Abstract
A vertebrate homologue of the Fox-1 protein from C. elegans was recently shown to bind to the element GCAUG and to act as an inhibitor of alternative splicing patterns in muscle. The element UGCAUG is a splicing enhancer element found downstream of numerous neuron-specific exons. We show here that mouse Fox-1 (mFox-1) and another homologue, Fox-2, are both specifically expressed in neurons in addition to muscle and heart. The mammalian Fox genes are very complex transcription units that generate transcripts from multiple promoters and with multiple internal exons whose inclusion is regulated. These genes produce a large family of proteins with variable N and C termini and internal deletions. We show that the overexpression of both Fox-1 and Fox-2 isoforms specifically activates splicing of neuronally regulated exons. This splicing activation requires UGCAUG enhancer elements. Conversely, RNA interference-mediated knockdown of Fox protein expression inhibits splicing of UGCAUG-dependent exons. These experiments show that this large family of proteins regulates splicing in the nervous system. They do this through a splicing enhancer function, in addition to their apparent negative effects on splicing in vertebrate muscle and in worms.
Alternative splicing allows the production of multiple mRNAs from a single pre-mRNA via selection of different splice sites. Regulated exons are controlled by splicing enhancer and silencer elements within the exon or in the adjacent introns. These RNA sequences bind to specific regulatory proteins that contribute to the tissue specificity of splicing. Most exons are controlled by combinations of both positive and negative regulators, and how tissue specificity of splicing is achieved is poorly understood (5, 44).
The N1 exon of the c-src gene serves as a model for an exon under both positive and negative control. In nonneuronal cells, the exon is repressed by the polypyrimidine tract binding protein (PTB) that binds to intronic splicing silencer elements flanking the N1 exon (1, 7, 9). In neurons, PTB-mediated repression is absent, and the exon is activated for splicing by an intronic splicing enhancer (4, 38). The enhancer region downstream of the N1 exon is complex, with binding sites for many proteins. However, the element most critical for enhancer activity is the sequence UGCAUG, which is flanked by PTB binding elements (4, 37, 38). Several proteins, including the hnRNPs F and H, the neuronal homologue of PTB, and the KH-type splicing regulatory protein, assemble onto this region in splicing extracts (8, 30, 34, 35). Immunodepletion and antibody inhibition experiments have indicated a role for these proteins in the splicing of N1 in vitro. However, none of these proteins specifically recognizes the UGCAUG element, and they do not positively affect an exon controlled by just a UGCAUG element in vivo (J. G. Underwood and D. L. Black, unpublished observations). Thus, they do not seem to mediate the function of the strongest enhancer element. Their function may be related to preventing PTB-mediated repression in neurons rather than true positive control of splicing. The proteins responsible for the UGCAUG-dependent enhancer activity are not known.
The UGCAUG hexanucleotide has been identified as controlling many alternative exons in addition to src N1 (11, 12, 18, 20, 24). This element has been studied extensively as a regulator of fibronectin EIIIB exon splicing, which is highly dependent on a group of UGCAUG elements dispersed throughout the downstream intron (29). Interestingly, these elements act at some distance from the upstream, activated exon, and their wide spacing is conserved between vertebrate species. Similarly, the UGCAUG element is found downstream of the c-src N1 exon in all vertebrates (4, 36, 45). These elements also play an important role in regulating the splicing of a neuron-specific exon in nonmuscle myosin heavy chain, as well as a neuronal pattern of processing in the calcitonin/calcitonin gene-related peptide (CGRP) transcript (18, 24, 39). The element UGCAUG was also identified in a computational study as the most common hexanucleotide found in the introns downstream of a set of neuron-specific exons (6). Thus, this element is a hallmark of many systems of neuronal splicing regulation.
Recently, several groups identified vertebrate homologues of the Caenorhabditis elegans protein Fox-1 (22, 46). The Feminizing locus on X (Fox-1) gene encodes a protein with a single RNA recognition motif (RRM)-type RNA binding domain and is a numerator element in reading the X-to-autosome ratio in C. elegans sex determination (19, 32, 33, 40, 43). Fox-1 protein controls expression of the Xol-1 gene (XO lethal), a key switch in determining male-versus-hermaphrodite development. Jin et al. identified homologues of Fox-1 in zebra fish and mouse and showed that they specifically recognize the element GCAUG (22). The zebrafish Fox-1 mRNA was specifically expressed in muscle, whereas the mouse mRNA was abundant in muscle, heart, and particularly brain. It was shown in cotransfection assays that this protein functioned as a repressor of certain exons in muscle but also enhanced the splicing of the fibronectin EIIIB exon (22).
