Abstract
RNA-binding motif protein 4 (RBM4) has been implicated in the regulation of precursor mRNA splicing. Using differential display analysis, we identified mRNAs that associate with RBM4-containing messenger RNPs in vivo. Among these mRNAs, α-tropomyosin (α-TM) is known to exhibit a muscle cell type-specific splicing pattern. The level of the skeletal muscle-specific α-TM mRNA isoform partially correlated with that of RBM4 in human tissues examined and could be modulated by ectopic overexpression or suppression of RBM4. These results indicated that RBM4 directly influences the expression of the skeletal muscle-specific α-TM isoform. Using minigenes, we demonstrated that RBM4 can activate the selection of skeletal muscle-specific exons, possibly via binding to intronic pyrimidine-rich elements. By contrast, the splicing regulator polypyrimidine tract binding protein (PTB) excluded these exons; moreover, RBM4 antagonized this PTB-mediated exon exclusion likely by competing with PTB for binding to a CU-rich element. This study suggests a possible mechanism underlying the regulated alternative splicing of α-TM by the antagonistic splicing regulators RBM4 and PTB.
Alternative splicing of pre-mRNA is a fundamental mechanism for gene regulation in metazoans (2, 4). This process leads to the production of two or more mRNA isoforms from a single gene which may encode functionally diverse proteins (1). Thus, cell type-specific or developmental-stage-specific protein isoforms can be generated via regulated alternative splicing (1, 2, 4). The misregulation of splicing may result in aberrant mRNA products or an imbalance in isoform levels, which can impair normal cell function (4).
The choice of splice sites is determined by interplay between cis-acting elements within the pre-mRNA and trans-acting regulatory factors (2, 4, 9, 24). The recognition of regulated splice sites can be modulated by adjacent cis elements present within exons or introns (2, 18). Exonic splicing enhancers (ESEs) may promote the use of a nearby weak splice site through the exon definition mechanism (2). Some of the purine-rich positive regulatory elements are recognized by members of the serine/arginine dipeptide-rich protein (SR protein) family (2). Splicing repressors may interfere with the access of the activators to ESEs via binding to adjacent exonic silencing elements (2). Likewise, the binding of repressors to intronic negative regulatory elements near the splice sites or the branch point may exclude essential spliceosomal factors (2). Some negative elements are located both upstream and downstream of the regulated exon; splicing repressors that recognize these elements may form multimers, thereby insulating the exon from activating factors (2, 30). Nevertheless, regulated exons often use intricate combinations of regulatory elements that are recognized by a variety of trans-acting factors, which may act in a synergistic or antagonistic manner to fine-tune the use of particular splice sites (2, 24).
Many lines of evidence indicate that SR proteins bind to ESEs and thereby promote the interactions of spliceosomal components or stabilize their binding to adjacent splice sites (2). Moreover, various splicing regulatory proteins bind to intronic splicing enhancers. For example, CELF family members, such as CUG-BP and ETR-3 proteins, are involved in a number of muscle- or neuron-specific splicing regulation events via CUG- or UG-rich elements (5, 17, 19). Neuron-specific Nova proteins activate the exon inclusion of several pre-mRNAs encoding neurotransmitter receptors by binding to adjacent intronic UCAU repeated sequences (9, 25). Many abundant hnRNP proteins play a role in the repression of regulated splicing (2). The hnRNP A1 protein binds to intronic as well as exonic inhibitory elements and represses splicing through different mechanisms. In the human immunodeficiency virus Tat exon 3, binding of hnRNP A1 to an exonic element can induce nonspecific associations of multiple copies of hnRNP A1 with upstream RNA, thereby interfering with spliceosome assembly (32). While hnRNP A1 binds to intronic sequences on both sides of the regulated exon, its dimerization may result in looping out of the exon (2). Polypyrimidine tract binding protein (PTB/hnRNP I) is involved in the repression of a wide range of tissue-specific exons (2, 30). For example, PTB preferentially binds to pyrimidine-rich intronic silencers present upstream or downstream of the regulated exon. PTB may outcompete U2AF for binding to the polypyrimidine tract or act similarly to hnRNP A1 to loop out the repressed exon (2, 30).
Control of tissue-specific alternative splicing can be achieved by regulated expression of splicing activators in restricted cell types or by counteracting the activity of ubiquitously expressed splicing repressors that act on regulated transcripts in most tissues (2, 9). For example, the muscle-specific Fox-1 protein activates exon skipping of the mitochondrial ATP synthase γ-subunit (F1γ) pre-mRNA and antagonizes the suppressive activity of the ubiquitous PTB (15). On the other hand, PTB can also act as an activator. Notably, alternatively spliced PTB isoforms and tissue-specific PTB paralogs may help modulate alternative splicing in particular cell types (8, 28, 31). Indeed, several tissue-specific regulators, including Fox-1, Nova, and CELF family proteins, can play both positive and negative roles in alternative exon selection (5, 6, 11, 15, 19). Thus, the mechanism by which cell type-specific and general regulators are integrated in regulated alternative splicing remains largely unclear.
Our previous report showed that the RNA binding motif protein 4 (RBM4) modulates alternative 5′ splice site and exon selection of model reporter pre-mRNAs, implying that RBM4 is a splicing regulatory factor (20). RBM4 shuttles rapidly between the nucleus and the cytoplasm, suggesting its additional roles in postsplicing events of mRNA metabolism (20). In this study, we initiated characterizing RBM4 function by searching for its mRNA targets. We identified mRNAs associated with RBM4-containing messenger ribonucleoproteins. RBM4 indeed regulated the alternative splicing of one of its target mRNAs, α-tropomyosin (α-TM), likely via direct binding to intronic elements, and antagonized the effect of PTB on regulation of α-TM pre-mRNA splicing.
MATERIALS AND METHODS
Plasmid construction.
The vector pCDNA-RNPS1-FLAG was previously described (22). The expression vectors for FLAG-tagged ASF and RBM4 were constructed by placing the ASF or RBM4 coding sequence in frame with the FLAG tag into pCDNA3.1 (Invitrogen). The PTB expression vector, pCDNA-FLAG-PTB-1, was a kind gift of D. L. Black (University of California, Los Angeles, CA). The pαTM-8/9a/9b minigene reporter was constructed by sequential insertion of three human α-TM genomic fragments to replace the β-galactosidase gene of pCH110 (Amersham Pharmacia). These α-TM fragments were as follows: 290-bp exon 8-intron 8 (nucleotides [nt] 34144180 to 34144469 of chromosome 15q22.1; accession number NM_000366, NCBI build 35.1), 657-bp intron 8-intron 9a (34145313 to 34145969) and 641-bp intron 9a-intron 9b (34147240 to 34147880). The mutant pαTM-8/9a/9b vectors containing changed nucleotides within intron 9a were made using the QuikChange site-directed mutagenesis system (Stratagene). The α-TM intron 8-intron 9a fragment was inserted into pSV40-p2, a rat β-tropomyosin minigene (3), to replace a fragment containing exons 6 and 7 and their flanking regions; the resulting chimeric minigene was named pβTM-α9a. The pαTM-1/2b/3 (pTS3st) (7) plasmid was a generous gift of C. W. J. Smith (University of Cambridge, Cambridge, United Kingdom).
Isolation of mRNAs associated with the FLAG-tagged RBM4.
