TTF-1 N-terminal activation domain and the Pax-8 C-terminal region (14). A glutathione transferase pull-down assay with glutathione transferase-Pax-8 immobilized on glutathione-Sepharose beads and protein extracts prepared from clone 1 and clone 3 cells showed no differences versus pcDNA control cells (see Fig. S2D in the supplemental material).
Overall, these data show that although TTF-1 is functionally impaired, it retains its ability to interact with Pax-8, and they support the finding that the TTF-1 homeodomain, which is not involved in binding Pax-8, is the region involved in N188K interaction.
N188K significantly down-regulates several thyroid differentiation markers including Tg and pendrin.
To assess the effect of N188K-mediated TTF-1 depletion on specific gene expression in FRTL-5 cells, we measured the expression levels of 10 thyroidal genes using real-time PCR. The expression of 4 of the 10 genes was significantly (twofold) reduced in clone 1 versus controls (Table 2). Interestingly, pendrin was among the genes affected by TTF-1 depletion. This is the first indication that TTF-1 directly affects pendrin in thyroid cells.
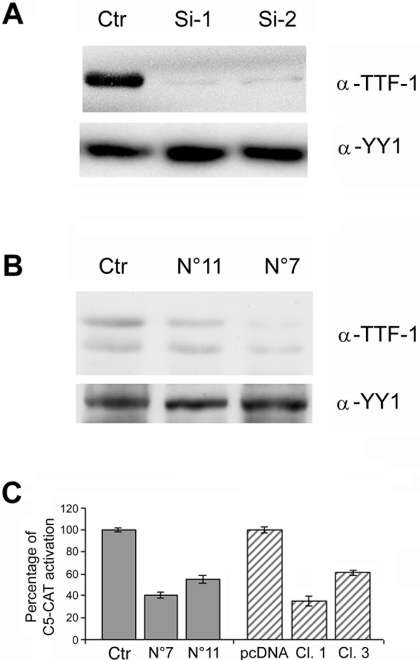
To verify this finding and to establish whether pendrin is indeed a TTF-1 target, we used short-interfering RNA to knock down TTF-1 in thyroid cells. To this end, we generated two plasmids containing a rat TTF-1-based hairpin short-interfering RNA expression cassette. To test the efficacy of the two constructs (Si1 and Si2), we cotransfected HEK-293 cells with rat CMV-TTF-1 plasmid plus either one of the two Si constructs or a control vector. Both plasmids interfered with TTF-1 synthesis (Fig. 3A), although endogenous TTF-1 was not affected in transiently transfected FRTL-5 cells (data not shown). The latter finding probably reflects the poor efficiency of transient transfections in FRTL-5 cells. To overcome this problem, we cotransfected the Si2 plasmid with a pPURO plasmid to generate FRTL-5/TTF-1-RNAi stably transfected clones. Among 30 clones identified, we selected two (7 and 11) whose TTF-1 levels were reduced versus cells transfected with control plasmid (Fig. 3B) and expanded these for further analyses. Consistent with the reduced TTF-1 levels, C5-CAT promoter activity was reduced in both clones, and the degree of TTF-1 functional impairment was similar to that observed in FRTL-5/N188K clones (Fig. 3C). We then evaluated Tg and Pds levels in the FRTL-5 TTF-1-RNAi clones. Northern blot (Fig. 4A, B) and real-time PCR (Fig. 4C) analyses revealed a significant reduction in Tg and Pds levels, which supports the finding that these two genes are bona fide TTF-1 targets in vivo.
FIG. 3.
Analysis of activity of the generated TTF-1/Si constructs and establishment and characterization of FRTL-5 cell lines with TTF-1 depletion by RNAi. (A) HEK-293 cells were cotransfected with CMV-TTF-1 plasmid (4 μg) and one silencing construct (2 μg each of TTF-Si1, TTF-Si2, or control vector[Ctr] as indicated). Nuclear extracts were immunoblotted with antibody against TTF-1. (B) FRTL-5 cells were transfected with TTF-Si2 (5 μg) or Ctr-Si and pPURO (0.5 μg) and grown with 1 μg/ml puromycin for 2 weeks. Nuclear extracts from clones 11 and 7 were analyzed for TTF-1 expression by immunoblotting. The membrane was subsequently stripped and reprobed with anti-YY-1-specific antibodies for loading control. Note that, as previously described (14), TTF-1 appears as two bands in thyroidal cells (B), whereas only the higher-molecular-weight band is visible in transfected nonthyroid cells (A). (C) FRTL-5 clones stably expressing N188K or the TTF-1 Si2 constructs were transiently transfected with C5-CAT and CMV-Luc as internal control. The C5-CAT/CMV-Luc ratio in control cells (pcDNA) is arbitrarily reported as 100%. Values ±SD are the mean of at least three experiments.
