Abstract
The alternative splicing of the mek5 gene gives rise to two isoforms. MEK5β lacks an extended N terminus present in MEK5α. Comparison of their activities led us to identify a novel mitogen-activated protein kinase (MAPK) docking site in the N terminus of MEK5α that is distinct from the consensus motif identified in the other MAPK kinases. It consists of a cluster of acidic residues at position 61 and positions 63 to 66. The formation of the MEK5/extracellular signal-regulated kinase 5 (ERK5) complex is critical for MEK5 to activate ERK5, to increase transcription via MEF2, and to enhance cellular survival in response to osmotic stress. Certain mutations in the ERK5 docking site that prevent MEK5/ERK5 interaction also abrogate the ability of MEKK2 to bind and activate MEK5. However, the identification of MEK5α mutants with selective binding defect demonstrates that the MEK5/ERK5 interaction does not rely on the binding of MEK5α to MEKK2 via their respective PB1 domains. Altogether these results establish that the N terminus of MEK5α is critical for the specific organization of the components of the ERK5 signaling pathway.
The mitogen-activated protein kinase (MAPK) signaling pathways regulate numerous physiological processes during development and pathogenesis (10). They consist of the sequential activation of protein kinases that include a MAPK, a MAPK/extracellular signal-regulated kinase (ERK) kinase (MEK or MKK) and a MEK kinase (MEKK) (10). At least four MAPK subfamilies have been identified: ERK1/2, ERK5, c-Jun NH2-terminal protein kinases (JNKs), and p38 MAPKs. MAPK activators include MEK1 and MEK2 for ERK1/2, MEK5 for ERK5, MKK4 and MKK7 for JNKs, and MKK3 and MKK6 for p38 MAPKs. MEKs/MKKs activate MAPK by dual phosphorylation on threonine (T) and tyrosine (Y) residues within a T-X-Y motif. A conserved cluster of two to five basic residues identified in the N terminus of MEK1 and -2 and MKK3, -4, -6, and -7 constitutes a consensus MAPK binding site that is necessary for the transmission of the signal (1, 28). A similar motif has been identified by sequence homology in MEK5, but its function as a docking site for ERK5 has not been proven (28).
The ERK5 signaling pathway regulates by phosphorylation the activity of a number of transcription factors including myocyte enhancer factor 2 (MEF2) (12, 13). Consistent with its role in stimulating gene expression through the regulation of MEF2 activity, ERK5 contributes in vitro to regulating muscle differentiation and neuronal survival (6, 16, 24). The analysis of mutant mice in which the erk5 gene can be conditionally deleted has provided in vivo genetic evidence that ERK5 mediates the survival of endothelial cells (9). The loss of endothelial survival may be responsible for the cardiovascular defects observed in erk5−/− and mek5−/− embryos (21, 25, 29, 32). Being twice the size (815 amino acids) of the other MAPKs, ERK5 possesses a unique C-terminal tail that contains a MEF2-interacting domain and a potent transcriptional activation domain (11). The C-terminal tail has been shown to regulate the nuclear shuttling of ERK5, but its role in mediating ERK5 activation remains controversial (3, 31). In contrast to the other MAPK cascades, only one activator of ERK5, MEK5, has been cloned (7, 33).
We have recently provided genetic evidence that MEK5 is a nonredundant component of the ERK5/MEF2-dependent cell survival pathway (29). Alternative splicing of the mRNA gives rise to two isoforms with different N termini, MEK5α (50 kDa) and MEK5β (40 kDa) (7). The N-terminal extension of MEK5α has been implicated in its restricted localization to the particulate fraction, while MEK5β is ubiquitously distributed and is primarily cytosolic (7). In addition, it contains a phox and Bem1p (PB1) domain that mediates the binding interaction of MEK5α with the MEK kinases, MEKK2 and MEKK3 (17). Blocking the PB1-dependent formation of the MEKK2/MEK5 complex prevents MEK5 activation of ERK5 (17). Consistent with this study, MEK5β that lacks the PB1 domain has been identified as a kinase-dead dominant-negative variant that can suppress ERK5 signaling (4). However, this is disputed by the ability of a MEK5β transgene to activate ERK5 in vivo and to promote eccentric cardiac hypertrophy that progresses to dilated cardiomyopathy and sudden death (18).
To elucidate the functional consequence of differential splicing of the mek5 gene, we have investigated the regulation of ERK5 by MEK5 isoforms. Our data demonstrate that both MEK5α and MEK5β are catalytically active enzymes, but the physical interaction of ERK5 with the N-terminal domain of MEK5α is critical for ERK5 activation. The docking site identified in the N terminus of MEK5α is unique since it does not match the consensus motif present in the other MAPK activators (28). Further studies demonstrate that the docking of ERK5 to MEK5 is essential for the function of MEK5 as a cell survival factor.
MATERIALS AND METHODS
Cell culture, transfection, and preparation of lysates.
COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (Invitrogen), 2 mM l-glutamine, 10 U/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere with 5% CO2. Mouse embryonic fibroblasts (MEFs) established from mek5−/− embryos were described previously (29). Transfection was performed using the Lipofectamine (Invitrogen) or Metafectene (Biontex, Munich, Germany) reagents, following the manufacturer's instructions. A total of 3 μg of each plasmid DNA was used unless indicated otherwise. Proteins were extracted from cells in triton lysis buffer (TLB; 20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 10% glycerol, 1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin). Extracts were clarified by centrifugation (14,000 × g for 10 min at 4°C). The concentration of soluble proteins was quantified by the Bradford method (Bio-Rad).
Plasmid constructs.
The epitope-tagged MEKK2 and MEKK3 (2), the dominant-active mutant MEK5α (MEK5D) (12), and the glutathione S-transferase (GST)-ERK5 (33) expression vectors were provided by C. Widmann, J.-D. Lee, and J. E. Dixon, respectively. Wild-type MEK5 was created by replacing D311 and D315 with S311 and T315 using a QuikChange XL Site-Directed Mutagenesis kit (Stratagene). N-terminal Flag-tagged MEK5α and MEK5β constructs were obtained by subcloning the cDNAs into p3*Flag-CMV-10 (where CMV is cytomegalovirus) vector (Sigma) using HindIII and NotI. Mammalian expression plasmids encoding full-length wild-type or mutant MEK5α, MEK5β, or N-terminal fragments of MEK5α fused to GST were created by in-frame ligation of the cDNAs into the pEBG vector (22) using SpeI and NotI. The ERK5 and MEK5 cDNAs were subcloned into pRSETA (BamHI/HindIII) and pGEX4T-1 (SalI/NotI) vectors to produce histidine (His) and GST fusion proteins, respectively. All constructs were confirmed by sequencing using the Big Dye kit (Applied Biosystems) on an ABI 377 sequencer.