HEK 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). The FLAG-RBM4 expression vector was transfected into HEK 293 cells using Lipofectamine 2000 (Invitrogen) for 24 h. Cells were collected and resuspended in radioimmunoprecipitation assay buffer containing 10 mM sodium phosphate (pH 7.2), 150 mM sodium chloride, 2 mM EDTA, and 1% NP-40. FLAG-RBM4 was immunoprecipitated from the cell lysate using anti-FLAG agarose (Sigma), and coprecipitated RNAs were subsequently recovered from the resin. Mock-transfected cells were used as a negative control.
Differential display-PCR.
Differential display-PCR was performed using the RNA imaging kit essentially according to the manufacturer's instructions (GenHunter). In brief, anti-FLAG-coprecipitated RNAs (see above) were reverse transcribed with an A-, C-, or G-anchored 11-mer deoxyribosylthymine (dT) oligonucleotide primer. The resulting cDNAs were subjected to PCR with the same 3′ primer and a 5′ arbitrary primer (AP1 to AP8; GenHunter) in the presence of [α-33P]dATP. The PCR products were resolved on denaturing polyacrylamide gels. For cloning purposes, the fragments representing RBM4-associated candidates were excised from the gels and reamplified with the same set of primers. To verify the differential display-PCR results, FLAG-RBM4-coprecipitated RNAs or FLAG-ASF-coprecipitated RNAs prepared from transiently transfected HEK 293 cells were subjected to reverse transcription (RT)-PCR analysis using gene-specific primers (primer sets 5 and 9 through 14) (Table 1).
TABLE 1.
Characteristics of sets of PCR primersa
| Set no. | Gene product | Accession no. | Forward primer | Reverse primer | PCR product size |
|---|---|---|---|---|---|
| 1 | α-Tropomyosin | NM_000366 | 725-741b | 1009-1024b | 347 bp |
| 2 | 725-741 | 51-69 of exon 9db | 162 bp | ||
| 3 | 725-741 | 17-34 of intron 9ab | 206 bp | ||
| 4 | 820-838b | 1009-1024 | 254 bp | ||
| 5 | 820-838 | 17-34 of intron 9a | 113 bp | ||
| 6 | SV40c | 1009-1024 | |||
| 7 | SV40c | β-TM P4c | |||
| 8 | SV40c | 241-264b | |||
| 9 | Calmodulin 1 | NM_006888 | 77-94 | 393-409 | |
| 10 | Flotillin 1 | NM_005803 | 20-35 | 360-376 | |
| 11 | Caldesmon 1 | NM_033138 | 62-77 | 531-547 | |
| 12 | Rho C | NM_175744 | 25-41 | 432-448 | |
| 13 | RPL27a | NM_000990 | 10-27 | 429-446 | |
| 14 | RIPK2 | NM_003821 | 34-51 | 761-777 | |
| 15 | RBM4 | NM_002896 | 587-601 | 1018-1037 | |
| 16 | GAPDH | NM_002046 | 18-41 | 300-325 |
PCR primers were designed from the sequences of the genes having the NCBI accession number listed. The sequences of α-TM exon 9d and intron 9a were derived from NCBI accession number NT_010194.
Primers used for detection of α-TM were derived from exon 8 (725-741 of NM_000366; AACAATTAAGAATAATG), exon 9a (820-838 of NM_000366; TCAGCTTGTCGGAAAGGAC), exon 9b (1009-1024 of NM_000366; GGTGTAAGCAGGCAGA), exon 9d (51-69 of exon 9d; CTGCGCCACATTCTCTCG), intron 9a (17-34 of intron 9a; CATGCCTTCCTTGCTCCC), and exon 3 (241-264 of NM_000366; see reference 12).
The SV40 and β-TM P4 primers were previously described (20).
Northern blot analysis.
A human multiple-tissue Northern blot (Clontech) was hybridized with a uniformly 32P-labeled DNA probe against the 3′ untranslated region (3′ UTR; nucleotides 1016 to 1560 of NCBI accession number BC000307) of the human RBM4a gene according to the manufacturer's instructions.
Establishment of stable clones and RNA interference.
HEK 293 cell-derived clones that stably expressed FLAG-tagged RBM4 were established essentially according to Lai et al. (21). For RNA interference experiments, the hairpin RNA against human RBM4a (nucleotides 520 to 540 of the protein coding sequence) was inserted into the pShuttle vector (Stratagene). The resulting plasmid was cotransformed with the pAdEASY-1 vector (Stratagene) into Escherichia coli strain BJ5183 to generate the recombinant adenovirus. The virus titer was determined by a plaque assay according to the manufacturer's instructions. To knock down the expression of RBM4a, 1 × 105 HEK 293 cells were incubated with the recombinant virus at a multiplicity of infection (MOI) of 10 to 30 for 2 h. Subsequently, infected cells were washed with phosphate-buffered saline and cultured for 2 days.
RT-PCR.
In general, oligo(dT) primers and SuperScript III (Invitrogen) were used in RT reactions, whereas gene-specific primers (Table 1) and Prozyme (Protec) were used in PCR. To examine tissue-specific expression of α-TM mRNA isoforms, 1 μg of total cellular RNA from various human tissues (Clontech) was subjected to RT-PCR using primer sets 1 through 3 (Table 1). Quantitative RT-PCR was conducted using the SYBR green I RNA amplification kit in a Roche light cycler according to the instructions provided by Roche. Briefly, 40 cycles of cDNA amplification were performed in which primer annealing was at 52°C. The amplification process was monitored in real-time via fluorescence data acquisition at the end of each amplification cycle as recommended (Roche).
In vivo splicing assays.
To analyze the expression of α-TM isoforms in FLAG-RBM4-expressing or anti-RBM4a shRNA-expressing HEK 293 cells, 5 μg of total cellular RNA was subjected to RT-PCR using primer sets 2, 4, and 5 (Table 1). The PCR products were either analyzed by electrophoresis on agarose gels or fractionated on 8% denaturing polyacrylamide gels followed by blotting. The blots were probed with 32P-labeled forward primer using standard methods (20).
To examine the splicing of the reporter minigenes, HEK 293 cells were grown to ∼60% confluence in 6-well plates and transfected with 2 μg of expression plasmid encoding the effector protein (FLAG-tagged RBM4, RNPS1, or PTB) and 1 μg of reporter plasmid (pαTM-8/9a/9b, pβTM-α9a, or pαTM-1/2b/3, respectively). At 48 h after transfection, total RNAs were isolated using the TRIzol reagent (Invitrogen), followed by RT-PCR analysis using primers sets 6 through 8 (Table 1). The resulting products were analyzed as described above. A densitometer coupled with the MetaMorph imaging system (Universal Imaging) was used to quantify the signals for agarose gels and a Typhoon 9410 (Amersham Biosciences) was used for blots. For quantitative analysis, the level of the E8-9a-9b mRNA was normalized to that of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in individual transfectants. For both pβTM-α9a′ and pαTM-1/2b/3 reporters, exon inclusion was measured by determining the ratio of internal exon-containing RNA to total splicing products (exon containing plus exon skipping).
RNA-protein interactions.
The RNA probes were in vitro transcribed using T7 RNA polymerase from a PstI-digested pGEMT vector (Promega) containing a 39-bp DNA insert containing a fragment of α-TM intron 9a; the probe sequence is 5′-gggcgaattgggcccgacgtcgcatgctcccggccgccatggccgcgggattCTCACCACCATGCCTTCCTTGCTCCCTAATCTCaatcactagtgcggccgcctgca (the vector and α-TM sequences are in lowercase and uppercase, respectively; underlined T's were changed to guanine in the CU1 mutant). The probe was uniformly labeled with 32P and gel purified. His-tagged mouse RBM4 (mRBM4) was overexpressed in E. coli strain BLR and purified essentially according to Lai et al. (20). E. coli lysate without recombinant protein was passed through nickel agarose, and eluates were used as control. For UV cross-linking, 1.2 pmol of RNA probe (1 × 106 cpm) was incubated with 1 μg (24 pmol) of purified mRBM4 or 10 μl cell lysate in a 20-μl mixture under in vitro splicing conditions (20) at 30°C for 20 min. The samples were irradiated on ice under 254-nm light for 10 min. The probes were digested with 1 μg of RNase A (Sigma) at 30°C for 20 min.