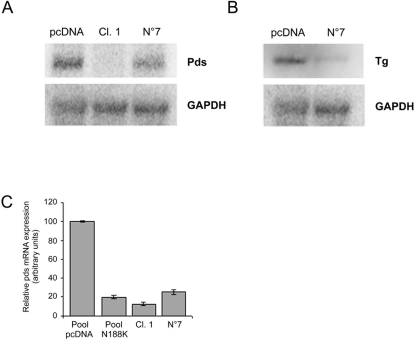
FIG. 4.
The levels of Pds and Tg expression decrease parallel to decreases in TTF-1 activity. (A) Northern blot analyses with 20 μg (for Tg expression) or 5 μg of poly(A)+ (for Pds expression) from the indicated clones. The blots were stripped and hybridized with a corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe for equivalence of RNAs. (B) Pds expression levels in pcDNA and N188K-transfected parental FRTL-5 cells (mass populations) in clones Cl.1 and N 7 measured by real-time PCR. For each reaction, standard curves for the reference gene were constructed by using six fourfold serial dilutions of cDNA. All samples were run in triplicate and are reported as amounts of gene expression calculated with β-actin expression as the internal standard.
The Pds promoter region is responsive to TTF-1, contains a TTF-1 binding site, and associates with TTF-1 by chromatin immunoprecipitation.
In an attempt to gain further insight into the mechanisms governing Pds gene expression in thyroid cells, we isolated and characterized the rat pendrin promoter, the structure of the rPds mRNA, and its TSS. Three alternative TSSs (Fig. 5B, lane 1) were identified in the Pds gene in FRTL-5 cells by both 5′ RACE and RT-PCR analyses (Fig. 5A and B). The presence of an alternative spliced intron (Fig. 5A, box) detected by RACE was confirmed by RT-PCR (data not shown). Based on sequence data available in the gene bank (UCSC Genome Browser, chromosome 6: 49430143-49433215), we used PCR with oligonucleotides rPDS1s and rPDS2r (Table 1) to generate a 3,074-nucleotide DNA fragment corresponding to the rat Pds genome that spans nucleotides −3085 to −38 from the initiator ATG. We then subcloned this fragment into the pGL3-basic reporter vector (Promega, Madison, WI), upstream to the luciferase gene. Interestingly, the full-length Pds promoter (rPDSLuc3.0) significantly induced reporter expression in both nonthyroid (Fig. 5C, HeLa) and thyroidal cells (FRTL-5; data not shown) versus the empty vector. In HeLa cells, the rPDSLuc3.0 construct response to TTF-1 (>6.0-fold) was comparable to that observed with the TPO promoter (6) and was strongly inhibited when we used a TTF-1-specific RNAi plasmid (Fig. 5C).
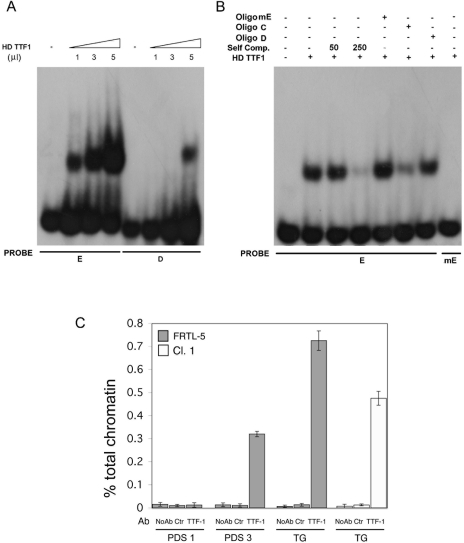
A 5′ truncation of the genomic region showed that when base pairs from −3140 to −2200 were deleted (pPDSLuc2.2), the reporter response to TTF-1 declined (Fig. 5C). Therefore, we attempted to identify the putative TTF-1-binding motif within the deleted region. Using MatInspector (35) (Genomatix Suite 3.1.0) and TRANSFAC (38) software, we identified a putative TTF-1-binding site, which we designated “E” site. The E site (ACT CAAG TGC) is very similar to the B site (ACT CAAG TAG) (boldface indicates the core motif) previously characterized on the rat Tg promoter (6). Using EMSA, we examined the binding of recombinant TTF-1 to the E oligonucleotide. As shown in Fig. 6A, not only did E specifically bind recombinant TTF-1, but it did so with a higher affinity than the D site. TTF-1 binding to E is specific since it was displaced by an excess of unlabeled “self” and of C or D oligonucleotides (Fig. 6B). The latter two correspond to the high-affinity TTF-1 binding sites isolated from the proximal rat Tg (6) and hDio2 promoters (21), respectively. Conversely, the TTF-1/E complex was not competed by the mutated E oligonucleotide (mE, which contains a mutation [underlined] of CAAG to CGTG in the TTF-1 binding core motif) (Fig. 6B). Moreover, radiolabeled mE did not form a specific complex when incubated with recombinant TTF-1 (Fig. 6B).