Immunoblot analysis.
Cell extracts (20 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12% polyacrylamide gel) and electrophoretically transferred to Immobilon-P membrane (Millipore). Membranes were incubated with 3% nonfat dry milk for 30 min at room temperature and then probed overnight at 4°C with primary antibodies to Flag (Sigma), hemagglutinin (HA; Covance), ERK5 (Sigma), GST (Amersham Biosciences), or MEK5 (29). Immunocomplexes were detected by enhanced chemiluminescence (Pierce or Amersham Biosciences) with goat (Jackson Laboratories), rabbit, or mouse (Amersham Biosciences) immunoglobulin G coupled to horseradish peroxidase as the secondary antibody. Immunoblot signals were quantified with the ImageQuantifier software (BioImage, Jackson, MI) to normalize for protein expression prior to protein kinase assays.
Binding assays.
Cell extracts (100 to 500 μg) were incubated at 4°C for 2 h with 1 to 2 μg of antibody and protein A agarose beads or with 1 to 5 μg of His- or GST-fusion protein and Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) or glutathione (GSH)-Sepharose (Amersham) beads, respectively. Complexes were washed three times with TLB and analyzed by immunoblotting. Affinity chromatography was performed as described previously (8).
Protein kinase assays.
ERK5 and MEK5 protein kinase activity was measured in cell lysates following incubation with antibodies to ERK5, Flag, or GST and protein A agarose beads for 2 to 3 h at 4°C. Immunocomplexes were washed three times with TLB and twice with kinase buffer (25 mM HEPES, pH 7.4, 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM dithiothreitol, 0.1% orthovanadate) prior to incubation for 30 min at 30°C in kinase buffer containing 50 μM [γ-32P]ATP (10 Ci/mmol) and 1 μg of GST-MEF2 and GST-ERK5 or myelin basic protein (MBP) for ERK5 and MEK5 assays, respectively. The radioactivity incorporated into recombinant proteins was quantitated after SDS-PAGE by PhosphorImager analysis (Fuji FLA 3000).
Reporter gene expression assay.
The reporter plasmid pG5E1bLuc (23) was transiently cotransfected together with a construct encoding the fusion protein GAL4-MEF2A (Hung-Ying Kao, Case Western University, Cleveland, Ohio) with or without expression vectors encoding wild-type or mutant MEK5α or MEK5β. A pRL-Tk plasmid encoding Renilla luciferase was cotransfected to monitor transfection efficiency. Aliquots of cell lysates were assayed for firefly and Renilla luciferase activities using a dual-luciferase reporter assay kit (Promega) on a Turner Designs 20/20 luminometer.
Cell survival assays.
Cell viability was quantified by luciferase activity following transfection with the pCMV luciferase plasmid (15) or by flow cytometry using an anti-GST antibody coupled to Alexa Fluor 488 (Molecular Probes). Fluorescent cells were counted on a DakoCytomation Cyan flow cytometer equipped with a coherent Sapphire laser (488 nm; 20 mW) using a 530/40 nm band-pass filter.
RESULTS
Comparison of protein kinase activities of MEK5 isoforms.
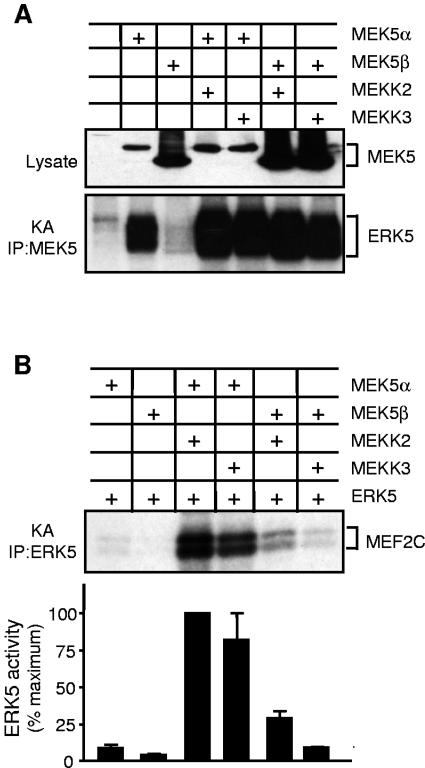
To further understand how MEK5 regulates ERK5, we compared the protein kinase activities of MEK5 isoforms. Lysates of COS-7 cells overexpressing ERK5 and dominant-active mutants of MEK5α or MEK5β were obtained. The activation of ERK5 by MEK5 was examined following immunoprecipitation of ERK5 using an anti-ERK5 antibody and recombinant MEF2 as a substrate (Fig. 1A and B). This assay demonstrated that MEK5α was a stronger activator of ERK5 than MEK5β regardless of the amount of MEK5 protein expressed. The analysis of the immune complexes by immunoblotting indicated that MEK5β interacted with ERK5 with a weaker affinity than MEK5α (Fig. 1A). Control experiments showed similar levels of expression of MEK5 isoforms. These observations suggested that the different activities of MEK5 isoforms were due to their distinct ability to interact with ERK5. Indeed, when MEK5 was coimmunoprecipitated with ERK5 using an anti-Flag antibody prior to the protein kinase assay, MEK5β activated ERK5 to a level similar to activation by MEK5α (Fig. 1A and B, IP:ERK5+MEK5). Similarly, ERK5 activation in cells transfected with 0.3 or 0.03 μg of MEK5α cDNA appeared higher than in experiments where only ERK5 was immunoprecipitated (Fig. 1A). In vitro activation of ERK5 by MEK5 contributes to increasing the levels of MEF2 phosphorylation, although we cannot totally exclude the presence of other protein kinases associated with the immunoprecipitated complexes.
