Abstract
AU-rich-element (ARE)-mediated mRNA regulation occurs in Saccharomyces cerevisiae in response to external and internal stimuli through the p38 mitogen-activated protein kinase (MAPK)/Hog1p pathway. We demonstrate that the ARE-bearing MFA2 3′ untranslated region (UTR) controls translation efficiency in a p38 MAPK/Hog1p-dependent manner in response to carbon source growth conditions. The carbon source-regulated effect on MFA2 3′-UTR-controlled translation involves the role of conserved ARE binding proteins, the ELAV/TIA-1-like Pub1p, which can interact with the cap/eIF4G complex, and the translation/mRNA stability factor poly(A) binding protein (Pab1p). Pub1p binds the MFA2 3′-UTR in a p38 MAPK/Hog1p-regulated manner in response to carbon source growth conditions. Significantly, the p38 MAPK/Hog1p is also required to modulate Pab1p in response to carbon source. We find that Pab1p can bind the MFA2 3′-UTR in a regulated manner to control MFA2 3′-UTR reporter translation. Binding of full-length Pab1p to the MFA2 3′-UTR correlates with translation repression. Importantly, Pab1p binds the MFA2 3′-UTR only in a PUB1 strain, and correlating with this requirement, Pub1p controls translation repression of MFA2 in a carbon source/Hog1p-regulated manner. These results suggest that the p38 MAPK/Hog1p pathway regulates 3′-UTR-mediated translation by modulating recruitment of Pab1p and Pub1p, which can interact with the translation machinery.
AU-rich elements (AREs) control gene expression through multiple posttranscriptional processes. AREs function to regulate mRNA stability, export, and translation depending on the stimulus (3, 7, 9, 14, 19, 37, 41). The ARE-bound mRNP complex is subject to modulation by altered cellular conditions via signaling pathways including the conserved extracellular signal-regulated kinase, Jun N-terminal kinase, and p38 mitogen-activated protein kinase (MAPK) pathways and by heat shock and calcium signaling (5, 6, 20, 24, 44, 56).
Different AREs respond to changing conditions by modulating either translation or mRNA stability. These transcripts may be stored as translationally repressed stable mRNPs, which are further processed by additional signals to the cells (21, 22). The tumor necrosis factor alpha (TNF-α) ARE, apart from functioning in mRNA decay, can regulate gene expression through translation control (27, 57). When the cell is quiescent, translation of specific ARE-bearing transcripts is repressed. However, on stimulation of myeloid cells, the p38 and Jun N-terminal kinase MAPK pathways are activated and translation is enhanced while the repressive signals are removed (25, 33). This translational control imparts further efficiency to ARE-dependent regulation of specific gene expression.
Since the rate-limiting step of ARE-dependent decay is deadenylation, the ARE-bound mRNP complex must interact with the general decay machinery to stimulate deadenylation (3, 12, 53). These interactions may also influence translation levels in an ARE-specific manner. The poly(A) tail and poly(A) tail binding protein can have a marked influence on assembly and initiation/reinitiation of translation (10, 11, 30, 36). Interaction of Pab1p with the poly(A) tail promotes translation through eukaryotic initiation factor 4G (eIF4G)/cap-poly(A) interaction and can also function to repress or promote translation via interactions with the 3′-untranslated region (3′-UTR) binding proteins as well as with the release factors (10, 11, 47, 48). Pab1p controls the stability and translation efficiency of an mRNA through interactions with eIF4G/cap complex, as well as through a C-terminal regulatory domain that interacts with several critical translation factors and promotes Pab1p to bind the poly(A) tail in a cooperative manner (26, 28, 29, 30, 32, 36, 48). Interestingly, the poly(A) tail has been shown to be required for the repressive effect of the ARE in the case of the granulocyte-macrophage colony-stimulating factor and beta interferon transcripts (9, 19). Deletion of the poly(A) tail eliminates ARE repression and allows for increased translation in cell-free systems, which correlates with the inability of a protein to bind to this ARE in the presence of the poly(A) tail (9, 19, 52). ePab in Xenopus laevis embryo extracts is 72% identical to PABP, was identified as an ARE binding protein, and can regulate deadenylation rates of both ARE-bearing and non-ARE-bearing transcripts in an in vitro Xenopus extract-based deadenylation system (50). Furthermore, human PABP was identified as an ARE binding protein that binds the 3′-UTR AU-rich elements of transcripts such as that of granulocyte-macrophage colony-stimulating factor and human papillomavirus type 1 late mRNA h1 ARE (2, 52). Therefore, PABP is recruited by the ARE to perform ARE-specific functions in mRNA stability and translation regulation. This is possibly an efficient adaptation by the ARE to utilize a powerful regulatory protein that can affect the transcript at multiple levels of posttranscriptional control.
Saccharomyces cerevisiae offers an exceptional genetic and biochemical system to define posttranscriptional mechanisms (16, 31, 39). In particular, we have developed a yeast system to elucidate the ARE-mediated mRNA turnover pathway that appears to mimic many aspects of the mammalian system (49). There are at least two classes of AREs in yeast that are differentially regulated. MFA2 bears an AU-rich element that represents a class of AREs in yeast whose mRNA turnover is immune to carbon source regulation (49). In this paper we investigated the regulation of the MFA2 3′-UTR element and determined that the p38 MAPK pathway directly or indirectly modulates Pab1p and Pub1p recruitment to the MFA2 3′-UTR to regulate MFA2 translation.
MATERIALS AND METHODS
Yeast strains.
Cells were grown according to standard protocols (1) and as described previously (15, 49). Yeast strains used in this study are as described in Table 1. The hog1Δ and pub1Δ strains were obtained from Research Genetics as described in reference 55.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Y497 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| Y519 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Winzeler et al., 1999 (55) |
| Y600 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 hog1::Kanr | |
| Y601 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 hog1::Kanr | |
| Y602 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pub1::Kanr | |
| Y603 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pub1::Kanr | |
| Y511 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pbp1::Kanr | |
| Y532 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pbp1::Kanrpbp1::HIS3 | This study |
| Y137 | MATaura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100(Am) ade2-1(Oc) | Wilusz et al., 2001 (54) |
| Y485 | MATaura3-1 ade2-1 leu2-3,112 trp1-1 his3-11,15 can1-100 spb2::URA3 | |
| Y487 | MATaura3-1 ade2-1 leu2-3,112 trp1-1 his3-11,15 can1-100 spb2::URA3 pab1::HIS3 |
DNA manipulations and plasmids.
Methods for general DNA manipulations followed standard protocols (1, 40). The following plasmids were used in this study. p4680 for expression of MFA2 under the PGK1 promoter and p4738 for expression of mini-PGK1 with the MFA2 3′-UTR were described earlier (42). p5071 was constructed by replacing the MFA2 3′-UTR of p4680 with the PGK1 3′-UTR as a BamHI-HindIII fragment excised from p5042 (49). The firefly luciferase coding region was inserted into the BamHI site of p4680 and p5071 to create the Luc-MFA2 (p5072) and Luc-PGK1 (p5073) reporters. MFA2-DII (p4680-DII) was constructed by PCR insertion into p4680 of an XbaI-HindIII fragment (oligonucleotides 418 and 290 [42]), amplified from pRP324 to get the AU-deletion MFA2 reporter (35). The firefly luciferase coding region was inserted into the BamHI site of p4680-DII to create the Luc-MFA2-DII (ΔAU) reporter and into previously constructed TNF-α and TIF51A reporters to create Luc-TNF-α and Luc-TIF51A, respectively (49). The MFA2 3′-UTR was cloned as an EcoRI/BamHI fragment into p5027, which is the pGEM4 vector (Promega), to get p5080.
mRNA measurements.
mRNA levels and half-life analysis were determined by Northern blotting or RNase protection assays as described earlier (15, 49). Transcription was inhibited by adding 20 μg/ml of the fungal transcription inhibitor thiolutin (a gift from Pfizer, Groton, CT). DNA and RNA probes were prepared as described earlier (15), and the results were quantitated using a PhosphorImager (Molecular Dynamics PSI-PC; Sunnyvale, CA). The mRNA levels were normalized for loading to the U3 RNA levels. All measurements are an average of at least three experiments.