RESULTS
Identification of mRNAs associated with RBM4.
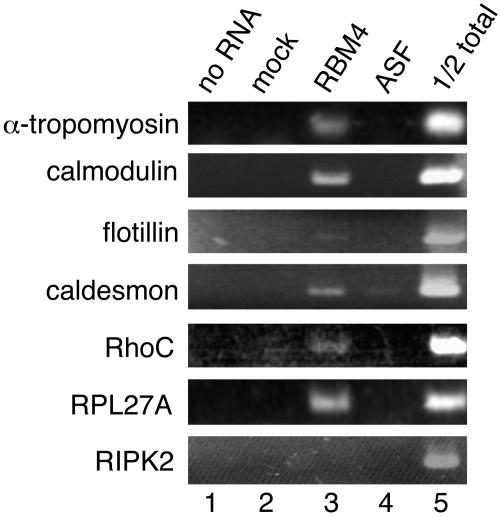
To explore the cellular function of RBM4, we set out to identify mRNAs that associate with RBM4-containing RNP complexes. Initial attempts to search for the candidate mRNA targets for RBM4 were performed through immunoprecipitation using polyclonal antibodies against RBM4. However, this approach did not reproducibly yield specific cDNA fragments in anti-RBM4 precipitates (data not shown). We reasoned that this was probably due to inefficient immunoprecipitation of RBM4 RNPs by anti-RBM4. Therefore, we transiently transfected a vector expressing FLAG-tagged RBM4 into HEK 293 cells. Like endogenous RBM4, overexpressed FLAG-RBM4 directly bound to poly(A)+ RNA and formed RNP complexes (data not shown). Monoclonal anti-FLAG immunoprecipitated FLAG-RBM4 from total cell lysates with higher efficiencies than anti-RBM4 did. Coprecipitated RNA samples from mock and RBM4-expressing transfectants were subjected to differential display-PCR analysis. PCR-amplified cDNA fragments were generated from 24 pairs of primers and labeled with 3P (see Materials and Methods). Each set of RBM4 and mock PCR products was resolved on denaturing gels in parallel for comparison. Approximately 20 bands were reproducibly observed with FLAG-RBM4 precipitates from several independent experiments, but these bands were not detected in mock precipitates (data not shown). Thirteen cDNA fragments were subsequently reamplified for cloning; among them, 10 showed sequence homology to known genes. As detected by using specific primers, six mRNA species indeed associated with transiently expressed FLAG-RBM4 but much more weakly, or not at all, with FLAG-ASF, and one (RIPK2) appeared to be a false-positive signal (Fig. 1). Interestingly, several of the RBM4 target cDNAs encode cytoskeletal proteins or exhibit muscle-specific expression patterns (Table 2), which is consistent with its higher expression in muscles and heart (Fig. 2A). One of the cDNAs, encoding α-TM, undergoes regulated alternative splicing in different muscle types (27). We thus investigated whether RBM4 regulates splicing of its candidate target mRNAs using α-TM as a model.
FIG. 1.
mRNAs associated with RBM4-containing messenger RNPs. FLAG-tagged RBM4 was transiently expressed in HEK 293 cells and then immunoprecipitated from cell lysates using anti-FLAG antibody. RNAs recovered from the immunoprecipitates were subjected to differential display-PCR. To verify the association of candidate mRNAs with RBM4, HEK 293 cells were transfected with an expression vector encoding FLAG-RBM4 (lane 3) or FLAG-ASF (lane 4) or with an empty vector (lane 2, mock). Anti-FLAG immunoprecipitation was performed with the lysates, followed by RT-PCR analysis using gene-specific primers (primer sets 5 for α-TM and 9 through 14 for others) (Table 1). Lane 1 shows RT-PCR without RNA templates, and the sample in lane 5 contained total RNA equivalent to one-half of the amount of the mock-lysate input. RIPK2 was a false positive signal.
TABLE 2.
Characteristics of candidate mRNA targets of RBM4
| mRNA target | Function of encoded protein | PCR-amplified regiona | CU-rich motifb |
|---|---|---|---|
| α-Tropomyosin | Contractile system of muscles and the cytoskeleton of nonmuscle cells | 972-1040 | GCCUUCCUUG (intron 9a)c |
| GCCUUUCCUG (intron 2a)d | |||
| Calmodulin 1 | Calcium-modulated protein binding to caldesmon in muscle and nonmuscle cells | 178-405 | GCUCCCUCUG (5′ UTR)c |
| GCCUUCCUUG (5′ UTR)c | |||
| Flotillin 1 | Caveolae-associated, integral membrane protein | 1638-1745 | GCUCCCCUUG (3′ UTR)c |
| Caldesmon 1 | Binding to calmodulin, actin, tropomyosin, and myosin; regulation of smooth muscle and nonmuscle contraction | 3051-3124 | GCUCCUCCUG (3′ UTR)e |
| Rho C | Small GTPase | 971-1086 | GCCUUUCCUA (3′ UTR)e |
| RPL27a | Ribosomal protein L27a | 450-508 |
PCR-amplified fragments correspond to the region of the genes with an accession number as in Table 1.
The CU-rich elements listed are composed of eight consecutive C/U residues anchored by one G (or A) at each end; the importance of the guanine residues was not characterized in this study.
The CU-rich sequence present in the cDNA fragments obtained from differential display-PCR of the candidate mRNAs.
The CU-rich sequence present in the cDNA fragments found in the intron of the candidate mRNA.
The CU-rich sequence present in the cDNA fragments found in the 3′ UTR of the candidate mRNAs.
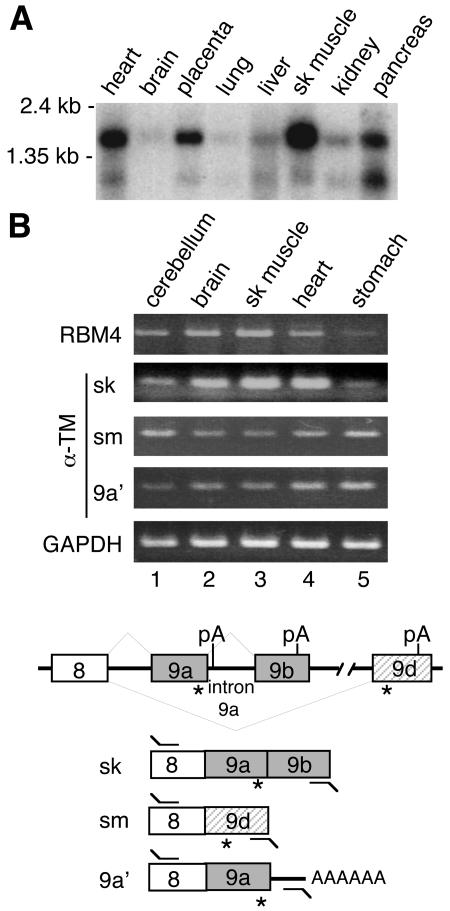
FIG. 2.