FIG. 1.
Activation of ERK5 by MEK5 isoforms. (A and B) COS-7 cells were cotransfected with expression vectors encoding Flag epitope-tagged ERK5 and MEK5α or MEK5β. The expression of MEK5 and ERK5 in cell lysates and the presence of MEK5 in the complexes immunoprecipitated with an anti-ERK5 antibody were examined by immunoblot analysis (IB) using an anti-Flag antibody (A). ERK5 activity was measured by protein kinase assays (KA) following immunoprecipitation using an anti-ERK5 (IP:ERK5) or an anti-Flag (IP:ERK5+MEK5) antibody (A and B). The radioactivity incorporated into GST-MEF2C was quantitated after SDS-PAGE by phosphorimager analysis and expressed as a percentage of the maximum value. (B). Data of three independent experiments are shown (means ± standard error of the means). (C) The reporter plasmid pG5E1bLuc was transiently cotransfected with a construct encoding the fusion proteins GAL4-MEF2A without (Cont) or with MEK5α or MEK5β. MEF2 transcriptional activity was measured by a dual-luciferase reporter assay system. Values are expressed as ratios of firefly to Renilla luciferase activity. The data correspond to the means ± standard error of the means of three independent experiments (***, P < 0.001).
The ability of MEK5 isoforms to regulate ERK5 in vivo was determined by testing their effect on the transcriptional regulation of MEF2 (Fig. 1C). mek5−/− fibroblasts were cotransfected with MEK5α or MEK5β together with the reporter plasmid pG5E1bLuc and a construct encoding GAL4-MEF2A. A pRL-Tk plasmid encoding Renilla luciferase was employed for monitoring transfection efficiency. The results demonstrated that only MEK5α increased the transcriptional activity of MEF2A as determined by the luciferase reporter assay (Fig. 1C).
To determine whether similar differences could be detected following activation of MEK5, we examined the effect of MEKK2 and MEKK3, two strong activators of the ERK5 signaling pathway (5, 26). COS-7 cells were cotransfected with wild-type MEK5α or MEK5β together with or without MEKK2 or MEKK3 and ERK5. MEK5 and ERK5 activity was measured following immunoprecipitation using recombinant ERK5 and MEF2 as substrates, respectively. At the basal level, MEK5α was more active than MEK5β, but no marked difference was detected in the ability of MEKK2 and MEKK3 to activate either isoform (Fig. 2A). However, the activation of ERK5 by MEKKs mediated via MEK5α was more potent than via MEK5β (Fig. 2B).
FIG. 2.
Effect of MEKK2 and MEKK3 on ERK5 signaling. COS-7 cells were cotransfected with expression vectors encoding Flag-tagged MEK5α or MEK5β and HA-tagged MEKK2 or MEKK3 without (A) or with ERK5 (B). MEK5 and ERK5 activities were measured by protein kinase assays (KA) following immunoprecipitation using an anti-Flag (A) or an anti-ERK5 (B) antibody, respectively. The radioactivity incorporated into GST-MEF2C was quantitated after SDS-PAGE by phosphorimager analysis and expressed as a percentage of the maximum value (B). Data of three independent experiments are shown (means ± standard error of the means).
Together these data provide evidence that MEK5α and MEK5β are catalytically active enzymes and that the binding interaction between ERK5 and MEK5 contributes to mediating ERK5 activation.
MEK5α interacts with ERK5 via its N-terminal domain.
The distinct affinity of MEK5 isoforms for ERK5 was confirmed by affinity chromatography. ERK5 protein produced in COS-7 cells was loaded onto GSH-agarose columns packed with equal amounts of bacterially expressed GST-MEK5α or GST-MEK5β. The elution was performed using increasing concentrations of KCl (0.01 to 1 M). The presence of ERK5 was detected in the eluates by immunoblot analysis. Whereas 10 mM KCl was sufficient to dissociate the ERK5/MEK5β complex, a significant proportion of ERK5 remained bound to MEK5α until the highest 1 M KCl concentration was used (Fig. 3A). Consistently, less ERK5 was detected in the first and the second washes and in the 10 mM KCl fraction of the MEK5α compared to the MEK5β column. These studies clearly demonstrated that MEK5α displayed a higher affinity for ERK5 than MEK5β. To confirm the interaction between MEK5α and ERK5, bacterially expressed His-tagged ERK5 incubated with lysates of fibroblasts was precipitated, and the presence of MEK5 in the precipitates was detected by immunoblot analysis (Fig. 3B). Endogenous MEK5α expressed in wild-type and erk5−/− MEFs coprecipitated with ERK5. A control experiment shows that no protein was detected when lysate of mek5−/− MEFs was used (Fig. 3B).
FIG. 3.
Interaction of ERK5 with MEK5 isoforms. (A) Lysates (500 μg) of COS-7 cells expressing Flag-ERK5 were passed through a GST-MEK5α or a GST-MEK5β affinity chromatography column. Columns were washed twice prior to being eluted with increasing concentrations of KCl. The presence of ERK5 in the fractions was detected by immunoblot analysis using an anti-Flag antibody. (B) MEF extracts (500 μg) were incubated with bacterially expressed His-tagged ERK5 (1 μg). ERK5 was isolated by incubation with Ni-NTA agarose beads. The binding of MEK5 was examined by immunoblot analysis with an antibody to MEK5. (C) Lysate (250 μg) of COS-7 cells expressing HA-tagged ERK5 was incubated with 5 μg of GST, GST-tagged MEK5α, MEK5β, or α-Nter. MEK5 was isolated by incubation with GSH-agarose beads. The binding of ERK5 was examined by immunoblot analysis with an antibody to the HA epitope tag. (D) COS-7 cells were cotransfected with the expression vectors encoding Flag-ERK5 without (Cont) or with GST, MEK5α, or fragments of the N-terminal domain of MEK5α (amino acids 1 to 89, 1 to 69, and 1 to 29) fused to GST. Immunoblot analysis using an anti-Flag and an anti-GST antibody confirmed similar expression of ERK5 and MEK5 in cell lysates, respectively. ERK5 was isolated by incubation with anti-ERK5 antibody. The binding of MEK5 was examined by immunoblot analysis with an antibody to GST.