Cross-linking analysis.
Cytoplasmic extracts were prepared as described earlier (4). To generate radiolabeled MFA2 3′-UTR, p5080 was linearized with HindIII for in vitro transcription by SP6 polymerase. The transcription reactions were carried out as described earlier using [α-32P]UTP (54). Five hundred micrograms of extract was incubated for 5 min with 30 fmol of radiolabeled RNA. Following UV irradiation for 10 min, samples were treated with both 30 μg of RNase A and 3 U of RNase I for 10 min prior to separation on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Immunoprecipitation was performed on the RNase-treated reaction mixtures as follows. For immunoprecipitation of cross-linked Pub1p, monoclonal antibody 4C3 against Pub1p (obtained from M. Swanson) was used as described previously (49). Polyclonal antibody against Pab1p or glyceraldehyde-3-phosphate dehydrogenase (GAPDH)/Tdhp was used to immunoprecipitate Pab1p as described elsewhere (S. Vasudevan et al., submitted). The samples were precleared for 20 min with 20 μl of a 50% slurry of protein A Sepharose. The beads were then washed three times with binding buffer and then run on a 10% SDS-polyacrylamide gel, dried, and visualized with a phosphorimager.
Translation assay.
Cytoplasmic extracts were prepared in buffer A [30 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2] from 1-liter cultures grown to an optical density at 600 nm of 0.8 as described earlier (18). Increasing concentrations of the extract were mixed with 200 μl of the reconstituted luciferase substrate (Promega), and luminescence was measured over 15 seconds on a TD-20/20 luminometer (Turner Designs). Each extract was assayed in triplicate, and each strain was tested at least three times to give an average luciferase activity. The luminescence values were normalized to RNA levels as measured by Northern blotting and PhosphorImager analysis.
RESULTS
MFA2 3′-UTR represses translation.
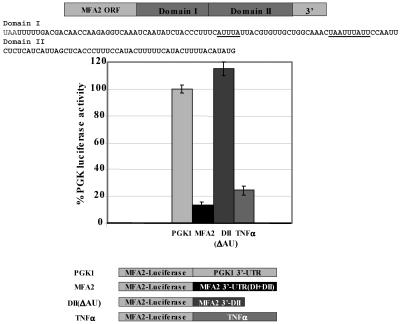
AREs control both mRNA stability and translation efficiency of transcripts (27). We therefore tested whether the MFA2 3′-UTR regulates translation efficiency, independent of its effects on mRNA stability. For these experiments we utilized the firefly luciferase gene as a reporter that was fused either to the MFA2 3′-UTR (Luc-MFA2) or to the PGK1 3′-UTR (Luc-PGK1) as a control. PGK1 is a constitutively stable mRNA whose decay and translation are independent of ARE-regulated mechanisms. The reporters were expressed in yeast grown in glucose medium, and the luciferase activity was measured in extracts and normalized to the abundance of each mRNA by Northern blotting analysis to determine the translation effects of the 3′-UTR, independent of effects on mRNA levels. Strikingly, the results demonstrated that translation of the Luc-MFA2 reporter was repressed at least 10-fold compared to the Luc-PGK1 control cells (Fig. 1).
FIG. 1.
The MFA2 3′-UTR can regulate translation through its AU-rich region. Luc-MFA2, Luc-DII, and Luc-TNF-α reporter translation compared to the Luc-PGK1 reporter in wild-type cultures. The Luc-MFA2 reporter bears the entire 3′-UTR, the Luc-TNF-α reporter bears the 34-nucleotide AU-rich region of TNF-α 3′-UTR (49), and the domain II reporter lacks the AU-rich region of MFA2 3′-UTR (35). All luciferase values were normalized to mRNA abundance and compared to the Luc-PGK1 luciferase activity in the wild-type strain, and the assay was repeated at least three times.
To determine whether translation repression required the AU-rich element of the MFA2 3′-UTR, we constructed a luciferase reporter (Luc-DII) that lacked the 60-nucleotide AU-rich sequences of the MFA2 3′-UTR (Fig. 1) (35). The Luc-DII reporter was transformed into yeast cells, and the luciferase activity was measured as described above. Remarkably, the Luc-DII reporter demonstrated complete derepression and translated as well as the PGK1 control reporter upon normalizing for mRNA levels (Fig. 1).
To determine whether translation regulation is mediated through an AU-rich domain, we utilized a Luc-TNF-α reporter that harbors the 3′-UTR of MFA2 in which the MFA2 AU-rich element was replaced by the TNF-α ARE. The TNF-α ARE reporter transcript was previously demonstrated to be stable when cells with the reporter transcripts were grown in glucose medium (49), as in these experiments. Therefore, this reporter allows us to determine the effect of an ARE on translation efficiency when the mRNA is stable. The reporters were transformed into yeast cells, which were then grown in glucose medium, and the luciferase activity was monitored as described above. The results demonstrated that translation of the Luc-TNF-α reporter was repressed fivefold compared to the Luc-PGK1 control after normalization for RNA levels (Fig. 1). These observations are consistent with the hypothesis that AREs repress translation in yeast cells grown in glucose medium apart from their effect on mRNA stability.
PABP interacts with the MFA2 3′-UTR element.
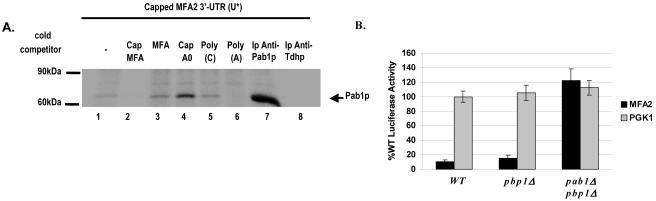
Previous studies suggested that human poly(A) binding protein (PABP), as well as Xenopus embryonic PABP, ePAB, can bind AREs in a specific manner (2, 50) and plays an essential role in mRNA stability and translation (10, 11, 30, 32, 36, 47, 49, 51, 54). We performed UV-cross-linking analysis with 32P-labeled in vitro-transcribed capped MFA2 3′-UTR with various unlabeled cold competitors, or the reactions were followed by immunoprecipitation with antibody against Pab1p or preimmune sera or polyclonal anti-GAPDH (Tdhp) as controls. Interestingly, analysis of the cross-linking results with capped MFA2 3′-UTR identified Pab1p as a 68-kDa poly(A)-sensitive factor present in extracts that interacts with the MFA2 3′-UTR (Fig. 2A). MFA2 3′-UTR-bound Pab1p could be immunoprecipitated with antibody against Pab1p (Fig. 2A, lane 7, compared to the control immunoprecipitation with anti-Tdh in lane 8). Pab1p bound capped MFA2 3′-UTR specifically, as the band was competed by either a 100-fold excess of cold capped MFA2 3′-UTR or 100 ng of poly(A) (Fig. 2A, lanes 2 and 6). A 100-fold excess of unlabeled uncapped MFA2 or excess unlabeled capped pGEM4 polylinker (Cap A0) or 100 ng of poly(C) failed to compete Pab1p bound to capped MFA2 3′-UTR (Fig. 2A, lanes 3 to 6 compared to lanes 2 and 6). Instead the capped non-ARE (Cap A0) competitor reproducibly enhanced the binding of Pab1p to the labeled RNA, as did the cap analog (Fig. 2A, lane 4, and data not shown), possibly by competing away cap-bound complexes, thereby breaking up endogenous Pab1p mRNPs to liberate Pab1p, which can then bind the labeled in vitro-transcribed capped MFA2 3′-UTR. These results indicate that Pab1p interaction with MFA2 3′-UTR or the stability of this interaction is dependent on the cap/cap-bound complex. Taken together, these data suggest that Pab1p can bind capped MFA2 3′-UTR in extracts.