Expression profiles for RBM4 and α-TM mRNA isoforms across selected human tissues. (A) Northern blot analysis of human RBM4. (B) Total RNAs from different human tissues were subjected to RT-PCR analysis using primers specific to RBM4 (primer set 15) (Table 1), three α-TM isoforms (primer set 1 for sk, 2 for sm, and 3 for 9a′) and GAPDH (primer set 16). The lower panel depicts gene structure of human α-TM and its splice isoforms and primers used in RT-PCR. Closed and hatched boxes represent sk- and sm-specific exons, respectively. *, translation stop codon; pA, polyadenylation signal.
The majority of the cDNA fragments identified, including that encoding α-TM, contained the 3′-terminal sequence of the target genes. This was probably because oligo(dT) primers were used for reverse transcription and amplification. The α-TM cDNA fragment we obtained possibly represented an isoform, herein termed α-TM 9a′, generated using the polyadenylation signal immediately downstream of the 5′ splice site of intron 9a (the intron between exons 9a and 9b) (Fig. 2B). We found that FLAG-RBM4 bound only to the α-TM 9a′ mRNA, not to any of the muscle cell-specific isoforms (data now shown). A similar result was observed with endogenous RBM4 through immunoprecipitation using anti-RBM4 in HEK 293 cell lysates. This intriguing observation is probably a consequence of the binding of RBM4 to intron 9a during the splicing of α-TM pre-mRNA (see below).
An α-TM isoform from Xenopus laevis analogous to 9a′ is predominately expressed in embryonic somites (14). The 9a′ mRNA may encode a protein identical to the skeletal muscle-specific isoform, and its expression was detected evenly in a variety of human cell lines (data not shown) and tissues, including skeletal muscle (Fig. 2B). However, the physiological function of α-TM 9a′ in mammalian species remains to be determined.
Expression of RBM4 correlates with the level of the skeletal muscle isoform of α-TM.
The skeletal (sk) and smooth (sm) muscle isoforms of α-TM differ by selective inclusion of exon 2 and terminal exon 9 (27). We therefore investigated whether RBM4 expression correlates with any of the cell type-specific isoforms. RT-PCR was performed with total RNA from human tissues using a specific 3′ primer. Figure 2B shows that the sk form of α-TM was highly expressed in skeletal muscle where RBM4 was abundant, and its levels were concurrently lowered in stomach (lanes 3 and 5). However, inverse correlations were observed between the levels of the α-TM sm mRNA and RBM4 in these tissues. Therefore, among tissues examined, the yield of the sk muscle-specific α-TM mRNA, at least in part, correlated with RBM4 abundance.
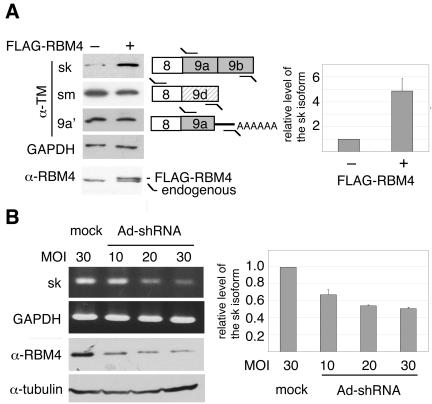
Next, we examined whether RBM4 directly influences the expression of the sk-specific α-TM isoform. We first established a stable HEK 293 clone that ectopically expressed FLAG-tagged RBM4 (Fig. 3A). In this cell line, the level of the α-TM sk isoform was elevated by approximately fivefold, as estimated by quantitative RT-PCR (Fig. 3A, right panel). In contrast, inclusion of the sm-specific exon 9d was slightly suppressed in FLAG-RBM4-expressing cells (a decrease of the sm form by ∼40%) (Fig. 3A). The yield of α-TM 9a′ was apparently not altered by RBM4 overexpression (Fig. 3A). This result suggested that inclusion of exons 9a and 9b as a cell type-specific α-TM isoform can be mediated directly or indirectly by RBM4.
FIG. 3.
Expression levels of RBM4 correlate with the inclusion of skeletal muscle-specific terminal exons of α-TM. (A) Total RNAs from HEK 293 cells that stably expressed FLAG-tagged RBM4 (+) or from mock cells (-) were analyzed by RT-PCR using primers (Table 1) specific to three α-TM isoforms (primer set 4 for sk, 2 for sm, and 5 for 9a′) and GAPDH (primer set 16). The PCR products were subjected to hybridization using a 32P-labeled forward primer as probe. Western blotting was performed using anti-RBM4. The right panel shows quantitative RT-PCR analysis of the α-TM sk isoform from three independent experiments. The level of the sk mRNA was normalized to that of GAPDH in individual experiments. The bar scale with standard deviation shows activation (n-fold) of sk isoform expression upon ectopic expression of RBM4. (B) HEK 293 cells were infected with recombinant adenovirus expressing an shRNA targeting RBM4a or with mock adenovirus at the MOIs indicated above the gel panels. RT-PCR and Western blotting were performed as in panel A. Quantitative RT-PCR shows relative levels of the α-TM sk isoform in individual infections with the shRNA-expressing adenovirus compared with the mock infection; average values with standard deviations were obtained from three independent experiments.
To test whether RBM4 is crucial for cell type-specific expression of the α-TM isoforms, we used RNA interference to suppress RBM4 expression. RBM4 proteins are encoded by two genes, the RBM4a and RBM4b genes (20). We tested several small hairpin RNAs (shRNAs) that targeted either the RBM4 gene or both genes, and only one shRNA against RBM4a effectively suppressed the level of RBM4 proteins (data not shown). Therefore, HEK 293 cells were infected with recombinant adenovirus expressing this RBM4a shRNA. Western blot analysis showed an ∼65% reduction of RBM4 protein (α-RBM4 at an MOI of 30) (Fig. 3B). From these results, it was unclear whether RBM4a was completely depleted and thereby the majority of the residual RBM4 was probably RBM4b. Nevertheless, the level of the sk-specific α-TM isoform was reduced by ∼50% at higher MOI values of the RBM4a shRNA-expressing adenovirus (Fig. 3B, right panel). Suppression of RBM4 expression, however, did not significantly affect the level of the sm-specific α-TM mRNA (data not shown).
Together, the above results indicated that alteration of RBM4 levels in cells influences the expression of the sk isoform of α-TM mRNA.
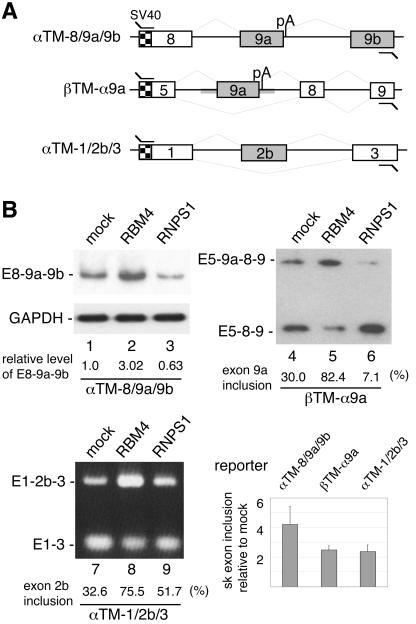
RBM4 promotes the inclusion of alternative exons in α-TM minigenes.