To determine how the binding affinity of MEK5 isoforms is affected by alternative splicing, we investigated the interaction of ERK5 with the N terminus of MEK5α (amino acids 1 to 89 [α-Nter, where α indicates MEK5α]). ERK5-transfected COS-7 cells were incubated with an equal amount of bacterially expressed GST-MEK5α, GST-MEK5β, or GST-α-Nter. Immunoblot analysis of the complexes pulled down by GSH beads showed that the binding affinity of ERK5 for α-Nter was similar to that for MEK5α and significantly higher than for MEK5β (Fig. 3C). To identify more specifically the region of MEK5α responsible for the binding to ERK5, COS-7 cells were cotransfected with ERK5 together with different N-terminal fragments of MEK5α (Fig. 3D). ERK5 was immunoprecipitated, and the presence of MEK5 in the precipitates was detected by immunoblot analysis. Fragments 1 to 89 and 1 to 69 displayed affinity for ERK5 similar to that of the full-length MEK5α. In contrast, fragment 1 to 29 was unable to bind ERK5 (Fig. 3D). These studies indicate that amino acids 29 to 69 within the PB1 domain of MEK5 (17) are critical for mediating a strong binding interaction between MEK5 and ERK5.
N terminus of MEK5α contains a docking site for ERK5.
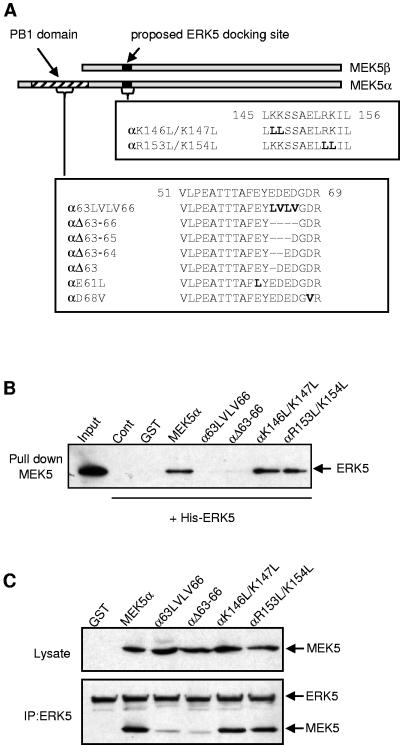
The analysis of the N terminus of MEK5α revealed the presence of four acidic residues, EDED (amino acids 63 to 66), which are highly conserved in PB1 domains found in other proteins (14). To determine the role of these residues in mediating the binding of MEK5α to ERK5, we generated two mutants in which the EDED motif was either deleted (αΔ63-66, MEK5α with a deletion of amino acids 63 to 66) or replaced with aliphatic amino acids (α63LVLV66) (Fig. 4A). Mutants αK146L/K147L and αR153L/K154L were engineered to test whether the putative docking site present in both MEK5 isoforms and identified by sequence homology with other MAPKKs (28) was implicated in mediating the binding to ERK5.
FIG. 4.
Identification of a novel MAPK docking site in the N terminus of MEK5α. (A) Schematic representation of the MEK5α and the MEK5β sequences. The residues mutated in the PB1 domain and in the proposed ERK5 docking site are indicated in bold. The deletion is indicated by dashes. (B) Bacterially expressed His-ERK5 (1 μg) was incubated without (Cont) or with 1 μg of GST or GST-tagged wild-type and mutant MEK5α. MEK5 was isolated by incubation with GSH-agarose beads. The binding of ERK5 was examined by immunoblot analysis with an antibody to ERK5. One-tenth of the His-ERK5 used is shown (input). (C) COS-7 cells were cotransfected with the expression vectors encoding Flag-tagged ERK5 and GST or wild-type or mutant MEK5α fused to GST. ERK5 was isolated by incubation with an anti-Flag antibody. The binding of MEK5 was examined by immunoblot analysis with an antibody to the GST epitope tag. Immunoblot analysis of the lysates and of the immunoprecipitates with an anti-GST and an anti-ERK5 antibody shows similar levels of expression of wild-type and mutant MEK5α and ERK5, respectively.
An in vitro binding assay was performed using bacterially expressed His-tagged ERK5 and GST-fused wild-type and mutant MEK5α. MEK5 was precipitated, and the presence of ERK5 in the complexes was detected by immunoblot analysis (Fig. 4B). This assay revealed that the ability of MEK5α to bind ERK5 was dramatically affected following the deletion or the mutation of the EDED motif, while mutations in the proposed docking site had no effect. To confirm in vivo the role of the EDED motif, lysates of COS-7 cells overexpressing ERK5 and wild-type or mutant MEK5α were obtained. The presence of MEK5 was detected by immunoblotting following the immunoprecipitation of ERK5 (Fig. 4C). A control experiment shows equal amounts of ERK5 in the immune complexes. Consistent with the in vitro data, the αK146L/K147L and the αR153L/K154L mutants, but not the αΔ63-66 or the α63LVLV66 mutants, bind ERK5 as well as the full-length MEK5α.
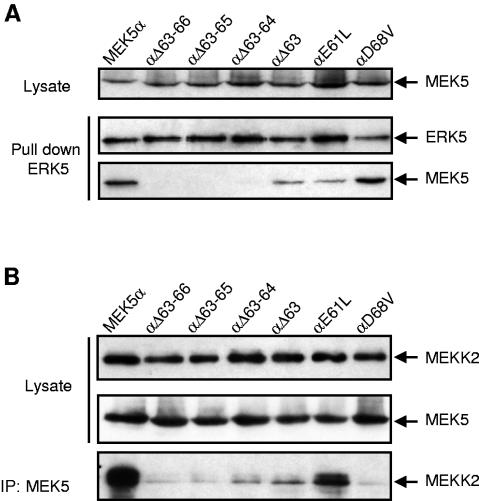
The MEK5/ERK5 interaction was further characterized by engineering more specific mutations in the N terminus of MEK5α (Fig. 4A). Lysates of COS-7 cells overexpressing wild-type or mutant MEK5α were incubated with bacterially expressed His-tagged ERK5. ERK5 was precipitated, and the presence of MEK5 in the complexes was detected by immunoblot analysis (Fig. 5A). The result showed that the deletion of two acid residues in the EDED motif (Δ63-64) was sufficient to completely abolish the binding of ERK5 to MEK5 (Fig. 5A). The αΔ63 mutant in which one acidic residue was deleted displayed a lower affinity for ERK5 compared to the wild-type MEK5α. Outside of the EDED motif, a glutamic acid and an aspartic acid residue at positions 61 and 68 were replaced by a leucine and a valine residue, respectively (Fig. 4A). The E61L mutation significantly decreased the ability of MEK5 to bind ERK5, while the D68V mutation had no effect. This analysis demonstrates that acidic residues within the EDED motif and at position 61 are critical for mediating the MEK5α/ERK5 interaction.