We examined the role of Fox proteins in mammalian neuron-specific splicing, and in c-src N1 exon splicing in particular. We found several homologous genes for these proteins in mammals that each give rise to a large family of proteins through extensive alternative splicing. Within the brain, these proteins are specifically expressed in neurons and not glia. Through both loss-of-function and gain-of-function experiments, we show that both of these proteins are the mediators of UGCAUG-mediated splicing enhancement in neurons.
MATERIALS AND METHODS
Northern blot analysis.
A commercial mouse poly(A)+ Northern blot (Ambion) was probed according to the manufacturer's instructions. Northern probes were sequences from the first constitutive exon (∼100 to 125 nucleotides [nt]) of each Fox gene PCR amplified with a T7 promoter in the antisense orientation. cRNA probes were synthesized by T7 transcription using the Ambion Strip-EZ T7 kit and [α-32P]UTP. β-Actin levels were assayed with the template provided with the blot.
Antibodies.
Rabbit polyclonal antibodies (Alpha Diagnostics, Inc.) were raised to an N-terminal peptide of each protein shared by all known isoforms. (Fox-1 NT peptide, MAQPYASAQFAPPQN, and Fox-2 NT peptide, TTTPDAMVQPFTTIP). No cross-reactivity was observed on Western blots with recombinant proteins. Specific antibodies were purified using the cognate peptide coupled to Sulfo-Link resin (Pierce). Briefly, serum was mixed 1:1 with Tris-buffered saline (TBS), passed through the peptide column, and washed with 5 column volumes of TBS and 5 column volumes of TBS plus 0.05% sodium dodecyl sulfate (SDS) and 0.1% Triton X-100. After buffer strength was reduced with 5 volumes of 20 mM Tris-HCl, pH 7.5, specific immunoglobulin G (IgG) was eluted with 100 mM glycine-HCl and fractions were neutralized with 1/10 volume of 1 M Tris-HCl, pH 8.0.
Tissue culture.
All cell lines were grown according to ATCC-recommended protocols. Neuronal N2A and N1E-115 cells were differentiated in Dulbecco's modified Eagle's medium (DMEM) plus 1% fetal bovine serum and 2% dimethyl sulfoxide (DMSO) for 5 days (17). Myoblast C2C12 cells were differentiated with DMEM in low sodium bicarbonate and 2% horse serum for 5 days (27).
Extract preparation and cross-linking.
Preparation of HeLa nuclear extracts, site-specific labeling, and cross-linking were performed as described previously (9).
Western blot analysis.
Cells were lysed in RIPA buffer containing Complete protease inhibitors (Roche) and 30 U/ml benzonase (Sigma) and incubated on ice for 10 min to allow lysis and nucleic acid fragmentation. Protein concentrations were normalized using a Bradford assay. Normalized protein samples were added to SDS loading buffer and heated to 90°C for 10 min. For the cell line Western analysis, proteins were resolved on 4 to 20% Tris-glycine gels. All other protein gels were 10% NuPage Bis-Tris gels (Novex). After transfer to nitrocellulose, Western blots were carried out with Fox-1 NT (1:200), Fox-2 NT (1:250), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Covance; 1:400,000), and FLAG (Sigma; 1:3,000) antibodies. Detection used horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia) with ECL or Femto Supersignal reagents (Pierce).
Immunohistochemistry.