FIG. 6.
Interaction of the TTF-1 protein with the E binding site within the rat Pds promoter by EMSA and ChIP analysis on the Tg and pendrin promoter regions. (A) The recombinant TTF-1 was added to radiolabeled oligonucleotides as indicated. (B) The specificity of complexes was tested by competition analysis. A constant amount (250-fold excess or as indicated) of cold oligonucleotides was added to the reaction mixture as indicated in Materials and Methods. The mutated E probe (mE), which contains the CAAG to CGTG mutation (underlined), did not compete for the binding of E to recombinant TTF-1 protein (lane 5), nor did it bind recombinant TTF-1 protein when radiolabeled (last lane). (C) TTF-1 associated with both the rat pendrin and Tg promoters in vivo. Chromatin extracted from cross-linked FRTL-5 cells was immunoprecipitated in parallel using no antibodies (No Ab), anti-ERK (Ctr), or antibodies against TTF-1. The immunoprecipitates were analyzed by real-time PCR with oligonucleotide primers corresponding to the rat Tg promoter (TG) or rat pendrin promoter (PDS1, spanning the E site, or PDS3, spanning a distal genomic region about 15 kb upstream to the initiator ATG). Parallel PCRs were performed with total input DNAs obtain from unprecipitated aliquots of similarly treated chromatin samples. The amount of precipitated DNA was calculated relative to the total input chromatin and expressed as percentage of the total according to the formula: % total = 2ΔCt × % input chromatin, where ΔCt = Ctinput − Ctimmunoprecipitation (20). Ct is the cycle threshold within the linear range of all reactions. Standard deviations are indicated, and the experiment is representative of at least three independent experiments.
In summary, TTF-1 binds to the E site in the pendrin promoter. We measured the functional contribution of this site to the transactivation of the rPds promoter by disrupting it in the context of the −3140-bp promoter construct. Mutation of the E site (rPDSLuc3.0m) reduced the transactivation response to TTF-1 (Fig. 5C), which indicates that this motif plays a functional role in the promoter. To verify that the E site is able to confer TTF-1 responsiveness to a minimal promoter region, we generated a plasmid in which three-E tandem motifs were subcloned upstream from the thymidine-kinase minimal promoter (3E-TKLuc). Whereas the parental plasmid did not respond to TTF-1 nor to the equivalent plasmid containing the corresponding mutant mE, there was a 2.7-fold response in the presence of the multimerized E sites (Fig. 5C).
To probe further the mechanism by which TTF-1 promotes Pds expression, we examined the association in vivo between endogenous TTF-1 and the Pds promoter using ChIP. In this experiment, protein-DNA complexes were immunoprecipitated with control antibodies (Ctr-anti-ERK) or antibodies specific for TTF-1. The DNA content of immunoprecipitates was then analyzed by real-time PCR. The results of this set of experiments are shown in Fig. 6. Interestingly, TTF-1 physically interacted with the promoter regions of both Tg and Pds. In fact, these regions (TG and PDS1, respectively) were enriched in immunoprecipitates obtained with TTF-1 antibodies, whereas there was no enrichment when we analyzed a distal genomic region 15 kb downstream from the ATG region in the Pds gene (PDS3) or when we used control antibodies. Consequently, the association between TTF-1 and the Tg (TG) and Pds (PDS1, spanning the E site) DNA regions indicated above is specific. Interestingly, quantitative ChIP showed that enrichment in Tg immunoprecipitates was consistently reduced in clone 1 (Fig. 6C), which is in accordance with the impaired activity and DNA binding of TTF-1 in these cells due to N188K expression.