FIG. 2.
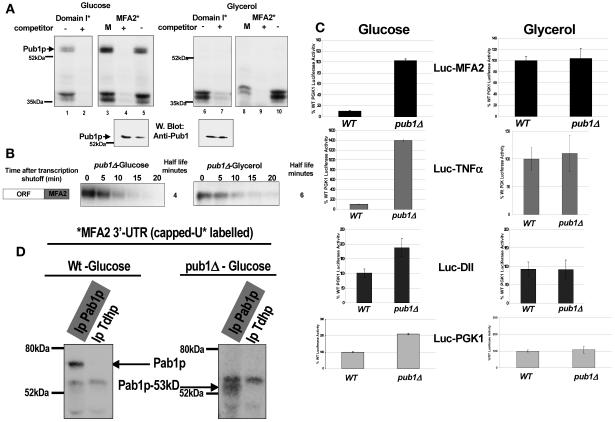
Pab1p binds the MFA2 3′-UTR in a cap-dependent manner and mediates translation repression. A. UV-cross-linking analysis performed with [α-32P]UTP-labeled capped MFA2 3′-UTR (U*) and wild-type yeast extracts. Competition was with a 100-fold excess of unlabeled capped and uncapped MFA2 3′-UTR in lanes 2 and 3, respectively; a 100-fold excess of nonspecific unlabeled capped polylinker RNA (Cap A0) in lane 4; and 100 ng of poly(C) and poly(A) in lanes 5 and 6, respectively. Pab1p reproducibly binds more strongly in the presence of a nonspecific RNA competitor (compare lane 1 to lanes 3 to 5). Two sets of the cross-linked reaction mixtures were subjected to immunoprecipitation with anti-Pab1p (lane 7) and anti-Tdh (lane 8) antibodies. B. Translation in the wild-type, pbp1Δ, and pbp1Δ pab1Δ strains. Black bars, translation of the Luc-MFA2 reporter. Gray bars, translation of the PGK1 reporter. All values are compared to the Luc-PGK1 reporter activity in the WT strain and are normalized for RNA levels. The luciferase experiment was repeated three times.
Pab1p mediates MFA2 3′-UTR-dependent translational control.
The results above suggest that Pab1p specifically controls ARE-mediated translation repression. This hypothesis predicts that the absence of Pab1p would lead to derepression of the MFA2 reporter transcript. To ascertain this, we examined translation of the Luc-MFA2 reporter in strains expressing or lacking PAB1. Although PAB1 is an essential gene, a spb2Δ pab1Δ strain (where Spb2p is a large ribosomal subunit protein) or a pbp1Δ pab1Δ strain (where Pbp1p is a Pab1p-interacting factor that controls polyadenylation rates) is able to grow (29, 30). Luciferase activity was measured in the pbp1Δ pab1Δ strain as described above and compared to luciferase activity in both the wild-type (WT) and pbp1Δ strains. The luciferase activity was normalized to mRNA levels in order to account for effects on mRNA abundance, as absence of Pab1p leads to increased poly(A) tails and increased mRNA levels as well as other processing defects that may contribute to the effect on translation. The results demonstrated that the MFA2 3′-UTR repressed translation 10-fold in both the wild-type and pbp1Δ strains, indicating that Pbp1p does not alter MFA2 3′-UTR reporter translation. Luciferase expression of the Luc-MFA2 reporter, however, increased by at least 10-fold in the pbp1Δ pab1Δ strain (Fig. 2B; compare WT and pbp1Δ strains to pbp1Δ pab1Δ strain) and was similar to that of the Luc-PGK1 reporter. In contrast, translation of the Luc-PGK1 reporter did not vary significantly between the three strains (Fig. 2B). It is difficult to analyze the ARE-specific effects of a pab1Δ strain (which does not grow in the absence of a compensating suppressor as the pbp1Δ pab1Δ strain does and affects multiple mRNA processing steps); however, together with the lack of a specific effect of the pbp1Δ strain and the fact that there is a derepression of MFA2 3′-UTR-specific translation beyond the effect on general translation (controlled by normalization to PGK1 reporter translation) and beyond effects on mRNA stability, since all luciferase values are normalized for luciferase mRNA levels, these results indicate that Pab1p directly or indirectly plays a specific role in MFA2 3′-UTR-regulated translational control.
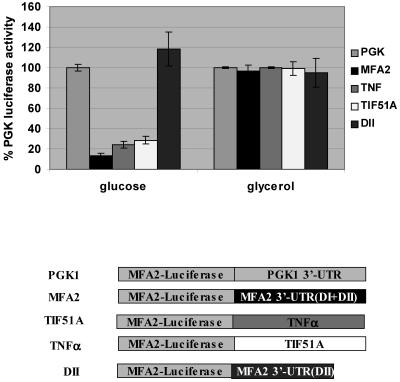
MFA2 3′-UTR regulates translation in response to carbon source.
In yeast cells, growth in the nonfermentable carbon source glycerol acts as a comparable ARE-modulating stimulus in a process controlled by the p38 MAPK/Hog1p pathway (49). To examine translation regulation by carbon source, the Luc-MFA2 and the Luc-PGK1 reporters were expressed in yeast and the luciferase activity was measured in extracts and normalized for abundance of each mRNA by Northern blotting. In glucose medium as described above, the translation of the Luc-MFA2 reporter was repressed at least 10-fold compared to the Luc-PGK1 control. We therefore assessed the effect of glycerol growth conditions on translation of the Luc-MFA2 reporter. In contrast to the 10-fold repression observed in glucose conditions, in cells grown in glycerol, translation of the Luc-MFA2 reporter was equivalent to that of the Luc-PGK1 control (Fig. 3). Translation of the PGK1 control construct remained unchanged between cells grown in the two carbon sources, glucose and glycerol (Fig. 3). Therefore, the MFA2 3′-UTR can repress mRNA translation in response to glucose as a carbon source and alternatively activate translation in non-glucose-grown cultures. The 3′-UTRs of TIF51A and the TNF-α reporter, Luc-TNF-α or Luc-TIF51A, also demonstrated a similar translation regulation of at least a fourfold difference between wild-type glucose- and glycerol-grown cultures compared to the Luc-PGK1 reporter in wild-type glucose-grown cultures or the Luc-DII construct that lacks the AU domain of the MFA2 3′-UTR. Both the Luc-TNF-α and Luc-TIF51A reporters stabilize the transcript in response to glucose growth conditions (49) and yet show translation repression under glucose conditions, as does the unstable MFA2 3′-UTR reporter, suggesting that this translation control is a distinct mechanism and not merely a readout of increased mRNA levels. Therefore, translation regulation via an AU-rich region is conserved in yeast and is controlled by carbon source growth conditions.