Adult CD1 mice were perfused transcardially with ice-cold phosphate-buffered saline (PBS), followed by ice-cold 4% paraformaldehyde in PBS. The brains were removed, fixed in 4% paraformaldehyde overnight, sunk in 20% sucrose-PBS, frozen in 4-methyl-butane, and stored at −80°C until use. Forty-micrometer sections were cut on a cryostat and stored in PBS plus 0.1% azide at 4°C until use. Free-floating sections were incubated overnight in 24-well plates on a rotator at room temperature in the presence of 0.1% azide, 0.25% Triton X-100, and 5% normal goat serum in 500 μl PBS and primary antibody at the following concentrations: Fox-1 NT, 1:100; Fox-2 NT, 1:500; GFAP, 1:1,000 (Chemicon AB1540); NeuN, 1:100 (Chemicon MAB377). Secondary antibodies were diluted 1:1,000 and included Cy3-conjugated anti-rabbit IgG and Cy5-conjugated anti-guinea pig IgG (Jackson Immunoresearch) and Alexa 488-conjugated anti-mouse IgG (Molecular Probes) antibodies. Nuclei were counterstained with Hoechst dye (Molecular Probes) and mounted with a Prolong AntiFade kit (Molecular Probes). In all cases, controls with no primary antibody yielded no labeling. For Fox-1 and Fox-2 double labeling, sections were incubated overnight with Fox-1 antibody and then for 1 h with Cy3 anti-rabbit IgG and blocked for 30 min in normal rabbit serum and then for 20 min in 1:50 donkey anti-rabbit Fab fragment (Jackson Immunoresearch), followed by 5 h of incubation with Fox-2 antibody and 1 h of incubation with Alexa goat anti-rabbit IgG (Molecular Probes). Nuclei were counterstained with ToPro 3 iodide (Molecular Probes) and mounted with a Prolong AntiFade kit (Molecular Probes). Lack of cross-reaction was confirmed by identification of nuclei in the cerebellum and olfactory bulb that labeled exclusively with either Fox-1 or Fox-2. Confocal microscopy was performed on a Zeiss LSM 510 META confocal microscope, and pseudocolored images were overlaid with Zeiss software or Adobe Photoshop. Three-dimensional reconstructions were used to confirm overlap of signals. Infrared wavelengths were most often pseudocolored blue.
Plasmids.
The DUP reporter plasmids were described previously (37, 38). Additional plasmids were constructed by similar methods. The synthetic enhancer constructs (see Fig. 5) were constructed by ligating phosphorylated annealed oligonucleotides into the unique Bgl2 site in the second intron of DUP 4-1. This placed the test enhancer 45 nucleotides away from the alternative exon 5′ splice site. The Fox cDNA expression plasmids were constructed in pcDNA3.1 (Invitrogen). Each carried an N-terminal FLAG tag.
FIG. 5.
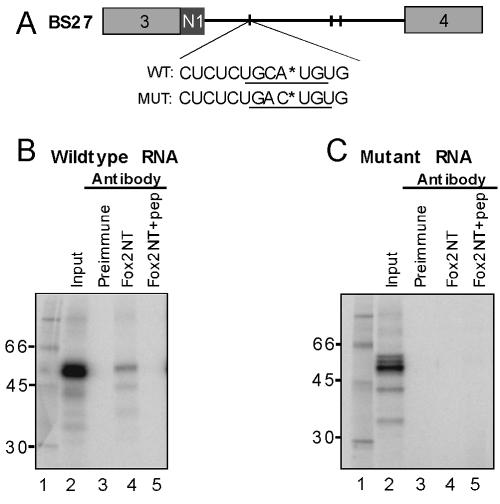
Fox-2 cross-links specifically to the UGCAUG element within the N1 exon intronic splicing enhancer. (A) Location of the labeled phosphate within the wild-type (WT) and mutant (MUT) BS27 RNAs. The labeled phosphates are indicated by asterisks. The wild-type and mutant UGCAUG elements are underlined. (B) Cross-linking to the wild-type UGCAUG element. (C) Cross-linking to the mutant UGACUG element (MUT). Lane 1 contains molecular weight markers and lane 2 the total cross-linking reaction (20% of the input to the immunoprecipitations in lanes 3 to 5). Lanes 3 to 5 are immunoprecipitations with the following antibodies: preimmune serum (lane 3); affinity purified anti-Fox-2 NT (Lane 4); anti-Fox-2 NT preblocked with the cognate Fox-2 NT peptide (lanes 5).
siRNAs.
The small interfering RNAs (siRNAs) were designed against target sequences that were identical in the human and mouse Fox genes and used the following established protocols (http://www.rockefeller.edu/labheads/tuschl/sirna.html; 1). siRNAs were synthesized from DNA oligonucleotide templates containing a T7 promoter and encoding one strand of the desired siRNA after a G-rich 5′ flap sequence. Each siRNA strand was separately transcribed with T7 RNA polymerase and then annealed to the cognate strand. Treatment with DNase I removed the template DNA, and treatment with RNase T1 removed the unhybridized G-rich flap to yield the siRNA with correct ends. The siRNAs were purified on G-25 resin and quantified by UV absorbance and/or serial dilutions on 4% agarose gels. Template oligonucleotides and product siRNAs for Fox-1 and Fox-2 were as follows: Fox-1 DNA template for the sense strand of siRNA, 5′-AATTAGTATGGAGCAAAACGGCCTGTCT; Fox-1 DNA template for the antisense strand of siRNA, 5′-AACCGTTTTGCTCCATACTAACCTGTCT; product strands of Fox-1 siRNA, CCGUUUUGCUCCAUACUAAUU and UUGGCAAAACGAGGUAUGAUU; Fox-2 DNA template for the sense strand of siRNA, 5′-AAAAATATTGCAATAGCCAGGCCTGTCT; Fox-2 DNA template for the antisense strand of siRNA, 5′-GGCCTGGCTATTGCAATATTTCCTGTC; product strands of Fox-2 siRNA, CCUGGCUAUUGCAAUAUUUUU and CCGGACCGAUAACGUUAUAAA; T7 promoter primer, 5′-TAATACGACTCACTATAGGGAGACAGG.