FIG. 5.
Characterization of the ERK5 docking site in the N terminus of MEK5α. (A) Bacterially expressed His-ERK5 (1 μg) was incubated with extracts of COS-7 cells transfected with the expression vectors encoding wild-type or mutant MEK5α fused to GST. ERK5 was isolated by incubation with Ni2+-NTA beads. The binding of MEK5 was examined by immunoblot analysis with an antibody to the GST epitope tag. (B) COS-7 cells were cotransfected with expression vectors encoding wild-type or mutant GST-MEK5α with HA-tagged MEKK2. MEK5 was isolated following precipitation with an anti-GST antibody. The binding of MEKK2 was examined by immunoblot analysis with an antibody to the HA epitope tag. The expression of MEK5 (A and B) and MEKK2 (B) in cell lysates and the presence of ERK5 (A) in the immune complexes were detected by immunoblot analysis using an anti-GST, an anti-HA, and an anti-ERK5 antibody, respectively.
Since the docking site is contained within the PB1 domain, we analyzed the ability of the mutants to bind MEKK2 (Fig. 5B). COS-7 cells were cotransfected with expression vectors encoding wild-type or mutant GST-MEK5α with HA-tagged MEKK2. MEK5 was precipitated, and the presence of MEKK2 in the complexes was detected by immunoblot analysis. Mutations in the EDED motif decreased the ability of MEK5 to interact with MEKK2. Similarly, the αD68V mutant that displayed normal affinity for ERK5 was unable to bind MEKK2. In contrast, the E61L substitution had no effect. These results indicate that the EDED motif and the D68 residue constitute an important sequence for mediating MEKK2/MEK5 interaction. The selective effect of the E61L and the D68V mutations on ERK5 and MEKK2 binding clearly demonstrates that the binding of MEK5 to ERK5 does not rely on the PB1 domain.
The docking site of MEK5 is essential for mediating ERK5 activation.
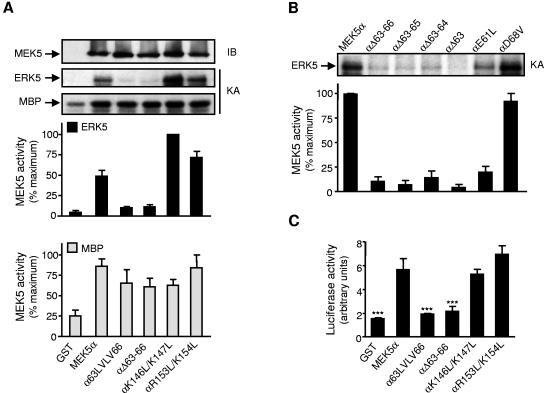
The role of the docking site in regulating MEK5 activity was investigated under basal conditions (Fig. 6). COS-7 cells were cotransfected with wild-type or mutant MEK5α. MEK5 activity was measured following immunoprecipitation using recombinant ERK5 or MBP as substrates (Fig. 6A and B). While the activity of the αK146L/K147L and the αR153L/K154L mutants remained comparable to the wild-type MEK5α, mutations in the EDED motif that affected the binding of MEK5 to ERK5 also prevented MEK5 from phosphorylating ERK5. Consistently, the αE61L mutant displayed a lower level of activity than the αD68V mutant or the wild-type MEK5α (Fig. 6B). A control experiment shows no marked difference in the ability of wild-type and mutant MEK5 to phosphorylate MBP, ruling out a deleterious effect of the mutations on protein kinase activity (Fig. 6A). The expression of MEK5α, αK146L/K147L, or αR153L/K154L in the mek5−/− fibroblasts caused a significant increase in luciferase activity. In contrast, αΔ63-66 and α63LVLV66 were unable to regulate MEF2-induced transcription (Fig. 6C). Together these data demonstrate that the binding of MEK5 to ERK5 via its docking site is required for activating the ERK5 cascade.
FIG. 6.
Mutations in the MAPK docking site affect MEK5 activity. (A and B) COS-7 cells transfected with expression vectors encoding GST or wild-type or mutant MEK5α fused to GST. MEK5 activity was measured by protein kinase assay (KA) following immunoprecipitation with an anti-GST antibody. The radioactivity incorporated into GST-ERK5 or MBP was quantitated after SDS-PAGE by phosphorimager analysis and expressed as a percentage of the maximum value. Data of three independent experiments are shown (means ± standard error of the means). The presence of MEK5 in the immune complexes was detected by immunoblot (IB) analysis using an anti-GST antibody (A). (C) The reporter plasmid pG5E1bLuc was transiently cotransfected together with a construct encoding GAL4-MEF2A with expression vectors encoding GST, wild-type or mutant GST-MEK5α. MEF2 transcriptional activity was measured by a dual-luciferase reporter assay system. Values are expressed as ratios of firefly to Renilla luciferase activity. The data are the mean ± standard error of the means of three independent experiments (***, P < 0.001).