FIG. 3.
ARE-controlled translation is regulated by carbon source growth conditions. The 3′-UTRs of PGK1, MFA2, and TIF51A and the domain II construct which lacks the AU-rich domain of the MFA2 3′-UTR were tested for luciferase activity using firefly reporters as described in Materials and Methods. Shown is Luc-PGK1, Luc-MFA2, Luc-TNF-α, Luc-TIF51A, and Luc-DII reporter translation in wild-type glucose- and glycerol-grown cultures compared to the Luc-PGK1 reporter in wild-type glucose-grown cultures. All values are compared to the Luc-PGK1 reporter activity in the WT strain grown in glucose conditions and are normalized for RNA levels. The luciferase experiment was repeated three times.
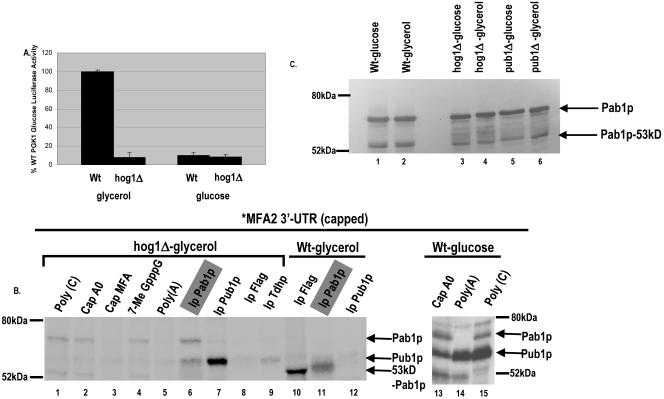
p38 MAPK/Hog1p is required for MFA2 3′-UTR translation regulation.
In conjunction with our previous results that Hog1p is required for carbon source-responsive ARE regulation of mRNA stability (49), the above results suggested that carbon source regulation of MFA2 3′-UTR translation may be controlled by the p38 MAPK/Hog1p pathway. In order to test this hypothesis, we examined translation of both Luc-MFA2 and Luc-PGK1 in glucose- and glycerol-grown cultures in a strain deleted of HOG1. We examined whether MFA2 translation activation in glycerol-grown cultures was maintained in the hog1Δ strain. Strikingly, while the Luc-PGK1 translation remained unaltered between wild-type and hog1Δ strains, the Luc-MFA2 translation was 10-fold repressed in the hog1Δ strain, similar to the repression observed in glucose-grown cultures (Fig. 4A). Translation was greatly repressed for the Luc-MFA2 reporter in glucose-grown hog1Δ strains, suggesting that translation activation could not occur in the absence of Hog1p in glucose and glycerol conditions due to deregulation of translation control (Fig. 4A). Together, these results demonstrate that Hog1p modulates translation activation through the MFA2 3′-UTR.
FIG. 4.
p38 MAPK/Hog1p controls carbon source-mediated ARE translation regulation. A. Translation in the hog1Δ strain grown in either glycerol or glucose medium. Shown is translation of the Luc-MFA2 reporter compared to the Luc-PGK1 reporter activity in the WT strain, normalized for RNA levels. The luciferase experiment is an average of three trials. B. UV-cross-linking analysis performed with [α-32P]UTP-labeled capped MFA2 3′-UTR (U*) and yeast extracts from the hog1Δ strain (lanes 1 to 9) or the wild-type strain grown in glycerol medium (lanes 10 to 12) or the wild-type strain grown in glucose medium (lanes 13 to 15). Shown is competition with a 100-fold excess of unlabeled cap in lane 4, a 100-fold excess of nonspecific unlabeled capped polylinker RNA (Cap A0) in lane 2 and lane 13, and 100 ng of poly(C) and poly(A) in lanes 1 and 5 and in lanes 15 and 14, respectively. Pab1p reproducibly binds more strongly in the presence of a nonspecific RNA competitor (compare lane 1 to lanes 3 to 5). Identical sets of the cross-linked reaction mixtures were subjected to immunoprecipitation with anti-Pab1p antibody (lane 6), anti-Pub1p antibody (lane 7), anti-Flag antibody (lane 8) as a monoclonal control antibody for the anti-Pub1p antibody, and anti-Tdh/GAPDH antibody (lane 9) as a polyclonal control antibody for the anti-Pab1p antibody. In wild-type glucose medium conditions, additional bands that are competed by poly(C) also appear that remain to be identified. C. Western blotting with anti-Pab1p polyclonal antibody with extracts used in this figure and Fig. 5 as marked.
p38 MAPK/Hog1p regulates Pab1p binding to the MFA2 3′-UTR in a carbon source-dependent manner.
Our previous results demonstrated that Pab1p binds the MFA2 AU-rich element to repress mRNA translation in a specific manner in glucose-grown cultures. All the pab1Δ strains that we have tested so far do not grow in glycerol conditions. Together, the above data suggest that p38 MAPK/Hog1p is required to regulate Pab1p in glycerol-grown cultures and thereby alter translation of ARE-bearing transcripts. We hypothesized that Hog1p would have to control Pab1p directly or indirectly in order to effect translation derepression in glycerol-grown cultures as shown in Fig. 4A. We performed UV-cross-linking analysis with 32P-labeled in vitro-transcribed capped MFA2 3′-UTR followed by immunoprecipitation with antibody against Pab1p or preimmune sera or polyclonal anti-GAPDH (Tdhp) and monoclonal anti-Flag as controls. In concurrence with our previous results that Pab1p binding in glucose-grown cultures results in translation repression, we observed no full-length Pab1p binding to the MFA2 3′-UTR in wild-type glycerol-grown cultures where translation is derepressed (Fig. 4B, lane 11). Remarkably, glycerol-grown cultures of the hog1Δ strain demonstrated Pab1p binding to the MFA2 3′-UTR in a specific manner which significantly was similar in pattern to that observed in glucose-grown wild-type yeast extracts (Fig. 2A, lanes 1 to 6, and 4B, lanes 13 to 15), correlating with the translation repression observed in this strain in these conditions (Fig. 4B, lanes 1 to 6). Interestingly, a 53-kDa proteolytic form of Pab1p that lacks the unique C terminus of Pab1p (30, 43) was observed to bind to the MFA2 3′-UTR in wild-type glycerol growth conditions where full-length Pab1p failed to bind (Fig. 4B, lane 11). The regulation of Pab1p binding to the MFA2 3′-UTR was not a result of altered protein levels, as the overall levels of the two forms of Pab1p remained similar in different carbon source conditions as well as in different strains as observed by Western blotting of the extracts used for cross-linking (Fig. 4C). These data suggest that Hog1p regulates MFA2 translation by either directly or indirectly controlling Pab1p in response to carbon source by either regulating Pab1p binding to the MFA2 3′-UTR or altering the form of Pab1p that can be recruited to the MFA2 3′-UTR.
Pub1p is required for translation repression by the MFA2 3′-UTR.