To examine whether RBM4 directly activates the inclusion of the sk muscle-specific α-TM exons, we constructed a minigene vector, pαTM-8/9a/9b, containing an α-TM genomic segment from exon 8 to exon 9b, with internally truncated introns (Fig. 4A). The pαTM-8/9a/9b plasmid was transiently cotransfected with effector-expressing vector into HEK 293 cells, followed by RT-PCR analysis of the splicing products. Using a 3′ primer against exon 9b, we observed an approximately fourfold (in average) increase of the fully spliced E8-9a-9b RNA analogous to the sk muscle isoform upon overexpression of FLAG-RBM4 (Fig. 4B, lane 2, bar scale). We also attempted to detect mRNA species generated by use of the alternative polyadenylation signal immediately downstream of the 5′ splice site of intron 9a. However, a fragment was detected which is similar to α-TM 9a′ but contains the unspliced intron between exons 8 and 9a (data not shown). Nevertheless, the level of this mRNA did not change when RBM4 was overexpressed (data not shown).
FIG. 4.
RBM4 promotes inclusion of the skeletal muscle specific exons of α-TM. (A) The diagram illustrates three minigene constructs. Checkered and closed boxes represent the SV40 promoter and regulated exons, respectively. Gray thick lines indicate the α-TM segment that was placed in the chimeric pβTM-α9a reporter. (B) Each minigene was cotransfected into HEK 293 cells along with an empty expression vector (lanes 1, 4, and 7) or vector that expressed FLAG-RBM4 (lanes 2, 5, and 8) or FLAG-RNPS1 (lanes 3, 6, and 9). The splicing products were analyzed by RT-PCR using primer sets as shown in Table 1 (primer set 6 for pαTM-8/9a/9b, 7 for pβTM-α9a, and 8 for pαTM-1/2b/3). For pαTM-8/9a/9b and pβTM-α9a minigenes, the PCR products were further subjected to blotting and hybridization using a 32P-labeled SV40 primer as probe. Below the gels are relative yields of E8-9a-9b RNA (normalized to that of GAPDH; lanes 1 through 3) and percentages of internal exon inclusion (lanes 4 through 9). The bar graph shows relative sk exon inclusion (n-fold) induced upon overexpression of FLAG-RBM4; average values with standard deviations were obtained from three independent experiments (see Materials and Methods).
Alternative selection of terminal exons occurs coordinately with differential transcription termination and polyadenylation (26). Therefore, we examined the possibility that RBM4 could prevent premature termination or modulate the use of alternative polyadenylation sites, thereby promoting sk exon inclusion. The chimeric construct pβTM-α9a was created from a rat β-TM minigene (3), in which the two internal tissue-specific β-TM exons were replaced by α-TM exon 9a along with its flanking introns. Using a 3′ primer derived from the β-TM terminal exon to detect the splicing products, we found that coexpression of FLAG-RBM4 significantly enhanced the use of α-TM exon 9a (Fig. 4B, lane 5). This result suggested that RBM4 acted through the activation of an internal exon during splicing rather than antitermination at the transcriptional level and that the α-TM exon 9a or its adjacent intronic sequence contains the cis elements sufficient for exon 9a inclusion even in a different gene context. Notably, our previous data showed that RBM4 induces the skipping of internal exons in the rat β-TM pre-mRNA (20). Thus, RBM4 may act as an activator as well as a repressor, but the factors that specify these RBM4 functions remain to be investigated.
Cell type-specific α-TM expression also involves mutually exclusive selection of exons 2a and 2b (27). To test whether RBM4 could activate inclusion of the sk muscle-specific exon 2b, the pαTM1/2b/3 (previously termed pTS3st) (7) reporter was cotransfected with the FLAG-RBM4-expressing vector into HEK 293 cells. RT-PCR analysis showed that RBM4 also enhanced inclusion of exon 2b (Fig. 4B, lane 8), thus emphasizing a role for RBM4 in the expression of the sk muscle-specific α-TM mRNA.
To evaluate the specificity of RBM4, we examined the effect of SR protein RNPS1 on the splicing of the above three minigenes. As shown in Fig. 4B, overexpression of FLAG-tagged RNPS1 suppressed the inclusion of α-TM exon 9a, but slightly enhanced exon 2b utilization (lanes 3, 6, and 9). Therefore, RNPS1 differentially affected exon selection of α-TM, reminiscent of its various activities in splicing (29). By contrast with RNPS1, RBM4 played a significant role in promoting sk-specific exon utilization.
RBM4 as well as PTB regulates exon selection via intronic CU-rich elements.
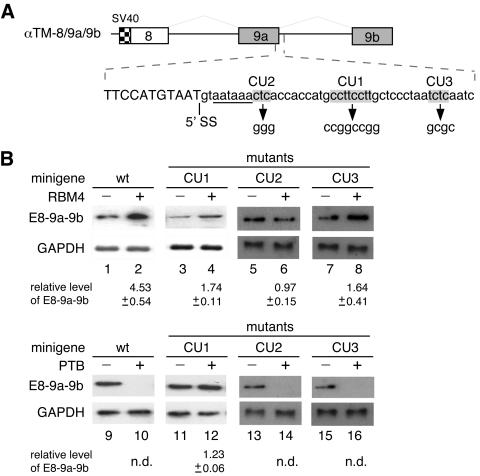
The immunoprecipitation experiments showed that RBM4 was steadily associated with the α-TM 9a′ mRNA (Fig. 1) but not with any cell type-specific α-TM isoforms (data not shown). A simple explanation for this intriguing observation is that RBM4 probably binds to a sequence within intron 9a which is spliced out of the muscle-specific mRNAs. We noted a set of CU-rich sequences present in at least three cDNA fragments derived from RBM4 target candidates, including α-TM; these CU-rich motifs are characteristic of eight consecutive pyrimidine residues flanked by a guanine at each end (Table 2). Therefore, we first investigated the functional role of such a CU-rich element (termed CU1) within intron 9a in α-TM alternative splicing. Point mutations were introduced in CU1 of the α-TM minigene to disrupt its pyrimidine-rich sequence (Fig. 5A). Transient transfection showed that the CU1 mutant was less responsive to coexpression of FLAG-RBM4 (Fig. 5B, lanes 3 and 4), thus suggesting a partial role of the CU1 element in RBM4-mediated α-TM splicing regulation.
FIG. 5.
RBM4 and PTB exert opposite effects on utilization of α-TM exon 9a via an intronic CU-rich element. (A) The diagram shows the sequence around the splice junction (5′ SS) of α-TM exon 9a. The mutant reporters contain pyrimidine-to-guanine changes in each CU-rich element (shown in gray). The polyadenylation signal is underlined. (B) The wild-type and mutant pTM-8/9a/9b minigenes were each cotransfected into HEK 293 cells along with an empty vector or with expression vector encoding FLAG-RBM4 (lanes 1 through 8) or FLAG-PTB (lanes 9 through 16). As in Fig. 4, the splicing products were examined by RT-PCR followed by blotting analysis. GAPDH was used as a normalization control. Relative levels of E8-9a-9b RNA are indicated below the gels. n.d., not detectable.
The splicing factor PTB has a characteristic preference for binding CU-rich sequences and is implicated in the repression of various tissue-specific exons (2, 9, 30). Therefore, we tested whether PTB could also regulate the selection of the α-TM sk muscle terminal exons. Figure 5B shows that overexpression of FLAG-tagged PTB drastically reduced the yield of the E8-9a-9b mRNA produced from the wild-type construct (lane 10). However, the CU1 mutation completely relieved the suppressive effect of exogenous PTB, leading to expression of the E8-9a-9b mRNA at levels comparable to that of mock-transfected cells (lanes 11 and 12). Thus, the effect of the CU1 mutation on PTB appeared to be greater than that on RBM4. Moreover, the CU1 element within intron 9a acted not only as an enhancer but also a silencer; its role is possibly determined upon the binding of trans-acting splicing regulatory factors.