Transfections.
DNA and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen). Briefly, 1 μg of plasmid DNA was diluted in 50 μl of Optimem and complexed with 2 μl of Lipofectamine 2000 in 50 μl of Optimem for 20 min. Trypsinized suspensions of N2A or HeLa cells (1.5 × 105 cells in 500 μl in DMEM plus 10% fetal bovine serum) were added to the lipid complex and rotated manually for 10 min. The mix was then aliquoted into 24-well plates, and the medium was changed after 4 h. For splicing reporter assays, 100 ng of reporter and 900 ng of total pcDNA plasmid were used. For siRNA transfections, the siRNA was added to 1 μg of pUC plasmid DNA during the lipid formulation phase. For siRNA reporter assays, 100 ng of reporter and 900 ng of pUC were cotransfected. The siRNA titration (see Fig. 6) was performed with 5, 10, and 20 pmol of siRNA. All other siRNA experiments utilized 20 pmol of siRNA.
FIG. 6.
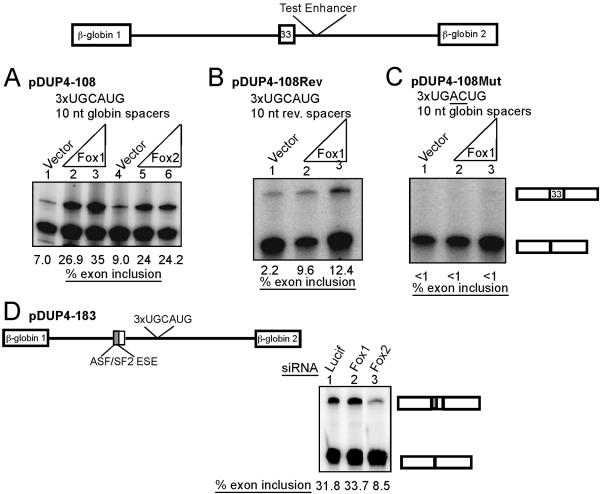
Fox enhances splicing through tandem UGCAUG sequences in HeLa cells. Test enhancers were inserted into the second intron of pDUP4-1 β-globin splicing reporter. (A) An enhancer construct with three copies of UGCAUG element separated by 10-nt globin intron spacers. pDUP4-108 (100 ng) was cotransfected with empty expression vector (lane 1) or 100 ng or 900 ng of Fox-1 or Fox-2 expression plasmid. Lanes 2 and 3 have increasing Fox-1, and lanes 5 and 6 have increasing Fox-2. RNA was harvested after 48 h, and inclusion was assayed by RT-PCR. (B) Conditions similar to those for panel A but using the pDUP4-108Rev reporter plasmid, which has the spacer sequences reversed. (C) Conditions similar to those for panel A but with the pDUP4-108Mut reporter plasmid carrying three copies of UGACUG. (D) Knockdown of Fox-2 in HeLa cells results in loss of enhancer activity. The pDUP4-183 reporter plasmid is similar to that in panel A but carries the 5′ half of the c-src N1 exon, a known ASF/SF2-dependent exonic splicing enhancer (ESE) (41a). HeLa cells were cotransfected with 100 ng of pDUP4-183, 900 ng pUC carrier DNA, and the indicated siRNAs. RNA was harvested after 60 h, and exon inclusion was assayed by RT-PCR.
Reverse transcription (RT)-PCR.
Total RNA was isolated from cells with TRI reagent (MRC) and reverse transcribed with random hexamers and Superscript II (Invitrogen). Splice products were PCR amplified (22 cycles) using an end-labeled primer in the 3′ constitutive exon and a cold primer in the 5′ constitutive exon. Primers for c-src and EWS were as published (17, 31). Products were separated by urea-polyacrylamide gel electrophoresis (PAGE) and visualized by phosphorimager. Exon inclusion was quantified using NIH ImageJ software.