The p38 MAPK pathway also regulates another MFA2 3′-UTR binding protein, Pub1p. Pub1p is homologous to the TIA-1/TIAR proteins and to members of the ELAV family, which have been observed to repress translation of ARE-bearing transcripts and to interact with yeast eIF4G (8, 17, 21, 22, 49). We had previously demonstrated that Pub1p is an ARE binding protein in yeast that can bind TNF-α ARE in glucose-grown culture extracts and stabilizes transcripts bearing the TNF-α ARE (49). We find that Pub1p can cross-link to either the AU domain I of MFA2 3′-UTR (Fig. 5A, lane 1) or the MFA2 3′-UTR (Fig. 5A, lanes 3 and 5) in a specific manner, as the cross-link can be competed off by unlabeled MFA2 3′-UTR but not by a 100-fold excess of the rest of the MFA2 transcript (MFA2 5′-UTR and coding region) or by a 100-fold excess of a nonspecific polylinker RNA (Fig. 5A, glucose panel, compare lane 2 with lane 1 and lane 4 with lane 3 or 5). Pub1p binds the MFA2 3′-UTR and domain I in glucose-grown cultures only and not in glycerol-grown cultures (Fig. 5A, compare lanes 1 to 5 with lanes 6 to 10); however, MFA2 mRNA stability is unaffected by the absence of Pub1p in a pub1Δ strain grown in either carbon source medium (Fig. 5B). Interestingly, Pub1p fails to bind the MFA2 3′-UTR or domain I and the TNF-α ARE in glycerol conditions (Fig. 5A, TNF-α ARE) (49), although Pub1p is present at similar levels in glycerol- and glucose-grown cultures (Fig. 5A, lower panel). Significantly, Pub1p can bind the MFA2 3′-UTR in glycerol-grown extracts from a hog1Δ strain, suggesting that Pub1p is regulated in a coordinate manner like Pab1p by the Hog1p/p38 pathway (Fig. 4B, lanes 6 and 7 compared to lanes 11 and 12).
FIG. 5.
Pub1p is required for MFA2 3′-UTR-mediated translation regulation. A. Pub1p binds the MFA2 3′-UTR and the AU-rich domain I in a carbon source-regulated manner, in yeast extracts from glucose-grown cultures but not in yeast extracts from glycerol-grown cultures. UV-cross-linking analysis was performed with [α-32P]UTP-labeled uncapped MFA2 3′-UTR (U*) (lanes 3 to 5 and 8 to 10) or with domain I (sequence as in Fig. 1, lanes 1 and 2 and lanes 6 and 7) as described in Materials and Methods. The uncapped MFA2 3′-UTR does not bind Pab1p (data not shown), and therefore Pub1p binding can be easily visualized around the 60-kDa marker. Shown is competition either with a 100-fold excess of MFA2 mRNA (5′-UTR and coding region without the 3′-UTR, M, lanes 3 and 8), with a 100-fold excess of unlabeled MFA2 3′-UTR (+) (lanes 2, 4, 7, and 9), or with nonspecific unlabeled polylinker RNA (−) (lanes 1, 5, 6, and 10) using extracts from glucose-grown cultures (left panel) or extracts from glycerol-grown cultures (right panel). The lower panel depicts Pub1p levels in the above extracts determined by Western analysis using 4C3 monoclonal anti-Pub1p antibody. B. Half-life analysis was performed in a pub1Δ strain grown in glucose or in glycerol growth medium as described in Materials and Methods. The Northern blot depicted was probed for MFA2 using an RNA probe as described in Materials and Methods and then normalized against U3 for loading and quantitation. C. First panel from top, Luc-MFA2 reporter translation compared to the Luc-PGK1 reporter in wild-type and pub1Δ strains grown in glucose and glycerol cultures; second panel, Luc-TNF-α ARE reporter translation; third panel, Luc-domain II reporter translation; fourth panel, translation of the control PGK1 reporter in wild-type and pub1Δ strains in glucose and glycerol media. All values are compared to the Luc-PGK1 reporter activity in the WT strain grown in glucose, are normalized for luciferase RNA levels, and were reproducible three times. D. Pab1p binds the MFA2 3′-UTR only in a PUB1 strain. UV-cross-linking analysis followed by immunoprecipitation was performed with [α-32P]UTP-labeled capped MFA2 3′-UTR (U*) as described in Materials and Methods. Each reaction mixture included a 100-fold excess of nonspecific unlabeled capped polylinker RNA, Cap A0, and was followed by immunoprecipitation using anti-Pab1p antibody or as a control anti-Tdh/GAPDH antibody using extracts from glucose-grown cultures from a PUB1 strain (left panel) or a pub1Δ strain (right panel). This experiment was reproduced five times.
We asked whether Pub1p may facilitate the repression mediated by Pab1p, possibly through the formation of a repressive translation complex between the ARE-bound Pab1p and the cap/eIF4G complex. If this hypothesis is true, then deletion of Pub1p must prevent translation repression. We examined translation of the MFA2 3′-UTR reporter in a PUB1 and a pub1Δ strain grown in glucose and glycerol media. While translation of the MFA2 3′-UTR reporter was repressed in the wild-type strain in glucose conditions, translation was fully derepressed in glycerol conditions of the same strain as shown previously. In the pub1Δ strain, however, translation was fully derepressed in both glucose and glycerol growth conditions, suggesting that Pub1p is required for translation repression (Fig. 5C, top panel). The Luc-TNF-α ARE reporter behaved in a similar manner (Fig. 5C, second panel from top). Interestingly, the pub1Δ strain grows very poorly in glycerol medium, suggesting that the absence of Pab1p in glycerol along with deletion of Pub1 affects essential functions. It can also be noted that the pub1Δ strain enhanced translation of the domain II and PGK1 reporters in glucose cultures by twofold reproducibly (Fig. 5C, lower two panels), suggesting that general translation is also affected in the absence of Pub1p, which may in part be due to the effect of pub1Δ on the levels of transcripts of ribosomal protein genes (13). If the Luc-MFA2 values are further normalized for this twofold increase in general translation, a pub1Δ strain grown in glucose still demonstrated a significant fivefold derepression of translation, suggesting that Pub1p specifically repressed the translation of the MFA2 3′-UTR reporter beyond its indirect effect on general translation.
We next questioned whether Pab1p mediated this effect on translation in a pub1Δ strain. We were unable to construct a pab1Δ pub1Δ strain (data not shown). Alternatively, to question this hypothesis, we examined the fate of Pab1p binding to the ARE in the pub1Δ strain to determine whether Pub1p is required. Cross-linking analysis was performed using capped in vitro-labeled MFA2 3′-UTR in extracts from wild-type and pub1Δ strains followed by immunoprecipitation using Pab1p antibody and anti-Tdh as a control. Pab1p binds capped MFA2 3′-UTR in extracts of glucose-grown cultures or in hog1Δ glycerol-grown extracts as shown above, correlating with translation repression conditions (Fig. 2A and 4B). Pab1p bound the capped ARE only in extracts from a PUB1 strain, where translation is indeed repressed, and did not bind in extracts from the pub1Δ strain, where ARE translation is derepressed (Fig. 5D). The regulation of Pab1p binding was not a result of altered protein levels, as the overall levels of the two forms of Pab1p remained similar in different carbon source conditions as well as in different strains as observed by Western blotting of the extracts used for cross-linking (Fig. 4C, lanes 1 and 2 and lanes 5 and 6). Therefore, Pab1p directly or indirectly requires Pub1p for binding the capped ARE to mediate translation repression. An increase in the binding of the truncated form of Pab1p, the 53-kDa proteolytic form that can be immunoprecipitated with Pab1p antibody, was observed in the pub1Δ strain extracts, as was observed earlier in the wild-type glycerol-grown extracts (Fig. 4B, lane 11). The binding of the 53-kDa Pab1p form correlates with conditions where translation is not repressed, suggesting that the ARE might be bound by this form in a regulated manner to provide altered translation regulation in the absence of Pub1p, as in the case of wild-type glycerol growth conditions or the pub1Δ strains (Fig. 6).