To examine whether any other CU-rich sequences also contribute to exon 9a utilization, we mutated two abbreviated CU elements (CU2 and CU3) within intron 9a (Fig. 5A). Figure 5B shows that the CU2 mutation severely affected RBM4-induced exon 9a inclusion, and the CU3 mutation also had a partial effect (lanes 5 through 8). This result was apparently intriguing but indicated that RBM4 regulates sk exon utilization probably through multiple CU-rich elements within intron 9a. In contrast, neither CU2 nor CU3 mutation had any effect on PTB-mediated exon 9a suppression (lanes 13 through 16). Therefore, the CU1 element acted as a major PTB responsive element for splicing regulation.
RBM4 antagonizes the function of PTB in alternative exon selection.
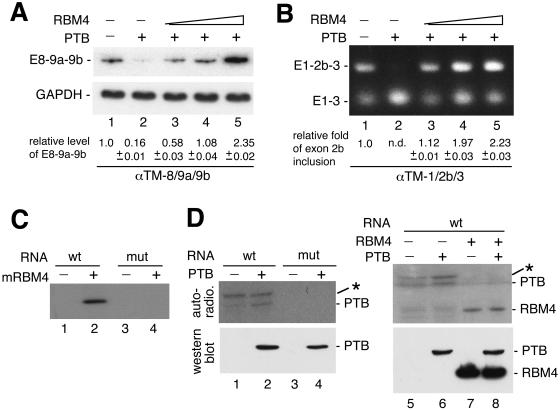
Since RBM4 and PTB modulated utilization of α-TM sk exons in opposite manners, we therefore examined whether RBM4 could alleviate the inhibitory effect of PTB. As shown above, overexpressed FLAG-tagged PTB greatly reduced the production of the E8-9a-9b mRNA (Fig. 6A, lane 2). Cotransfection of the FLAG-RBM4 vector reversed PTB-mediated suppression of terminal exon utilization in a dose-dependent manner (lanes 3 through 5). This result suggested that PTB activity is directly antagonized by RBM4. We also examined the antagonistic action of PTB and RBM4 in the utilization of α-TM exon 2b. Overexpression of FLAG-tagged PTB indeed suppressed exon 2b inclusion in the pre-mRNA produced from pαTM-1/2b/3 (lane 2), as reported previously (12, 31). Coexpression of FLAG-RBM4 counteracted the regulatory effect of PTB on exon 2b splicing, resulting in elevated levels of the exon 2b-included product (Fig. 6B, lanes 3 through 5). These data indicated that RBM4 was able to reactivate PTB-mediated splicing suppression of the sk muscle-specific exons in the α-TM pre-mRNA.
FIG. 6.
RBM4 acts antagonistically to PTB in alternative α-TM pre-mRNA splicing. The pαTM-8/9a/9b (A) or pαTM-1/2b/3 (B) minigene was cotransfected into HEK 293 cells along with the expression vector encoding FLAG-tagged PTB alone (lanes 2) or with increasing amounts of the vector encoding FLAG-RBM4 (lanes 3 through 5). Lanes 1 show cotransfection of the reporter with the empty effector vectors. Splicing of the α-TM transcripts was assayed by RT-PCR as in Fig. 4. GAPDH was used as a control for pαTM-8/9a/9b. (C) Recombinant mouse RBM4 (lanes 2 and 4) or mock eluate (lanes 1 and 3) was incubated with 32P-labeled α-TM intron 9a RNA probe (wild type or CU1 mutant) followed by UV cross-linking. The reactions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by autoradiography. The RNA probe contained a fragment corresponding to nucleotides 9 through 34 of α-TM intron 9a (Materials and Methods). (D) FLAG-PTB-containing cell lysate was prepared from transiently transfected HEK 293 cells and subsequently incubated with wild-type (lanes 1 and 2) or mutant (lanes 3 and 4) 32P-labeled α-TM intron 9a probe. For competition, the experiment was performed with the wild-type RNA probe using cell lysate prepared from mock-transfected HEK 293 cells (lane 5) or cells that transiently expressed either FLAG-PTB (lane 6) or FLAG-RBM4 (lane 7) or both proteins (lane 8). UV cross-linking and analysis were performed as in panel C. Western blotting using anti-FLAG is shown in the lower panel. The asterisk represents an unidentified protein of ∼70 kDa.
Next, we tested whether RBM4 reversed the inhibitory effect of PTB in α-TM sk exon inclusion by directly competing for binding to some cis elements. We first examined the binding of recombinant RBM4 to a CU-rich element-containing intron 9a fragment by using UV cross-linking. In this study, mouse RBM4 (mRBM4) was used because it showed higher expression levels and stability in E. coli than its human homolog. The mouse and human proteins share an identical sequence in RNA recognition motifs suggesting the same or similar RNA binding properties. The probe RNA of ∼110 nt containing a 39-nt intron 9a segment (see Materials and Methods) was radioisotope labeled. The mutation of CU1 was examined since it might be a common site involving RBM4 and PTB-mediated splicing regulation. UV cross-linking showed that His-tagged mRBM4 interacted with the wild-type probe but not the mutant probe (Fig. 5C). Gel mobility shift assay confirmed this result and, moreover, showed that RBM4 bound the intron 9a probe in a dose-dependent manner (data not shown). Thus, RBM4 promoted the inclusion of the sk muscle-specific terminal exons in α-TM mRNA likely via direct binding to at least one intronic CU-rich element, i.e., CU1. Although the CU1 element may be critical for RBM4 binding, the observation that its mutation only partially disrupted the activity of RBM4 in exon 9a inclusion was intriguing. We thus speculated that recognition of CU1 by RBM4 in vivo is assisted by additional cis elements or by other splicing factors.
We assumed that RBM4 binding to the CU1 element may disturb PTB function in α-TM splicing regulation. To test this possibility, we incubated a radiolabeled intron 9a RNA probe with FLAG-PTB-containing cell lysate or mock lysate and performed UV cross-linking. The signal of an ∼60 kDa-band was barely detected in the mock lysate but was enhanced when FLAG-PTB was overexpressed (Fig. 6D, autoradiogram, lanes 1 and 2). Evidence suggesting that this band is probably PTB also came from the observation that it migrated closely with the anti-FLAG-positive signal (Fig. 6D, Western blot) and with a doublet of PTB and its isoforms (31) detected by anti-PTB (data not shown). In addition, an ∼70-kDa protein of unknown identity was cross-linked to the intron 9a RNA, even in the mock lysate (lanes 1 and 2). Whether this ∼70-kDa band has a higher affinity for the α-TM intron 9a than the PTB candidate is uncertain. Nevertheless, neither band could be detected with the mutant RNA (lanes 3 and 4), and their binding to wild-type intron 9a was completely eliminated by coexpression of FLAG-RBM4 in cells (lanes 7 and 8) or by the addition of excess recombinant RBM4 in vitro (data not shown). Thus, our data suggested a possible interaction of PTB with the α-TM CU1 element, which contributes to the repression of sk-specific exons. Moreover, reversal of the inhibitory effect of PTB by RBM4 is likely due to competition between these two proteins for binding to the same cis element.
DISCUSSION
We identified several mRNAs, including α-TM, as RBM4 targets by using immunoprecipitation coupled with differential display-PCR analysis. These specific associations suggest a role for RBM4 in the posttranscriptional processing or regulation of these mRNAs. Our data show that selection of the sk muscle-specific exons in α-TM was indeed modulated by levels of RBM4 in cells. Moreover, using minigenes, we demonstrated that RBM4 and PTB were antagonistic with regard to utilization of the sk muscle-specific α-TM exons, probably through competition for intronic CU-rich elements. This study thus provides insight into the regulation of α-TM exon selection by the antagonistic action of two RNA binding proteins.