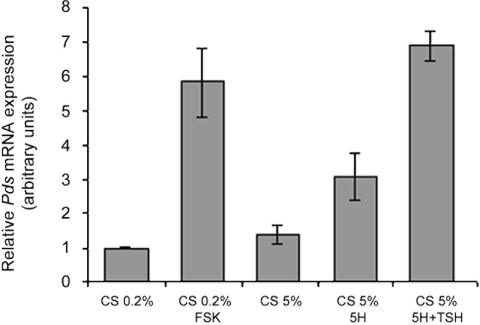
Finally, we asked whether the Pds gene is under the same transcriptional control as Tg, namely whether it is controlled by serum, cyclic AMP (cAMP), insulin, and TSH. We therefore stimulated FRTL-5 cells, starved of serum and hormones, with various compounds known to modulate thyroid cell function and analyzed the resulting mRNAs by real-time PCR (Fig. 7). Interestingly, Pds was induced by forskolin, five hormones, and TSH (Fig. 7), which coincides with the pattern observed with Tg under the same conditions (data not shown). In conclusion, we show that Pds is a novel thyroid differentiation marker that, like Tg, requires cAMP, serum, and TSH in addition to TTF-1 for its expression.
FIG. 7.
The Pds gene is induced by forskolin, serum, and TSH in thyroid cells. FRTL-5 cells were starved of calf serum (CS) and six hormones (6H) for 72 h (CS, 0.2%) and then stimulated with forskolin (FSK; 10 μM) for 12 h before harvesting or starved of six hormones (CS, 5%) and then stimulated with five hormones (5H) or five hormones and TSH for 12 h. Pds mRNA levels were monitored by real-time PCR as previously described. Data are the means of three separate experiments.
DISCUSSION
TTF-1 is essential for lung and thyroid development and is associated with the expression of tissue-specific genes. Elucidation of the intricate molecular mechanisms underlying TTF-1 action is critical for a deeper understanding of these events. TTF-1 is expressed in thyroid precursor cells throughout thyroid development, and its expression persists in adult life. Conversely, Nkx-2.5, which belongs to the same family of NK2 transcription factors as TTF-1, is expressed only in the thyroid primordium within a precise temporal/spatial window, i.e., between E8.5 and E12.5. Nkx-2.5, whose role in thyroid development is unknown, homo- and heterodimerizes to exert its function (28). We previously identified four missense mutations in the NKX-2.5 gene in 240 patients affected by congenital hypothyroidism due to thyroid dysgenesis; these mutations were absent in control euthyroid patients (M. Dentice et al., unpublished data). In the same study, we characterized a partial defect in thyroid differentiation in Nkx-2.5 knockout −/− mice that implicated Nkx-2.5 in thyroid development.
Protein-protein interactions with other transcription factors and coactivators are involved in TTF-1 action. We demonstrate that the TTF-1 homeodomain interacts with the Nkx-2.5 C-terminal domain. The homeodomain, which is critical for protein-DNA interactions, is also involved in protein-protein interactions (15, 22). The Nkx-2.5/TTF-1 interaction identified in this study is consistent with the observation that other NK2 transcription factors (e.g., Nkx-2.3 and Nkx-2.6) bind, with different affinities, to Nkx-2.5 (28). EMSA experiments with TTF-1 and wild-type Nkx-2.5 showed that both proteins were able to bind and compete in binding DNA (see Fig. S2E in the supplemental material). Overexpression of Nkx-2.5 did not superinduce Tg in either thyroid cells or human papillary thyroid carcinoma-derived cells (TPC-1) (unpublished results), which is consistent with the concept that multiple mechanisms control thyroid-specific gene expression. It is important to establish whether or not Nkx-2.5 and TTF-1 interact in thyroid cells during development. However, a cell model that mimics the thyroid precursor cell in terms of TTF-1 and Nkx-2.5 expression is not yet available.
The interaction between the N188K mutant and the TTF-1 homeodomain reduced TTF-1 binding to DNA but not binding to Pax-8, which is in agreement with the finding that the TTF-1 N-terminal domain is critical for interaction with Pax-8 (14). The data we obtained by using N188K to modulate the action of TTF-1 in thyroid cells in vivo demonstrate that TTF-1/DNA binding is a prerequisite for the activation of tissue-specific gene expression (i.e., Tg and PDS) in thyroid cells.
Various procedures have been used by others and by us to interfere with the effect exerted by TTF-1, albeit with scarce success. Using antisense technology, Rossi and coworkers showed that TTF-1 controls TSH-dependent growth in thyroid cells, although they did not examine its effects on thyroid-specific gene expression (41). RNAi is a highly efficient, specific method with which to knock down gene expression in eukaryotic cells, but it is sometimes associated with a general attenuation of transcription. Hence, an independent criterion is often required to validate RNAi-based results. Similarly, it is difficult to understand how protein N188K, which has a dominant-negative effect on TTF-1 action, actually functions in the cell. Therefore, we used the two different approaches (RNAi and N188K) to reduce TTF-1 activity in thyroid cells. The data obtained with the two methods show that reduced TTF-1 activity causes down-modulation of the expression of thyroid-specific genes including Tg and pendrin.