FIG. 6.
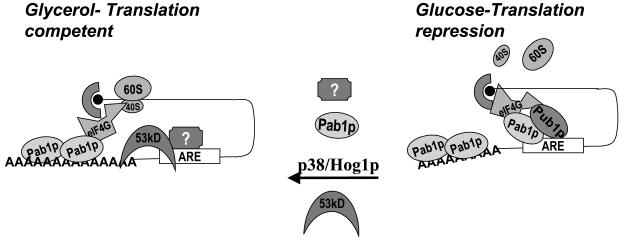
Pab1p mediates MFA2 translation regulation in a p38 MAPK/Hog1p-regulated pathway. p38 MAPK/Hog1p mediates translation regulation through the MFA2 3′-UTR in response to the presence of glucose medium growth conditions. In glucose-grown cultures, the 3′-UTR recruits Pub1p and Pab1p, which can interact with the eIF4G/cap complex as well. Pab1p may under these conditions be recruited to the AU element and interact with the eIF4G/cap complex. Such an mRNP would prevent the cap/poly(A) interaction that may be necessary to promote translation. Furthermore, the Pub1p-bound message may be localized to translationally silenced locations in the cell as described for the mammalian homolog TIA-1. Alternatively, in nonglucose conditions p38 MAPK/Hog1p may modulate Pub1p and Pab1p indirectly through unknown factors (represented as “?”) to prevent their recruitment to the MFA2 3′-UTR. The cap complex would not be involved in forming a translationally silenced complex through interactions with the 3′-UTR-3′-UTR-bound factors. This may be feasible due to the truncated form of Pab1p, the 53-kDa form, binding to the MFA2 3′-UTR instead of the full-length form (Pab1p). Pab1p may be modified such that the full-length form binds the poly(A) tail instead of the 3′-UTR, permitting a translationally favorable mRNP complex. Additionally, in the absence of Pub1p bound to the 3′-UTR, the transcript may be mobilized onto polysomes (depicted as 40S and 60S).
DISCUSSION
AU-rich elements have been shown to regulate expression of tightly controlled transcripts. The AU-rich 3′-UTR of MFA2 mediates rapid decay of the transcript (34). In this study, we elucidated the mechanism of translation regulation mediated by this ARE. Our results demonstrate a novel role for the MFA2 3′-UTR in controlling the efficiency of translation in response to the p38 MAPK/Hog1p pathway. Furthermore we find that the p38 MAPK control over ARE-mediated translation is achieved through modulation of Pab1p and Pub1p, two ARE binding proteins that can interact with and modulate the translation machinery.
Hog1p regulates MFA2 ARE-mediated translation in response to carbon source.
Several studies have shown that TNF-α mRNA translation is regulated by its AU-rich element in response to the p38 MAPK pathway (25, 38, 41). In yeast, AREs have been shown to regulate stability in response to carbon source as well as the p38 MAPK pathway. In glycerol, which mimics one such p38 MAPK-regulated condition, MFA2 mRNA stability remains unaltered (49). However, translation of a reporter bearing the yeast MFA2 3′-UTR is at least 10-fold repressed in glucose-grown cultures compared to a control PGK1 reporter (Fig. 1). This translation is regulated by carbon source and is completely derepressed in glycerol conditions in a Hog1p-dependent manner (Fig. 3). Additionally, in glucose growth conditions, a hog1Δ strain demonstrates a stabilized MFA2 3′-UTR reporter, although translation remains deregulated in this strain and greatly reduced in both carbon source conditions (Fig. 4A) (Vasudevan et al., submitted). Remarkably, complete response to mating-type repression or G1 arrest on pheromone signaling occurs only in glucose growth conditions while, in nonfermentable glycerol medium, the cells fail to repress the MAT locus expression (45). MFA2 expression is therefore also influenced by carbon source conditions by the Hog1p pathway. Therefore, the MFA2 3′-UTR AU-rich sequences, in addition to mediating mRNA stability, control translation in a p38 MAPK-regulated manner. Similar results were obtained upon testing the 3′-UTR of TIF51A and the TNF-α ARE reporter translation in wild-type glucose- and glycerol-grown cultures compared to the Luc-PGK1 or the Luc-domain II reporter in wild-type glucose-grown cultures. Our data suggest that the yeast system has conserved ARE regulation by the p38 MAPK pathway.
ARE binding proteins such as Pub1p modulate Pab1p recruitment to regulate MFA2 translation.
Pub1p, a yeast homolog of the ARE translation repressor protein TIA-1/ELAV family of proteins (21, 22, 49), binds the MFA2 3′-UTR specifically in extracts from glucose-grown cultures and not in extracts from glycerol-grown cultures (Fig. 5A). In glycerol conditions, binding of Pub1p to the AREs of MFA2, TIF51A, and TNF-α is strikingly absent and the stability of the TNF-α ARE reporter transcript is concomitantly altered (Fig. 5A) (49). However, this regulated binding of Pub1p to the MFA2 3′-UTR does not influence MFA2 mRNA stability (Fig. 5B). The results demonstrated that deletion of Pub1p enhanced translation of the MFA2 luciferase reporter by 10-fold in glucose-grown cultures (Fig. 5C). Translation in the pub1Δ strain remained at least fivefold higher when normalized for the PGK1 reporter translation, suggesting that Pub1p specifically repressed MFA2 3′-UTR-controlled translation. In glycerol conditions, MFA2 reporter translation is increased in wild-type cells, suggesting that Pub1p binding to the 3′-UTR leads to repression of MFA2 translation in glucose-grown cultures. Conversely, absence of Pub1p promotes translation as observed in glycerol-grown cultures. In the pub1Δ strain grown in glycerol, translation of the MFA2 reporter remained as high as in the wild-type strain (Fig. 5C). This conformed to our hypothesis that Pub1p binding to the MFA2 3′-UTR represses translation. Furthermore, we find that, in the pub1Δ strain, Pab1p no longer binds capped MFA2 3′-UTR in extracts, suggesting that an altered mRNP formed possibly with the truncated 53-kDa form of Pab1p. Pab1p can interact with Pub1p only in an RNA-dependent manner (data not shown), suggesting that the interaction between these ARE binding factors is indirect through a common mRNP complex such as the cap complex through eIF4G (8, 17) or the ARE, which can interact with both Pab1p and Pub1p. Interestingly, Pab1p does not bind MFA2 3′-UTR in a pub1Δ strain, suggesting that the mRNP complex formed by Pub1p is essential for Pab1p recruitment to the MFA2 3′-UTR. One likely possibility is that, similar to the mammalian TIA-1 protein, Pub1p is responsible for localizing the mRNA to a stress granule-like body to form a silenced mRNP (21, 22) where Pab1p is recruited to the ARE to repress translation. A second possibility that may concurrently occur is that Pub1p interacts with the cap complex/Pab1p (8, 17, 21, 22) to promote Pab1p interaction with the ARE, thereby preventing the translationally favorable closed loop complex between the poly(A) tail and the cap (47, 51) (Fig. 6). Several interesting questions arise from this study, such as what factor(s) is directly modulated by the p38 kinase and modulates Pub1p, as we do not observe any apparent change in phosphorylation status of Pub1p or in levels of Pub1p in either the hog1Δ strain or glycerol conditions.