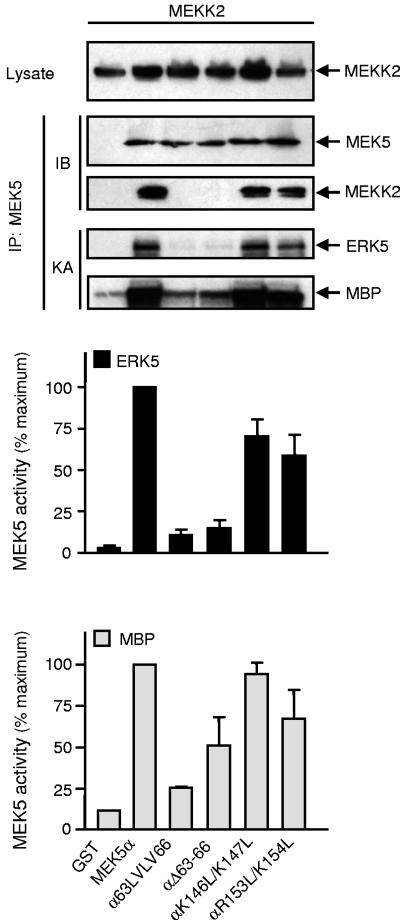
A similar defect was observed in the regulation of MEK5 by MEKK2 (Fig. 7). COS-7 cells were cotransfected with wild-type or mutant MEK5α together with or without MEKK2. Mutations in the docking site that affected the MEKK2/MEK5 interaction also abrogated the ability of MEKK2 to increase MEK5 activity. This is demonstrated using MBP as a substrate to exclude the effect of the mutations on MEK5 binding to ERK5. A similar result was obtained with MEKK3 (data not shown).
FIG. 7.
Effect of mutations in the docking site on the regulation of MEK5 by MEKK2. COS-7 cells cotransfected with expression vectors encoding GST, wild-type or mutant GST-MEK5α, and HA-tagged MEKK2. MEK5 activity was measured by protein kinase assay (KA) following immunoprecipitation with an anti-GST antibody. The radioactivity incorporated into GST-ERK5 or MBP was quantitated after SDS-PAGE by phosphorimager analysis and expressed as a percentage of the maximum value. Data of three independent experiments are shown (means ± standard error of the means). The expression of MEKK2 in cell lysates and the presence of MEK5 and MEKK2 in the immune complexes were detected by immunoblot analysis (IB) using an anti-GST and an anti-HA antibody.
The survival function of MEK5 is dependent on its docking domain.
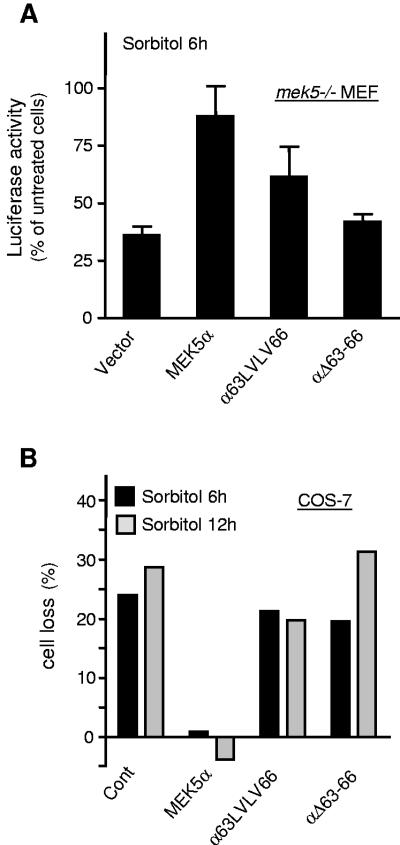
Consistent with our previous data (29), we found that the mek5−/− MEFs were more sensitive than the wild-type cells to the toxic effect of sorbitol (data not shown). Cell viability assays that employ a luciferase plasmid showed that the ectopic expression of MEK5α but not of αΔ63-66 or α63LVLV66 mutants was able to partially restore this defect. mek5−/− MEF transfected with an empty vector or a vector encoding MEK5α mutants defective in ERK5 binding displayed between 40% to 60% decrease in luciferase activity following sorbitol treatment (Fig. 8A). In contrast, no marked decrease was observed in cells overexpressing MEK5α (Fig. 8A). To confirm the requirement of the docking site in mediating the protective effect of MEK5α, we tested the effect of wild-type and mutant MEK5α on the survival of transfected COS-7 cells in response to sorbitol (Fig. 8B). The number of viable COS-7 cells expressing MEK5 was quantified by flow cytometry using a fluorescence-coupled antibody against MEK5. The results showed that cell loss was significantly prevented by wild-type MEK5α but not by the αΔ63-66 or α63LVLV66 mutants. These results demonstrate that, unlike wild-type MEK5α, MEK5α mutants defective in their ability to bind and activate ERK5 are unable to protect cells against stress-induced death.
FIG. 8.
The docking site is essential for the survival function of MEK5α. (A) mek5−/− fibroblasts were cotransfected with an empty vector (vector) or a vector encoding wild-type or mutant GST-MEK5α and luciferase (pCMV-luc) to monitor cell viability. At 36 h after transfection the cells were treated with 500 mM sorbitol for 6 h prior to being lysed, and the luciferase activity was measured. The values are normalized to the protein content. The data expressed as the percentage of treated versus untreated cells correspond to the means ± standard error of the means of three independent experiments. (B) COS-7 cells were transfected with an expression vector encoding wild-type or mutant GST-MEK5α and were treated with 500 mM sorbitol for the indicated times 24 h after transfection. The cells were fixed, stained with an anti-GST antibody coupled to Alexa Fluor 488, and analyzed by flow cytometry. The percentage of viable transfected cells identified by high fluorescence (>100-fold higher than baseline) was calculated. Cell loss represents the normalized values of treated versus untreated cells. The data are representative of two independent experiments.
DISCUSSION
Consistent with the analysis of mutant mice in which the erk5 gene has been deleted (9, 21, 25, 32), we have previously established the role of MEK5 in vivo as an activator of ERK5 and as an essential regulator of cell survival that is required for normal embryonic development (29). Here we demonstrate that the protective effect of MEK5 is dependent on its ability to bind ERK5. A novel MAPK docking site in the N terminus of MEK5 mediates the interaction between MEK5α and ERK5 that is critical for ERK5 activation. As purified recombinant MEK5 and ERK5 form a stable complex, the interaction between the two proteins is direct.
Based on sequence homology with the other MAPK activators, the docking site on MEK5 was predicted to correspond to a cluster of basic residues on positions 146 to 153 present in both MEK5 isoforms (28). Our data show that this was not the case. The substitution of lysine 146 and 147 or arginine 153 and lysine 154 to leucine did not affect the binding of MEK5α or MEK5β with ERK5 (Fig. 4B and C and 6A; data not shown). In addition, no marked differences were observed between the ability of these mutants and the wild-type proteins to activate ERK5 under basal conditions or following activation (Fig. 6A and 7 and data not shown). The region in the MEK5β isoform that contributes to the interaction with ERK5 (Fig. 1A and 3A and C) remains to be identified.