The finding that mice harboring complete knockout of TTF-1 lack a thyroid gland and die at birth implicates TTF-1 in the early stages of thyroid development. A challenge in thyroid physiology is to establish whether TTF-1 is required within the thyroid gland also in early postembryonic life. Our understanding of the role of TTF-1 in thyroid differentiation is largely based on indirect evidence. For instance, TTF-1 is not expressed in FRTL-5 cells transformed with various oncogenes where the thyroid phenotype is lost (19). However, in human thyroid neoplasms, Tg and TPO expression can be lost before the disappearance of TTF-1 (18). Consequently, the concept that TTF-1 plays a major role in thyroid cell differentiation was only inferred. Here we show that reduced TTF-1 activity affects the expression of some differentiation markers, which implicates TTF-1 in thyroid cell differentiation.
We also demonstrate that TTF-1 is a crucial component in the transcriptional regulation of the Tg and pendrin genes in the thyroid. In line with this finding, thyroid-derived FRT cells, which lack TTF-1, express neither Tg nor pendrin (34). Tg and pendrin belong to a large subset of genes that are differentially expressed after TTF-1 reduction. Whereas Tg dependence on TTF-1 activity was not surprising because TTF-1 is known to regulate the Tg promoter (6), the effect of TTF-1 on pendrin expression was unexpected. Pendrin is a recently characterized apical iodide transporter in thyroid follicular cells that causes the Pendred syndrome (11, 31, 32). Very little is known about its role in thyroid diseases.
Immunohistochemical data show that pendrin expression is more extensive and pronounced, especially in areas with a high rate of follicular cell proliferation, in Graves' disease than in normal thyroid. Pendrin is negatively regulated in follicular carcinomas, papillary carcinomas (1), and medullary carcinomas (4), whereas its levels are increased in toxic adenomas (4). These observations suggest that pendrin expression is a feature of the fully differentiated thyroid. The pattern of Pds expression in thyroid cells under different conditions (Fig. 7) supports the concept that Pds is a marker of thyroid differentiation.
Transfection assays showed that TTF-1 is a transcriptional activator of the Pds promoter. The TTF-1 DNA binding site E identified in this study is functionally relevant as demonstrated by EMSA, by functional analysis of the mutated promoter in which reporter activity was reduced by 20%, and by quantitative chromatin immunoprecipitation experiments. The residual TTF-1-dependent activity, present in both the deleted and E-mutated promoter constructs (Fig. 5C), is probably mediated by other TTF-1 binding site(s) present in the promoter. In this context, it is noteworthy that computer analysis identified several other putative TTF-1-binding sites, together with putative CRE(B) binding site(s), which may be critical for the TTF-1 and cAMP responsiveness, respectively, observed in vivo (Fig. 7). In conclusion, we demonstrate that TTF-1 controls the expression of Tg and Pds in thyroid cells in vivo. Our finding that Pds expression in thyroid is TTF-1 dependent and is associated with the fully differentiated thyroid state identifies this gene as a new marker useful in the treatment of human thyroid diseases. This study sheds light on the role of TTF-1 in the maintenance of thyroid differentiation in vivo and lays the groundwork for mechanistic and genetic studies aimed at establishing the role of Nkx-2.5 in thyroid development.
Supplementary Material
Acknowledgments
We are grateful to C. Missero and A. Leonardi for their critical reading of the manuscript. We also thank Jean Gilder for text editing.