Hog1p modulates the ARE binding protein Pab1p, directly or indirectly, to regulate MFA2 mRNA translation in response to growth stimuli.
We demonstrated that Pab1p binds the MFA2 3′-UTR in a cap-dependent manner and specifically mediates translation repression in glucose-grown cultures. Remarkably, full-length Pab1p failed to bind capped MFA2 3′-UTR in extracts from glycerol-grown cultures, suggesting that the Hog1p pathway regulates Pab1p binding to the MFA2 3′-UTR, which permits the translation observed in glycerol-grown cultures (Fig. 4B). PABP1 can be phosphorylated through the mammalian p38 MAPK pathway and was found to be an ARE binding protein (2, 50). We have not observed any changes in phosphorylation status of either Pub1p or Pab1p in the hog1Δ strain or in glycerol conditions, suggesting that these effects may be mediated indirectly through phosphorylation of additional unknown interacting factors. The pub1Δ strain grows very poorly in glycerol conditions, the pab1Δ strains do not grow in glycerol, and a pub1Δ pab1Δ strain was inviable (data not shown); therefore, we could not test the absence of Pab1p in translation derepression conditions as in the pub1Δ strain or in glycerol conditions.
Since the Hog1p pathway is essential for the 3′-UTR regulation between these two carbon sources and for Pab1p binding, we anticipated Hog1p regulation of Pab1p forms as well as its ability to bind the MFA2 3′-UTR. In conjunction with this theory we find an altered truncated form of Pab1p (43) binding the MFA2 3′-UTR in wild-type glycerol-grown cultures and in the translationally derepressed conditions in a pub1Δ strain grown in glucose medium (Fig. 4B, lane 11, and 5C, second panel from the top), although we observed no comparative changes in the levels of the two forms of Pab1p expressed in both glucose- and glycerol-grown cultures (Fig. 4C). Therefore, Pab1p is one essential translation factor that also functions as an ARE binding protein modulated directly or indirectly by the p38 MAPK/Hog1p pathway.
There are several observations outlined below that lead us to propose a model where the 53-kDa truncated form of Pab1p may play a regulated role in MFA2 translation, although it is possible that the regulated binding of full-length Pab1p between the two conditions is sufficient for translation regulation. As there is very little literature on the in vivo production and function of this form of yeast Pab1p, we have not been able to inhibit production of this form and observe effects on translation. First, the Sachs group demonstrated that yeast cells boiled in loading buffer also show this form, suggesting that it is unlikely to be an in vitro degradation product (23). We have also repeated this experiment and observed the same, although it cannot be ruled out that this form did not arise during the heating in SDS loading buffer.
The primary evidence that our data provide towards a regulated role for this form in vivo is the fact that the binding to labeled MFA2 3′-UTR is faithfully regulated in favor of the truncated form of Pab1p, strictly in translation upregulation conditions, although by Western blotting (Fig. 4C), there is little change in the ratio of the two Pab forms in glucose versus glycerol in a wild-type strain or a pub1 strain or a hog1 strain. For example, the truncated Pab1p alone binds the labeled MFA2 3′-UTR in wild-type extracts from glycerol-grown cultures where translation is activated (Fig. 4B, lane 11) compared to the hog1 glycerol-grown extract cultures (Fig. 4B, lane 6), where full-length Pab1p also binds and translation is repressed. This is not due to a global difference in proteolysis between extracts from glycerol- and those from glucose-grown cell cultures, since, in glucose-grown cell cultures, the truncated Pab1p alone binds the labeled MFA2 3′-UTR only in a pub1Δ glucose-grown cell extract where translation is activated (Fig. 5C, second panel from the top), in contrast to glucose-grown wild-type cell extracts (Fig. 5C, top panel), where full-length Pab1p predominantly binds and translation is repressed. In each case, as shown in Fig. 4C, the extracts were prepared identically and have similar levels of the two Pab1p forms in glucose and glycerol conditions. This suggests a functional or at least a regulated binding characteristic for this form. Since the regulation is in vivo, it suggests that this truncated form is an in vivo form.
The above correlations also indicate that it is the C-terminus-bearing form that is prevented from binding under the translation activation conditions. In conjunction with the role of the C terminus, both human and mouse systems express PABPC5, a distinct gene that produces such a truncated PABP without the C terminus (30). This domain is cleaved by viral proteases in mammalian cells to partially facilitate cap-independent viral mRNA translation (28). Therefore, there is a natural conservation of such a truncated form in higher species and during viral infections/altered translation conditions.
Together, these pieces of evidence support a model where the selective regulation of ARE translation may be mediated by the regulated recruitment of Pab1p to the ARE; recruitment of the full-length Pab1p leads to repression by perturbing the normal poly(A)-cap-mediated translation while inhibition of binding of full-length Pab1p and recruitment of the 53-kDa form as well as additional unknown factors correlate with translation activation since the regulatory C terminus that can interact with inhibitory factors such as PAIP2 is absent. Furthermore, the binding of the 53-kDa form in this case would exclude interaction with the full-length form since it cannot multimerize and perhaps affect the Pab1p interaction between the poly(A) tail and cap complex or compete for interaction with certain translation factors (26, 28, 29, 30, 32, 43, 46, 47).
In our conditions, the truncated form of Pab1p binds in extracts to poly(A) tails apparently just as well as the full-length form of Pab1p by UV cross-linking analysis (54; data not shown). This has been observed previously, and in vitro studies by Alan Sachs' group showed that there is a modest twofold increase in binding affinity of recombinant Pab1p with a deleted C terminus. Interestingly, G. Goodall's group demonstrated that the mammalian protein with a similar deletion bound threefold better to the AU element than the full-length form of human PABP (46), suggesting that this truncated form would have a unique advantage in binding the poly(A) tails of AU-bearing messages since Pab1p needs only the regions of RRM2 and to some extent RRM1 to bind the poly(A) tail and regions RRM3 and RRM4 to bind the AU element. The physiological implications of this strong binding affinity but lack of cooperative multimerization are not yet outlined, but from our data one can hypothesize that, while this could be restrictive for general translation of non-ARE messages, on an ARE-bearing transcript, such a Pab1p molecule would be better tethered by the ARE and the poly(A) tail without having the capability to inhibit translation through the absent C terminus. This may provide such transcripts with a translational advantage, as observed in the glycerol conditions, where full-length Pab1p fails to bind the ARE and only truncated Pab1p is observed to bind.
We propose that modulation of Pab1p recruitment to the ARE is one critical switch for ARE-mediated translation regulation (Fig. 6). The p38/MAPK pathway can regulate Pab1p through distinct unknown mechanisms involving the TIA-like protein Pub1p, to effect translation regulation in a stimulus-specific manner. In glucose growth conditions, Pub1p sequesters the ARE mRNP and may also directly affect Pab1p binding to the ARE, as Pub1p can interact with the eIF4G/cap complex (8, 17, 21, 22) and promote Pab1p recruitment to the ARE through the cap complex. This would switch the ARE mRNP from a translationally favorable cap complex-Pab1p bound to the poly(A) tail as proposed by the circularization model for translation (51). Alternatively, in glycerol conditions, Pub1p cannot bind the ARE and full-length Pab1p is excluded from the ARE as well. Additionally, factors that activate translation in such conditions may prevent translation repression either independently or through the role of Pab1p/truncated 53-kDa Pab1p itself since a pab1Δ strain that is viable in glucose conditions due to suppressor mutants does not grow in glycerol, suggesting a critical function for Pab1p in these conditions (Fig. 6). This is further supported by the fact that translation activation conditions such as a pub1Δ strain or wild-type glycerol-grown strains reveal that a truncated form of Pab1p can be recruited to the ARE complex in these altered conditions (Fig. 4B and 5D). Regulation of such a key factor that can mediate mRNA stability and translation in a global and specific manner provides a very efficient mechanism for ARE regulation of gene expression in response to the p38 MAPK pathway.