Are RBM4 target mRNAs regulated in a tissue-specific or coordinated manner?
It has been postulated that RNA binding proteins participate in posttranscriptional regulation of their associated mRNAs via binding to cis elements (16). Such elements may be common in a set of mRNAs that are functionally coherent, thus leading to the coordinated regulation of these mRNAs. Interestingly, several mRNAs identified in the RBM4 immunoprecipitates encode cytoskeletal or muscle-specific proteins, and some of them contain CU-rich elements (Table 2). At present, in addition to α-TM, it is not yet known whether RBM4 functionally targets any of the other mRNAs we identified. Our data show that RBM4 expression levels correlated with the sk isoform of α-TM in tissues (Fig. 2) and that RBM4 regulated α-TM pre-mRNA splicing via intronic CU-rich elements (Fig. 5). Therefore, RBM4 may possibly participate in the metabolism of some CU element-containing mRNAs in a tissue-specific manner.
Tropomyosin associates with actin filaments to form fibrous cytoskeletons (27). Tropomyosin-actin interactions are modulated in a calcium-sensitive manner by calmodulin/caldesmon or troponin complexes in different muscle cell types (27). Both α-TM and cardiac troponin T pre-mRNA undergo cell type-specific alternative splicing which involves some intronic CU-rich regulatory elements (18). We observed here that, in addition to α-TM, mRNAs encoding calmodulin and caldesmon are also potential targets of RBM4 and contain CU1-like elements (Table 2). It will be interesting to investigate whether these cytoskeletal protein-coding mRNAs as well as others that are not identified in this study, such as cardiac troponin T, are subjected to coordinated posttranscriptional regulation involving RBM4. Since RBM4 shuttles between the nucleus and the cytoplasm (20), such regulation may occur in either or both cellular compartments and at different steps of mRNA maturation.
Competition between splicing and polyadenylation.
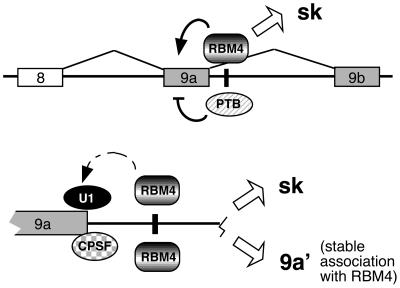
Immunoprecipitation of RBM4 followed by RT-PCR analysis revealed an α-TM cDNA fragment corresponding to a less-characterized mRNA isoform, 9a′. Association of RBM4 with the α-TM 9a′ mRNA is likely due to its direct binding to intron 9a (Fig. 6). Binding of RBM4 to intron 9a also explains the observation that neither of the completely spliced isoforms coimmunoprecipitated with RBM4. However, it appears that RBM4 binds but does not promote splicing under some circumstances, thus yielding the 9a′ mRNA. We note that the 5′ splice site sequence of intron 9a overlaps with a polyadenylation signal (Fig. 5) that is activated in the 9a′ mRNA. Thus, competition between splicing and polyadenylation at the 5′ splice/polyadenylation sites of intron 9a may modulate the relative levels of α-TM isoforms (Fig. 7). To generate the sk form, RBM4 may facilitate or stabilize U1 snRNP binding to this composite, yet weak, 5′ splice site, thereby overriding polyadenylation (Fig. 7). Meanwhile, RBM4 may activate splicing across intron 9a to include exon 9b in the fully spliced sk isoform. Nevertheless, the level of the 9a′ mRNA remained unchanged regardless of RBM4 overexpression (Fig. 3) or down-regulation (data not shown), suggesting that RBM4 does not influence polyadenylation per se.
FIG. 7.
Schematic representation of possible mechanisms involved in regulation of α-TM terminal exon utilization. The upper scheme shows that RBM4 and PTB act oppositely on exon 9a utilization of α-TM. Overexpression of RBM4 reverses the suppressive activity of PTB in exon 9a inclusion via the CU1 element (black vertical line), thus yielding the sk isoform. The lower scheme shows a possible competition between splicing and polyadenylation. We propose that the U1 snRNP and cleavage and polyadenylation specificity factor (CPSF) compete for the overlapping 5′ splice/polyadenylation site. Enhanced expression of the sk isoform by RBM4 suggests that RBM4 facilitates the interaction of the U1 snRNP with the 5′ splice site of intron 9a, thus precluding CPSF from binding to the overlapping polyadenylation signal. Expression of the 9a′ mRNA is not modulated by RBM4; however, its association with RBM4 suggests that RBM4 binds to intron 9a but is silent in splicing activation under some circumstances.
A Xenopus α-TM isoform equivalent to the 9a′ mRNA was previously identified in embryonic somites (14); however, its biological function is not yet clear. It is interesting to note that the 9a′ mRNA differs from the authentic sk isoform in terms of length and sequence of the 3′ UTR and in its stable association with RBM4. Therefore, the α-TM isoforms may be differentially regulated at the posttranscriptional steps. It is also reasonable to hypothesize that RBM4 is involved in postsplicing mRNA maturation events of the 9a′ isoform, such as stability or translational control. This possibility remains to be tested.
Intronic cis elements for α-TM splicing regulation.
Presence of a characteristic CU-rich sequence in several RBM4 targets hinted a possible RBM4 recognition site. A mutation in the α-TM CU1 element indeed disrupted RBM4 binding to intron 9a in vitro but retained a partial activity for RBM4-mediated exon 9a inclusion (Fig. 5 and 6). Perhaps in vivo binding of RBM4 to the CU1 site of the α-TM pre-mRNA is stabilized by its interaction with additional cis elements or with other splicing factors. In addition, two short CU-rich sequences also contributed to RBM4-mediated exon 9a inclusion. To our surprise, disruption of CU2 completely abolished RBM4-mediated splicing activity (Fig. 5). Given that RBM4 stabilizes U1 snRNP binding to the 5′ splice site of intron 9a (see above), mutations of its most adjacent CU element, i.e., CU2, may thus exhibit a severe effect on exon 9a inclusion. Future experiments are needed to test this possibility and address how each CU-rich RBM4-responsive element contributes to an intricate splicing regulation. Nevertheless, RBM4 effectively antagonized the activity of PTB in exon 9a suppression by competing PTB binding to CU1, a major site responsible for PTB function.
RBM4 as well as PTB regulated the selection of exon 2b in α-TM, probably also via binding to intronic element(s) similar to those within exon 9a. Indeed, intron 2b contained several CU-rich sequences, including one similar to the CU1 element (Table 2). It is reasonable to assume that alternative selection with regard to exons 2b and 9a is coordinated using the same set of splicing regulators; hence, corresponding cis elements should be located proximally to these two exons.
Antagonistic actions of RBM4 and PTB.