This work was supported by grants from AIRC 2004 and MIUR-FIRB RBAU01MEPC to D.S.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arturi, F., D. Russo, J. M. Bidart, D. Scarpelli, M. Schlumberger, and S. Filetti. 2001. Expression pattern of the pendrin and sodium/iodide symporter genes in human thyroid carcinoma cell lines and human thyroid tumors. Eur. J. Endocrinol. 145:129-135. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. W., G. M. Silberbach, A. Kavanaugh-McHugh, C. Cottrill, Y. Zhang, S. Riggs, O. Smalls, M. C. Johnson, M. S. Watson, J. G. Seidman, C. E. Seidman, J. Plowden, and J. D. Kugler. 1999. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Investig. 104:1567-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidart, J. M., L. Lacroix, D. Evain-Brion, B. Caillou, V. Lazar, R. Frydman, D. Bellet, S. Filetti, and M. Schlumberger. 2000. Expression of Na+/I− symporter and Pendred syndrome genes in trophoblast cells. J. Clin. Endocrinol. Metab. 85:4367-4372. [DOI] [PubMed] [Google Scholar]
- 4.Bidart, J. M., C. Mian, V. Lazar, D. Russo, S. Filetti, B. Caillou, and M. Schlumberger. 2000. Expression of pendrin and the Pendred syndrome (PDS) gene in human thyroid tissues. J. Clin. Endocrinol. Metab. 85: 2028-2033. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. Y., and R. J. Schwartz. 1995. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, Nkx-2.5. J. Biol. Chem. 270:15628-15633. [DOI] [PubMed] [Google Scholar]
- 6.Civitareale, D., R. Lonigro, A. J. Sinclair, and R. Di Lauro. 1989. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 8:2537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damante, G., and R. Di Lauro. 1994. Thyroid-specific gene expression. Biochim. Biophys. Acta 1218:255-266. [DOI] [PubMed] [Google Scholar]
- 8.Damante, G., G. Tell, and R. Di Lauro. 2001. A unique combination of transcription factors controls differentiation of thyroid cells. Prog. Nucleic Acid Res. Mol. Biol. 66:307-356. [DOI] [PubMed] [Google Scholar]
- 9.De Felice, M., G. Damante, M. Zannini, H. Francis-Lang, and R. Di Lauro. 1995. Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J. Biol. Chem. 270:26649-26656. [DOI] [PubMed] [Google Scholar]
- 10.De Felice, M., and R. Di Lauro. 2004. Thyroid development and its disorders: genetics and molecular mechanisms. Endocr. Rev. 25:722-746. [DOI] [PubMed] [Google Scholar]
- 11.Dentice, M., C. Luongo, A. Elefante, R. Romino, R. Ambrosio, M. Vitale, G. Rossi, G. Fenzi, and D. Salvatore. 2004. Transcription factor Nkx-2.5 induces sodium/iodide symporter gene expression and participates in retinoic acid- and lactation-induced transcription in mammary cells. Mol. Cell. Biol. 24:7863-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dentice, M., C. Morisco, M. Vitale, G. Rossi, G. Fenzi, and D. Salvatore. 2003. The different cardiac expression of the type 2 iodothyronine deiodinase gene between human and rat is related to the differential response of the dio2 genes to Nkx-2.5 and GATA-4 transcription factors. Mol. Endocrinol. [DOI] [PubMed]
- 13.Deshpande, V., and S. G. Venkatesh. 1999. Thyroglobulin, the prothyroid hormone: chemistry, synthesis and degradation. Biochim. Biophys. Acta 1430:157-178. [DOI] [PubMed] [Google Scholar]
- 14.Di Palma, T., R. Nitsch, A. Mascia, L. Nitsch, R. Di Lauro, and M. Zannini. 2003. The paired domain-containing factor Pax8 and the homeodomain-containing factor TTF-1 directly interact and synergistically activate transcription. J. Biol. Chem. 278:3395-3402. [DOI] [PubMed] [Google Scholar]
- 15.Durocher, D., F. Charron, R. Warren, R. J. Schwartz, and M. Nemer. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, L. A., B. Glaser, J. C. Beck, J. R. Idol, A. Buchs, M. Heyman, F. Adawi, E. Hazani, E. Nassir, A. D. Baxevanis, V. C. Sheffield, and E. D. Green. 1997. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet. 17:411-422. [DOI] [PubMed] [Google Scholar]
- 17.Everett, L. A., H. Morsli, D. K. Wu, and E. D. Green. 1999. Expression pattern of the mouse ortholog of the Pendred's syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA 96:9727-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbro, D., C. Di Loreto, C. A. Beltrami, A. Belfiore, R. Di Lauro, and G. Damante. 1994. Expression of thyroid-specific transcription factors TTF-1 and PAX-8 in human thyroid neoplasms. Cancer Res. 54:4744-4749. [PubMed] [Google Scholar]