Consistent with its low binding affinity for ERK5, MEK5β that lacks the N terminus extension of MEK5α is a weak ERK5 activator (Fig. 1 and 2). However, when both MEK5 and ERK5 were coimmunoprecipitated, MEK5 isoforms displayed a similar ability to activate ERK5 (Fig. 1). These results lead us to conclude that MEK5β is an active enzyme, thereby emphasizing the functional importance of the interaction between MEK5 and ERK5 to mediating ERK5 signaling. The different activities of MEK5 isoforms are due to their distinct ability to interact with ERK5. Overall, our data do not support the idea that MEK5β is an enzyme-dead variant and a dominant-negative regulator of the ERK5 signaling pathway (4). Further studies are required to elucidate the function of MEK5β in vivo in order to understand the physiological significance of alternative splicing of the mek5 gene.
The higher affinity of MEK5α compared to MEK5β for ERK5 (Fig. 3) suggested that the N-terminal extension of MEK5α was implicated in mediating the strong binding of MEK5 to ERK5. We found that the deletion of at least one acidic residue within the 63-EDED-66 motif or the mutation of a glutamic acid residue at position 61 in the N terminus of MEK5α dramatically decreased the binding interaction between MEK5 and ERK5 as well as the ability of MEK5 to activate ERK5 (Fig. 4, 5, and 6). These residues define a novel MAPK docking motif in the N-terminal domain of MEK5α within its PB1 domain (28). In contrast to the αE61L mutant, the αD68V mutant displayed normal affinity for ERK5 but was unable to interact with MEKK2 (Fig. 5). The identification of specific mutations in MEK5α that selectively affect the binding to ERK5 and MEKK2 clearly demonstrates that the interaction of MEK5α with its upstream and downstream kinases is mediated via distinct motifs. In particular, the formation of the MEK5/ERK5 complex does not rely on the PB1 domain of MEK5α. The region in the N terminus of ERK5 responsible for the binding to MEK5 corresponds to a stretch of 61 amino acids (amino acids 78 to 139) containing 11 basic residues (31). Altogether, these studies suggest that the interaction between ERK5 and MEK5 is triggered by opposite charges in the contact sites of the proteins.
The binding between MEKK2 or MEKK3 and MEK5α is mediated via their respective PB1 domains (17). The substitution of a conserved lysine residue at position 47 to an alanine residue in the N-terminal part of the PB1 domain of MEKK2 renders MEKK2 unable to bind MEK5 (17). In this study, we have identified residues 63 to 66 and 68 as critical amino acids of the PB1 domain of MEK5α (Fig. 5B and 7). Heterodimerization between PB1 domains occurs via the interaction between conserved basic and acidic residues (19). Thus, residues 63 to 66 and 68 of MEK5 may constitute the binding site for lysine 47 of MEKK2. This interaction between MEK5 and MEKK2 is unique among the other MEK/MKK families (MEK1, MKK3, MKK4, MKK6, and MKK7) that have been found to bind their upstream kinases through a DVD site located in their C termini (27).
Mutations that prevent the formation of the MEK5α/MEKK2 complex abolish the activation of MEK5 by MEKK2 (Fig. 7). This observation appears to be inconsistent with the ability of MEKK2 to increase MEK5β activity (Fig. 2A). Indeed, MEK5β that lacks the PB1 domain is unlikely to bind MEKK2. To explain this apparent discrepancy we propose that the N terminus of MEK5α contains an auto-inhibitory domain that masks the phosphorylation sites of MEK5 by upstream kinases. The interaction of MEK5 with MEKK2 affects the overall conformation of MEK5α so that S311 and T315 become accessible for phosphorylation. The recently solved structure of MEK1 (20) together with the identification of a conserved MEKK docking site in the C terminus of MEK1 (27) supports this general idea that the formation of the MEK/MEKK complex is not only required for mediating specificity but is also critical for making MEK receptive to phosphorylation by the bound MEKK.
In conclusion, as both MEKK2 and ERK5 interact with the N-terminal extension of MEK5α, we suggest that MEKK2 and ERK5 compete for binding to MEK5 rather than form a ternary complex. Based on a similar model proposed to explain the organization of the JNK signaling pathway (30), we hypothesize that MEKK2 and MEK5 form a complex that is dissociated upon activation. Activated MEK5 becomes free to interact with its substrate ERK5. Accordingly, the specific transmission of the signal within the ERK5 signaling pathway may not implicate a scaffold protein.
Acknowledgments
We are indebted to Jiing-Dwan Lee, Eric Olson, Jack Dixon, Hung-Ying Kao, Christian Widmann, and Alan Whitmarsh for kindly providing the MEK5D, GST-MEF2, GST-ERK5, GAL4-MEF2A, MEKK2/3, and ERK5 cDNAs, respectively. We thank A. Whitmarsh for critically reviewing the manuscript.
This work was supported in part by the Biotechnology and Biological Sciences Research Council and principally by the Medical Research Council, the Royal Society, and a Lister Institute of Preventive Medicine Research Fellowship to C.T.