Protein accession numbers.
The protein accession numbers of the isoforms used for expression are NP_067452 for Fox-1 and AAL67150 for Fox-2.
RESULTS
Mouse Fox-1 and Fox-2 are both expressed in neurons.
To test whether a vertebrate Fox-1 homologue was responsible for mediating the action of the N1 exon splicing enhancer, we isolated cDNAs encoding these proteins and also obtained plasmids from other laboratories and the Riken DNA Bank. There are several homologues in the mouse and human genomes to C. elegans Fox-1. In addition to human Fox-1 (hFox-1) (gene name, A2bp1; chromosome 16) (42), there are additional paralogous genes in the mammalian genomes. One gene we called Fox-2 (gene name, RBM9; also called fxh and RTA; human chromosome 22) (28, 41). There is also a third paralog (gene name, HRNbp3; chromosome 17) (46). However, we did not initially find evidence for expression of this gene in the cells used for our experiments (data not shown), and it was not studied further. Alignments of the cDNA, expressed sequence tag (EST), and genomic databases indicate that both the fox-1 and fox-2 genes are large in the mouse (mFox-1, 1.5 Mb, and mFox-2, 220 kb) and larger in humans (hFox-1, 2.5 Mb, and hFox-2, 280 kb). Both genes have multiple alternative promoters and multiple internal cassette exons that are variably included in the mRNA (Fig. 1A; see Table S1 in the supplemental material). Thus, alternative splicing gives rise to a large family of proteins from each gene.
FIG. 1.
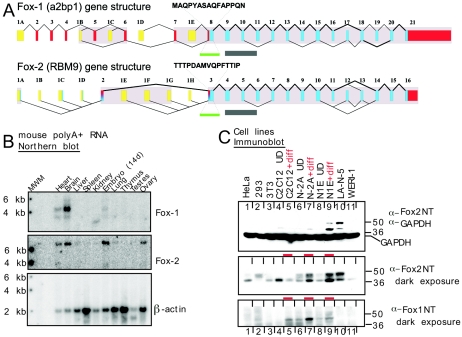
Mammalian Fox proteins are expressed in neuronal tissues and cell lines. (A) Human Fox genes are represented with first exons in yellow, untranslated regions in red, and coding exons in blue. Exons showing clear homology in the mouse are shaded. The exons are numbered to allow their identification in the table of genomic coordinates (see Table S1 in the supplemental material). This numbering does not necessarily correspond to previously reported exon numbers. The exon used for probing the Northern blot in panel B is marked with a green bar, and the peptides from the same exon used for antibody production are indicated. The exons encoding the RRM are marked with a gray bar. Both genes have multiple promoters and numerous alternative splicing events. The coding alternative exons shown all conform to splice site consensus rules and are conserved between human and mouse. Nearly all are predicted from at least two ESTs, and some were confirmed experimentally by RT-PCR (data not shown). Alternative splicing of exon 19 of Fox-1 and exon 13 of Fox-2 gives rise to two C-terminal peptide sequences. These two splice variants of Fox-1, Fox-2, and Fox-3 from human and mouse are aligned in Fig. S1 in the supplemental material, along with two worm homologues. (B) Northern blot analysis of mouse tissues. A blot (Ambion) carrying 2 μg of poly(A)+ RNA from each of the indicated mouse tissues was probed sequentially with Fox-1, Fox-2, and β-actin cRNAs. MWM, molecular weight markers; 14d, 14 days. (C) Western blot analysis of human and mouse cell lines. Protein (50 μg) from whole-cell lysates was separated by 4 to 20% gradient SDS-PAGE, transferred to nitrocellulose, and probed with the indicated antibodies. GAPDH is shown as a loading control. The top and middle panels are different exposures of the same blot. For the longer exposure, the GAPDH portion of the blot was cut off. The cell lines are HeLa, HEK293, 3T3, C2C12 without and with myotube differentiation (diff), mouse N2A and N1E-115 neuroblastomas without and with neuronal differentiation, human neuroblastoma LA-N-5, and human retinoblastoma WERI-1. Differentiated samples are marked with a red dash.