- 19.Francis-Lang, H., M. Zannini, M. De Felice, M. T. Berlingieri, A. Fusco, and R. Di Lauro. 1992. Multiple mechanisms of interference between transformation and differentiation in thyroid cells. Mol. Cell. Biol. 12:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gereben, B., D. Salvatore, J. W. Harney, H. M. Tu, and P. R. Larsen. 2001. The human, but not rat, dio2 gene is stimulated by thyroid transcription factor-1 (TTF-1). Mol. Endocrinol. 15:112-124. [DOI] [PubMed] [Google Scholar]
- 22.Hiroi, Y., S. Kudoh, K. Monzen, Y. Ikeda, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28:276-280. [DOI] [PubMed] [Google Scholar]
- 23.Hosoda, T., I. Komuro, I. Shiojima, Y. Hiroi, M. Harada, Y. Murakawa, Y. Hirata, and Y. Yazaki. 1999. Familial atrial septal defect and atrioventricular conduction disturbance associated with a point mutation in the cardiac homeobox gene CSX/NKX2-5 in a Japanese patient. Jpn. Circ. J. 63:425-426. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, Y., Y. Hiroi, T. Hosoda, T. Utsunomiya, S. Matsuo, T. Ito, J. Inoue, T. Sumiyoshi, H. Takano, R. Nagai, and I. Komuro. 2002. Novel point mutation in the cardiac transcription factor CSX/NKX2.5 associated with congenital heart disease. Circ. J. 66:561-563. [DOI] [PubMed] [Google Scholar]
- 25.Javaux, F., F. Bertaux, A. Donda, H. Francis-Lang, G. Vassart, R. DiLauro, and D. Christophe. 1992. Functional role of TTF-1 binding sites in bovine thyroglobulin promoter. FEBS Lett. 300:222-226. [DOI] [PubMed] [Google Scholar]
- 26.Kasahara, H., S. Bartunkova, M. Schinke, M. Tanaka, and S. Izumo. 1998. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ. Res. 82:936-946. [DOI] [PubMed] [Google Scholar]
- 27.Kasahara, H., B. Lee, J. J. Schott, D. W. Benson, J. G. Seidman, C. E. Seidman, and S. Izumo. 2000. Loss of function and inhibitory effects of human CSX/NKX2.5 homeoprotein mutations associated with congenital heart disease. J. Clin. Investig. 106:299-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasahara, H., A. Usheva, T. Ueyama, H. Aoki, N. Horikoshi, and S. Izumo. 2001. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J. Biol. Chem. 276:4570-4580. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, S., Y. Hara, T. Pineau, P. Fernandez-Salguero, C. H. Fox, J. M. Ward, and F. J. Gonzalez. 1996. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10:60-69. [DOI] [PubMed] [Google Scholar]
- 30.Komuro, I., and S. Izumo. 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. USA 90:8145-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix, L., C. Mian, B. Caillou, M. Talbot, S. Filetti, M. Schlumberger, and J. M. Bidart. 2001. Na(+)/I(−) symporter and Pendred syndrome gene and protein expressions in human extra-thyroidal tissues. Eur. J. Endocrinol. 144:297-302. [DOI] [PubMed] [Google Scholar]
- 32.Lazzaro, D., M. Price, M. de Felice, and R. Di Lauro. 1991. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development 113:1093-1104. [DOI] [PubMed] [Google Scholar]
- 33.Lints, T. J., L. M. Parsons, L. Hartley, I. Lyons, and R. P. Harvey. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:969. [DOI] [PubMed] [Google Scholar]
- 34.Mascia, A., M. De Felice, C. Lipardi, R. Gentile, G. Cali, M. Zannini, R. Di Lauro, and L. Nitsch. 1997. Transfection of TTF-1 gene induces thyroglobulin gene expression in undifferentiated FRT cells. Biochim. Biophys. Acta 1354:171-181. [DOI] [PubMed] [Google Scholar]
- 35.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missero, C., G. Cobellis, M. De Felice, and R. Di Lauro. 1998. Molecular events involved in differentiation of thyroid follicular cells. Mol. Cell Endocrinol. 140:37-43. [DOI] [PubMed] [Google Scholar]
- 37.Pendred, V. 1896. Deaf-mutism and goitre. Lancet 2:532. [Google Scholar]
- 38.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reardon, W., O. M. CF, R. Trembath, H. Jan, and P. D. Phelps. 2000. Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM 93:99-104. [DOI] [PubMed] [Google Scholar]
- 40.Rillema, J. A., and M. A. Hill. 2003. Prolactin regulation of the pendrin-iodide transporter in the mammary gland. Am. J. Physiol. Endocrinol. Metab. 284:E25-E28. [DOI] [PubMed] [Google Scholar]