RBM4 has various activities in splicing regulation, including the determination of 5′ splice site usage and the inclusion or exclusion of regulated exons (20 and this study). We demonstrate here that RBM4 activates sk exon utilization in α-TM via binding to intronic CU-rich elements. RBM4 expression is not restricted to skeletal muscle cells, but its higher abundance in these cells suggests that it participates in cell type-specific splicing activation. On the other hand, PTB is a key repressor of exon definition in a variety of cell types (2, 9, 25, 31). Negative regulation of α-TM exon 2b in part involves PTB in smooth muscle cells, which in turn allows inclusion of exon 2a in the sm isoform (10). More notably, PTB suppresses the yield of the Xenopus α-TM myotomic isoform in nonmuscle cells (13), which is equivalent to the 9a′ mRNA characterized in this report. This result suggests that PTB prevents the use of exon 9a, and thus is consistent with our observation. Suppression of exon inclusion in both cases was mediated in part through intronic PTB-responsive CUCC elements (10, 13). Such CUCC elements can be found in the intron immediately upstream of human α-TM exon 9a. Our data show that the CU1 element within intron 9a was necessary for PTB-mediated exon repression (Fig. 5). Since binding of PTB both upstream and downstream of a regulated exon is a common mechanism to achieve exon silencing (2, 25, 31), two types of cis elements may possibly act in a cooperative manner to inhibit α-TM exon 9a selection.
Using minigene assays, we demonstrated that RBM4 reversed the repressive effect of PTB, thereby facilitating the inclusion of exons 2b and 9a/9b (Fig. 6). The repressive activity of PTB on global splicing regulation can be antagonized by other tissue- or stage-specific factors, such as Fox-1 and several CELF proteins, to achieve spatial or temporal expression patterns of target mRNAs (2, 9, 25). Oppositely, a brain-specific PTB analog can abrogate the effect of neuron-specific Nova proteins on exon inclusion (28). Brain-specific PTB acts through an intronic site adjacent to Nova binding and possibly also through a direct interaction with Nova (28). Nevertheless, these antagonistic regulators often show distinct binding specificities to intronic cis elements and may compete with one another for binding to adjacent motifs. RBM4 did not interact directly with PTB in our experiments (data not shown), but it likely competed directly with PTB for the same or overlapping intronic elements, thus preventing PTB access to regulated exons (Fig. 7). In addition to acting via a derepression mechanism, RBM4 may actively promote interactions between the spliceosome and intron; this possibility needs further investigation.
RBM4 acts as both an activator and a repressor.
We previously reported that RBM4 modulates alternative 5′ splice site utilization of the adenovirus E1a pre-mRNA and activates skipping of alternative exons of rat β-TM (20). For these two pre-mRNAs, RBM4 had an effect opposite to that of the SR protein ASF; the underlying mechanism, however, was not characterized. Nevertheless, the effect of RBM4 on β-TM splicing established a role for RBM4 as a repressor of exon selection. Surprisingly, RBM4 activated rather than inhibited inclusion of the sk exons in human α-TM pre-mRNA (Fig. 4). Interestingly, when placed internally into β-TM, the penultimate sk-specific α-TM exon was still included upon overexpression of RBM4 (Fig. 4). Therefore, the segment encompassing this exon and flanking introns may possess sufficient elements for RBM4-mediated exon inclusion even in a heterologous context. It appears that RBM4 can act as both a splicing activator and a repressor. RBM4 is thus similar to CUG-BP and ETR-3, which activate the smooth muscle exon of α-actinin but repress its nonmuscle exon (11), and to Fox-1, which plays an opposite role in alternative exon selection (15). These splicing regulators may change their activity with regard to splice site selection, perhaps by interacting with different proteins (23). Thus, a search for protein ligands of RBM4 may provide clues as to how its role in splicing is determined.
Acknowledgments
We are grateful to C. W. J. Smith and D. L. Black for the generous gifts of cDNA clones. We acknowledge W. C. Chang for Northern blotting, M. C. Lai for RBM4-expressing cell lines, and P. J. Peng for recombinant RBM4 protein. We are grateful to J. Y. Wu (Vanderbilt University, Nashville, TN) for critical reading of the manuscript. We thank T. C. Taylor for editing the manuscript.
This work was supported by grant NHRI-EX92-9122BI from the National Health Research Institutes of Taiwan.
REFERENCES
- 1.Black, D. L. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367-370. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-mRNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 4.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 5.Charlet-B., N., P. Logan, G. Singh, and T. A. Cooper. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9:649-658. [DOI] [PubMed] [Google Scholar]
- 6.Dredge, B. K., G. Stefani, C. C. Engelhard, and R. B. Darnell. 2005. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 24:1608-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gooding, C., G. C. Roberts, G. Moreau, B. Nadal-Ginard, and C. W. J. Smith. 1994. Smooth muscle-specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 13:3861-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gooding, C., P. Kemp, and C. W. J. Smith. 2003. A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J. Biol. Chem. 278:15201-15207. [DOI] [PubMed] [Google Scholar]
- 9.Grabowski, P. J., and D. L. Black. 2001. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65:289-308. [DOI] [PubMed] [Google Scholar]
- 10.Gromak, N., and C. W. J. Smith. 2002. A splicing silencer that regulates smooth muscle specific alternative splicing is active in multiple cell types. Nucleic Acids Res. 30:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromak, N., A. J. Matlin, T. A. Cooper, and C. W. J. Smith. 2003. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA 9:443-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gromak, N., A. Rideau, J. Southby, A. D. Scadden, C. Gooding, S. Huttelmaier, R. H. Singer, and C. W. J. Smith. 2003. The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 22:6356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamon, S., C. Le Sommer, A. Mereau, M.-Rose Allo, and S. Hardy. 2004. Polypyrimidine tract-binding protein is involved in vivo in repression of a composite internal/3′-terminal exon of the Xenopus α-tropomyosin pre-mRNA. J. Biol. Chem. 279:22166-22175. [DOI] [PubMed] [Google Scholar]
- 14.Hardy, S., S. Hamon, B. Cooper, T. Mohun, and P. Thiébaud. 1999. Two skeletal α-tropomyosin transcripts with distinct 3′UTR have different temporal and spatial patterns of expression in the striated muscle lineages of Xenopus laevis. Mech. Dev. 87:199-202. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9:1161-1171. [DOI] [PubMed] [Google Scholar]
- 17.Ladd, A. N., N. Charlet, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladd, A. N., and T. A. Cooper. 23October2002, posting date. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 3:reviews0008.1- reviews0008.16. [Online.] http://genomebiology.com/2002/3/11/reviews/0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladd, A. N., N. H. Nguyen, K. Malhotra, and T. A. Cooper. 2004. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J. Biol. Chem. 279:17756-17764. [DOI] [PubMed] [Google Scholar]
- 20.Lai, M. C., H. W. Kuo, W. C. Chang, and W. Y. Tarn. 2003. A novel splicing regulator shares a nuclear import pathway with SR proteins. EMBO J. 22:1359-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2003. Differential effects of hyperphosphorylation on splicing factor SRp55. Biochem. J. 371:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, C., R.-I. Lin, M. C. Lai, P. Ouyang, and W. Y. Tarn. 2003. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via its interaction with RNPS1. Mol. Cell. Biol. 23: 7363-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matlin, A. J., F. Clark, and C. W. J. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 25.Musunuru, K. 2003. Cell-specific RNA binding proteins in human disease. Trends Cardiovasc. Med. 13:188-195. [DOI] [PubMed] [Google Scholar]
- 26.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 27.Perry. S. V. 2001. Vertebrate tropomyosin: distribution, properties and function. J. Muscle Res. Cell Motil. 22:5-49. [DOI] [PubMed] [Google Scholar]
- 28.Polydorides, A. D., H. J. Okano, Y. Y. L. Yang, G. Stefani, and R. B. Darnell. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 97:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakashita, E., S. Tatsumi, D. Werner, H. Endo, and A. Mayeda. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281- 3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wollerton, M. C., C. Gooding, F. Robinson, E. C. Brown, R. J. Jackson, and C. W. Smith. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]