REFERENCES
- 1.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonvin, C., A. Guillon, M. X. van Bemmelen, P. Gerwins, G. L. Johnson, and C. Widmann. 2002. Role of the amino-terminal domains of MEKKs in the activation of NF kappa B and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell Signal. 14:123-131. [DOI] [PubMed] [Google Scholar]
- 3.Buschbeck, M., and A. Ulrich. 2005. The unique C-terminal tail of the mitogen-activated protein kinase ERK5 regulates its activation and nuclear shuttling. J. Biol. Chem. 280:2659-2667. [DOI] [PubMed] [Google Scholar]
- 4.Cameron, S. J., J. Abe, S. Malik, W. Che, and J. Yang. 2004. Differential role of MEK5alpha and MEK5beta in BMK1/ERK5 activation. J. Biol. Chem. 279:1506-1512. [DOI] [PubMed] [Google Scholar]
- 5.Chao, T.-H., M. Hayashi, R. I. Tapping, Y. Kato, and J.-D. Lee. 1999. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J. Biol. Chem. 274:36035-36038. [DOI] [PubMed] [Google Scholar]
- 6.Dinev, D., B. W. Jordan, B. Neufeld, J. D. Lee, D. Lindemann, U. R. Rapp, and S. Ludwig. 2001. Extracellular signal-regulated kinase 5 (ERK5) is required for the differentiation of muscle cells. EMBO Rep. 2:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.English, J. M., C. A. Vanderbilt, S. Xu, S. Marcus, and M. H. Cobb. 1995. Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem. 270:28897-28902. [DOI] [PubMed] [Google Scholar]
- 8.Glossop, J. A., T. R. Dafforn, and S. G. Roberts. 2004. A conformational change in TFIIB is required for activator-mediated assembly of the preinitiation complex. Nucleic Acids Res. 32:1829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi, M., S. W. Kim, K. Imanaka-Yoshida, T. Yoshida, E. D. Abel, B. Eliceiri, Y. Yang, R. J. Ulevitch, and J. D. Lee. 2004. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Investig. 113:1138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 11.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, Y., V. V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J. D. Lee. 1997. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, Y., M. Zhao, A. Morikawa, T. Sugiyama, D. Chakravortty, N. Koide, T. Yoshida, R. I. Tapping, Y. Yang, T. Yokochi, and J.-D. Lee. 2000. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275:18534-18540. [DOI] [PubMed] [Google Scholar]
- 14.Lamark, T., M. Perander, H. Outzen, K. Kristiansen, A. Overvatn, E. Michaelsen, G. Bjorkoy, and T. Johansen. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568-34581. [DOI] [PubMed] [Google Scholar]
- 15.Lei, K., A. Nimnual, W. X. Zong, N. J. Kennedy, R. A. Flavell, C. B. Thompson, D. Bar-Sagi, and R. J. Davis. 2002. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol. Cell. Biol. 22:4929-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, L., J. E. Cavanaugh, Y. Wang, H. Sakagami, Z. Mao, and Z. Xia. 2003. ERK5 activation of MEF2-mediated gene expression plays a critical role in BDNF-promoted survival of developing but not mature cortical neurons. Proc. Natl. Acad. Sci. USA 100:8532-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura, K., and G. L. Johnson. 2003. PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J. Biol. Chem. 278:36989-36992. [DOI] [PubMed] [Google Scholar]
- 18.Nicol, R. L., N. Frey, G. Pearson, M. Cobb, J. Richardson, and E. N. Olson. 2001. Activated MEK5 induces serial assembly of sarcomeres and eccentric cardiac hypertrophy. EMBO J. 20:2757-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda, Y., M. Kohjima, T. Izaki, K. Ota, S. Yoshinaga, F. Inagaki, T. Ito, and H. Sumimoto. 2003. Molecular recognition in dimerization between PB1 domains. J. Biol. Chem. 278:43516-43524. [DOI] [PubMed] [Google Scholar]
- 20.Ohren, J. F., H. Chen, A. Pavlovsky, C. Whitehead, E. Zhang, P. Kuffa, C. Yan, P. McConnell, C. Spessard, C. Banotai, W. T. Mueller, A. Delaney, C. Omer, J. Sebolt-Leopold, D. T. Dudley, I. K. Leung, C. Flamme, J. Warmus, M. Kaufman, S. Barrett, H. Tecle, and C. A. Hasemann. 2004. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11:1192-1197. [DOI] [PubMed] [Google Scholar]
- 21.Regan, C. P., W. Li, D. M. Boucher, S. Spatz, M. S. Su, and K. Kuida. 2002. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. USA 99:9248-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez, I., R. T. Hughes, B. J. Mayer, K. Yee, J. R. Woodgett, J. Avruch, J. M. Kyriakis, and L. I. Zon. 1994. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature 372:794-798. [DOI] [PubMed] [Google Scholar]
- 23.Seth, A., F. A. Gonzalez, S. Gupta, D. L. Raden, and R. J. Davis. 1992. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 267:24796-24804. [PubMed] [Google Scholar]
- 24.Shalizi, A., M. Lehtinen, B. Gaudilliere, N. Donovan, J. Han, Y. Konishi, and A. Bonni. 2003. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J. Neurosci. 23:7326-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn, S. J., B. K. Sarvis, D. Cado, and A. Winoto. 2002. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J. Biol. Chem. 277:43344-43351. [DOI] [PubMed] [Google Scholar]
- 26.Sun, W., K. Kesavan, B. C. Schaefer, T. P. Garrington, M. Ware, N. Johnson, E. W. Gelfand, and G. L. Johnson. 2001. MEKK2 associates with the adapter protein Lad/RIBP and regulates the MEK5-BMK1/ERK5 pathway. J. Biol. Chem. 276:5093-5100. [DOI] [PubMed] [Google Scholar]
- 27.Takekawa, M., K. Tatebayashi, and H. Saito. 2005. Conserved docking site is essential for activation of mammalian MAP kinase kinases by specific MAP kinase kinase kinases. Mol. Cell 18:295-306. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue, T., and E. Nishida. 2003. Molecular recognitions in the MAP kinase cascades. Cell Signal. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., A. J. Merritt, J. Seyfried, C. Guo, E. S. Papadakis, K. G. Finegan, M. Kayahara, J. Dixon, R. P. Boot-Handford, E. J. Cartwright, U. Mayer, and C. Tournier. 2005. Targeted deletion of mek5 causes early embryonic death and defects in the extracellular signal-regulated kinase 5/myocyte enhancer factor 2 cell survival pathway. Mol. Cell. Biol. 25:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia, Y., Z. Wu, B. Su, B. Murray, and M. Karin. 1998. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 12:3369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 276:10870-10878. [DOI] [PubMed] [Google Scholar]
- 32.Yan, L., J. Carr, P. R. Ashby, V. Murry-Tait, C. Thompson, and J. S. C. Arthur. 2003. Knockout of ERK5 causes multiple defects in placental and embryonic development. BMC Dev. Biol. 3:11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270:12665-12669. [DOI] [PubMed] [Google Scholar]