Acknowledgments
This work is dedicated to the memory of P. Vasudevan.
We thank Pfizer for the generous gift of thiolutin.
This work was supported by a grant from the National Institutes of Health to S.W.P. (GM58276).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology, vol. 2, p. 13.1-13.13. John Wiley and Sons, New York, N.Y. [Google Scholar]
- 2.Bollig, F., R. Winzen, M. Gaestel, S. Kostka, K. Resch, and H. Holtmann. 2003. Affinity purification of ARE-binding proteins identifies poly(A)-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 301:665-670. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 4.Czaplinski, K., M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng, H. A. Perlick, H. C. Dietz, M. D. Ter-Avanesyan, and S. W. Peltz. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst, T. J., A. R. Ritchie, G. D. Demetri, and J. D. Griffin. 1989. Regulation of granulocyte- and monocyte-colony stimulating factor mRNA levels in human blood monocytes is mediated primarily at a post-transcriptional level. J. Biol. Chem. 264:5700-5703. [PubMed] [Google Scholar]
- 6.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Grafi, G., I. Sela, and G. Galili. 1993. Translational regulation of human beta interferon mRNA: association of the 3′ AU-rich sequence with the poly(A) tail reduces translation efficiency in vitro. Mol. Cell. Biol. 13:3487-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 11.Gray, N. K., J. M. Coller, K. S. Dickson, and M. Wickens. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19:4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, M. E., A. B. Shyu, and J. G. Belasco. 1990. Deadenylylation: a mechanism controlling c-fos mRNA decay. Enzyme 44:181-192. [DOI] [PubMed] [Google Scholar]
- 13.Grigull, J., S. Mnaimneh, J. Pootoolal, M. D. Robinson, and T. R. Hughes. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24:5534-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 15.Hagan, K. W., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 15:809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilleren, P., and R. Parker. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33:229-260. [DOI] [PubMed] [Google Scholar]
- 17.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 18.Iizuka, N., and P. Sarnow. 1997. Translation-competent extracts from Saccharomyces cerevisiae: effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods 11:353-360. [DOI] [PubMed] [Google Scholar]
- 19.Jarzembowski, J. A., and J. S. Malter. 1997. Cytoplasmic fate of eukaryotic mRNA: identification and characterization of AU-binding proteins. Prog. Mol. Subcell. Biol. 18:141-172. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 21.Kedersha, N., and P. Anderson. 2001. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151: 1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler, S. H., and A. B. Sachs. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koeffler, H. P., J. Gasson, and A. Tobler. 1988. Transcriptional and posttranscriptional modulation of myeloid colony-stimulating factor expression by tumor necrosis factor and other agents. Mol. Cell. Biol. 8:3432-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontoyiannis, D., A. Kotlyarov, E. Carballo, L. Alexopoulou, P. J. Blackshear, M. Gaestel, R. Davis, R. Flavell, and G. Kollias. 2001. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20:3760-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozlov, G., J. F. Trempe, K. Khaleghpour, A. Kahvejian, I. Ekiel, and K. Gehring. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 98:4409-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruys, V., O. Marinx, G. Shaw, J. Deschamps, and G. Huez. 1989. Translational blockade imposed by cytokine-derived UA-rich sequences. Science 245:852-855. [DOI] [PubMed] [Google Scholar]
- 28.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangus, D. A., N. Amrani, and A. Jacobson. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18:7383-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy, J. E. G. 1998. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 62:1492-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo, E. O., R. Dhalia, C. Martins de Sa, N. Standart, and O. P. de Melo Neto. 2003. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly(A) and efficient cap distal translational repression. J. Biol. Chem. 278:46357-46368. [DOI] [PubMed] [Google Scholar]
- 33.Mijatovic, T., V. Kruys, D. Caput, P. Defrance, and G. Huez. 1997. Interleukin-4 and -13 inhibit tumor necrosis factor-alpha mRNA translational activation in lipopolysaccharide-induced mouse macrophages. J. Biol. Chem. 272:14394-14398. [DOI] [PubMed] [Google Scholar]
- 34.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 35.Muhlrad, D., and R. Parker. 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6:2100-2111. [DOI] [PubMed] [Google Scholar]
- 36.Munro, D., and A. Jacobson. 1990. Tales of poly(A): a review. Gene 91:151-158. [DOI] [PubMed] [Google Scholar]
- 37.Myer, V. E., X. C. Fan, and J. A. Steitz. 1997. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16: 2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 39.Peltz, S. W., and A. Jacobson. 1993. mRNA turnover in Saccharomyces cerevisiae. Control of messenger RNA stability, p. 291-328. In J. Belasco and G. Brawerman (ed.), Control of messenger RNA stability. Academic Press, Inc., San Diego, Calif.
- 40.Philippsen, P., A. Stotz, and C. Scherf. 1991. DNA of Saccharomyces cerevisiae. Methods Enzymol. 194:169-182. [DOI] [PubMed] [Google Scholar]
- 41.Piecyk, M., S. Wax, A. R. P. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Echevarria, M. J., R. Munshi, J. Tomback, T. G. Kinzy, and S. W. Peltz. 2001. Characterization of a general stabilizer element that blocks deadenylation-dependent mRNA decay. J. Biol. Chem. 276:30995-31003. [DOI] [PubMed] [Google Scholar]
- 43.Sachs, A. B., M. W. Bond, and R. D. Kornberg. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45:827-835. [DOI] [PubMed] [Google Scholar]
- 44.Schuler, G. D., and M. D. Cole. 1988. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell 55:1115-1122. [DOI] [PubMed] [Google Scholar]
- 45.Shei, G.-J., and J. R. Broach. 1995. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol. Cell. Biol. 15:3496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sladic, R. T., C. A. Lagnado, C. J. Bagley, and G. Goodall. 2004. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 271:450-457. [DOI] [PubMed] [Google Scholar]
- 47.Tarun, S. Z. J., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 48.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 49.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 50.Voeltz, G. K., J. Ongkasuwan, N. Standart, and J. A. Steitz. 2001. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 15:774-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 52.Wiklund, L., M. Sokolowski, A. Carlsson, M. Rush, and S. Schwartz. 2002. Inhibition of translation by UAUUUAU and UAUUUUUAU motifs of the AU-rich RNA instability element in the HPV-1 late 3′ untranslated region. J. Biol. Chem. 277:40462-40471. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, T., and R. Treisman. 1988. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336:396-399. [DOI] [PubMed] [Google Scholar]
- 54.Wilusz, C. J., M. Gao, C. L. Jones, J. Wilusz, and S. W. Peltz. 2001. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7:1416-1424. [PMC free article] [PubMed] [Google Scholar]
- 55.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901-906. [DOI] [PubMed] [Google Scholar]
- 56.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, T., V. Kruys, G. Huez, and C. Gueydan. 2001. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 30:952-958. [DOI] [PubMed] [Google Scholar]