- 41.Rossi, D. L., A. Acebron, and P. Santisteban. 1995. Function of the homeo and paired domain proteins TTF-1 and Pax-8 in thyroid cell proliferation. J. Biol. Chem. 270:23139-23142. [DOI] [PubMed] [Google Scholar]
- 42.Royaux, I. E., K. Suzuki, A. Mori, R. Katoh, L. A. Everett, L. D. Kohn, and E. D. Green. 2000. Pendrin, the protein encoded by the Pendred syndrome gene (PDS), is an apical porter of iodide in the thyroid and is regulated by thyroglobulin in FRTL-5 cells. Endocrinology 141:839-845. [DOI] [PubMed] [Google Scholar]
- 43.Royaux, I. E., S. M. Wall, L. P. Karniski, L. A. Everett, K. Suzuki, M. A. Knepper, and E. D. Green. 2001. Pendrin, encoded by the Pendred syndrome gene, resides in the apical region of renal intercalated cells and mediates bicarbonate secretion. Proc. Natl. Acad. Sci. USA 98:4221-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salvatore, D., T. Bartha, J. W. Harney, and P. R. Larsen. 1996. Molecular biological and biochemical characterization of the human type 2 selenodeiodinase. Endocrinology 137:3308-3315. [DOI] [PubMed] [Google Scholar]
- 45.Santisteban, P., A. Acebron, M. Polycarpou-Schwarz, and R. Di Lauro. 1992. Insulin and insulin-like growth factor I regulate a thyroid-specific nuclear protein that binds to the thyroglobulin promoter. Mol. Endocrinol. 6:1310-1317. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt, T. L., C. R. Espinoza, and U. Loos. 2001. Transcriptional regulation of the human sodium/iodide symporter gene by Pax8 and TTF-1. Exp. Clin. Endocrinol. Diabetes 109:27-31. [DOI] [PubMed] [Google Scholar]
- 47.Schott, J. J., D. W. Benson, C. T. Basson, W. Pease, G. M. Silberbach, J. P. Moak, B. J. Maron, C. E. Seidman, and J. G. Seidman. 1998. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281:108-111. [DOI] [PubMed] [Google Scholar]
- 48.Scott, D. A., and L. P. Karniski. 2000. Human pendrin expressed in Xenopus laevis oocytes mediates chloride/formate exchange. Am. J. Physiol. Cell Physiol. 278:C207-C211. [DOI] [PubMed] [Google Scholar]
- 49.Scott, D. A., R. Wang, T. M. Kreman, V. C. Sheffield, and L. P. Karniski. 1999. The Pendred syndrome gene encodes a chloride-iodide transport protein. Nat. Genet. 21:440-443. [DOI] [PubMed] [Google Scholar]
- 50.Sinclair, A. J., R. Lonigro, D. Civitareale, L. Ghibelli, and R. Di Lauro. 1990. The tissue-specific expression of the thyroglobulin gene requires interaction between thyroid-specific and ubiquitous factors. Eur. J. Biochem. 193: 311-318. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki, K., I. E. Royaux, L. A. Everett, A. Mori-Aoki, S. Suzuki, K. Nakamura, T. Sakai, R. Katoh, S. Toda, E. D. Green, and L. D. Kohn. 2002. Expression of PDS/Pds, the Pendred syndrome gene, in endometrium. J. Clin. Endocrinol. Metab. 87:938. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka, M., H. Kasahara, S. Bartunkova, M. Schinke, I. Komuro, H. Inagaki, Y. Lee, G. E. Lyons, and S. Izumo. 1998. Vertebrate homologs of tinman and bagpipe: roles of the homeobox genes in cardiovascular development. Dev. Genet. 22:239-249. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, M., S. B. Wechsler, I. W. Lee, N. Yamasaki, J. A. Lawitts, and S. Izumo. 1999. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development 126: 1439-1450. [DOI] [PubMed] [Google Scholar]
- 54.Yan, C., A. Naltner, J. Conkright, and M. Ghaffari. 2001. Protein-protein interaction of retinoic acid receptor alpha and thyroid transcription factor-1 in respiratory epithelial cells. J. Biol. Chem. 276:21686-21691. [DOI] [PubMed] [Google Scholar]
- 55.Yan, C., Z. Sever, and J. A. Whitsett. 1995. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J. Biol. Chem. 270:24852-24857. [DOI] [PubMed] [Google Scholar]
- 56.Yan, C., and J. A. Whitsett. 1997. Protein kinase A activation of the surfactant protein B gene is mediated by phosphorylation of thyroid transcription factor 1. J. Biol. Chem. 272:17327-17332. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, W., I. Shiojima, Y. Hiroi, Y. Zou, H. Akazawa, M. Mizukami, H. Toko, Y. Yazaki, R. Nagai, and I. Komuro. 2000. Functional analyses of three Csx/Nkx-2.5 mutations that cause human congenital heart disease. J. Biol. Chem. 275:35291-35296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.