The RRMs for Fox-1 and Fox-2 are identical over their entire lengths in both human and mouse and presumably recognize the same sequence (see Fig. S1 in the supplemental material). The RRM of mFox-3 is slightly more divergent, differing from Fox-1 and Fox-2 at 4 of 72 residues. In addition to the C. elegans Fox-1, there is another homologous gene on worm chromosome 3. Within the RRM domains, these two proteins are remarkably similar to the mammalian homologues, differing from the mammalian proteins at 17 and 23 out of 72 residues, respectively. There is also a Drosophila gene encoding a nearly identical RRM, which apparently produces a much larger protein with extended N- and C-terminal domains (CG32062; chromosome 3L) (data not shown). All three mammalian proteins have homologous alternative C termini and N termini arising from alternative exons and promoters. Using similar termini, mammalian Fox-1 and Fox-2 share 66% identity over their entire lengths (see Fig. S1 in the supplemental material). The worm proteins are much more divergent in the N- and C-terminal domains. However, the extreme C termini of the worm proteins (RFTPY and RFAPY) clearly match one of the C termini of each of the mammalian homologues (see Fig. S1 in the supplemental material).
Alternatively spliced internal exons create additional changes in the protein. One exon-skipping event deletes essential amino acids from the RRM and presumably eliminates RNA binding. EST expression data and recent work from the Kawamoto laboratory indicate that some of these transcripts are expressed in a tissue-specific manner (reference 39 and data not shown). In the face of this complexity, we decided to characterize an equivalent isoform from each gene and chose those forms that have C termini similar to those of the worm protein (see Fig. S1 in the supplemental material).
To confirm the expression pattern of mFox-1 and determine that of mFox-2, we generated Northern blot probes from the first common exon of all the transcripts from each gene (Fig. 1B). When applied to a mouse tissue Northern blot, the observed pattern of mFox-1 mRNA expression was the same as seen previously (22, 26). Fox-1 expression was high in brain and heart, with lower levels in embryos. (Skeletal-muscle RNA was not on this Northern blot, but the expression of Fox-1 was observed in muscle cell lines.) Fox-2 showed higher expression than Fox-1 in the embryo relative to the adult, and expression in the ovary, but otherwise showed a pattern of expression very similar to that of Fox-1 (Fig. 1B).
To examine the expression of the Fox proteins in cell lines that we would use in later experiments, we raised antibodies to mFox-1 and mFox-2. Antibodies were raised to peptides within the first common exon of Fox-1 and Fox-2 (Fig. 1A). These antibodies reacted well with recombinant target protein but did not cross-react with the opposite homologue (data not shown). When applied to immunoblots, the Fox-2 antibody gave two prominent bands of approximately 48 and 40 kDa that presumably resulted from two of the many splice variants of the Fox-2 mRNA (Fig. 1C). These Fox-2 proteins were most highly expressed in the mouse neuroblastoma cell line N1E-115, which had been induced to differentiate with DMSO and low serum, and in the human neuroblastoma LA-N-5 (Fig. 1C, lanes 9 and 10). On longer exposure, these same Fox-2 proteins were seen in other cell lines, although the relative abundances of the two forms vary. In a second neuroblastoma, N2A, Fox-2 was also induced by the differentiation treatment (Fig. 1C, lanes 6 and 7). The Fox-1 antibody was reactive with one prominent band of about 43 kDa, as well as other less abundant species. Fox-1 expression was more limited than that of Fox-2. It was seen in the mouse neuroblastomas N1E-115 and N2A, but not in LA-N-5. Interestingly, the differentiation of the neuroblastomas into neurons again stimulated expression of Fox-1 (Fig. 1C, lanes 6 to 9). Differentiation of the muscle cell line C2C12 into myotubes also increased Fox-1 expression, but in this case it decreased Fox-2 expression (Fig. 1C, lanes 4 and 5).
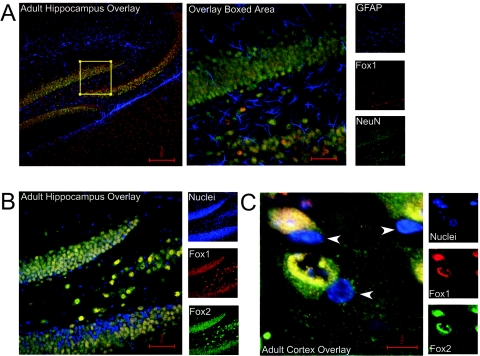
To examine what cell types in the brain express Fox proteins, we performed immunofluorescence staining with the Fox antibodies on sections of brain tissue. Both proteins were broadly expressed throughout the brain in largely, but not exactly, overlapping patterns. Both proteins show strong staining in the neurons of the dentate gyrus and CA3 region of the hippocampus (Fig. 2A and B). This staining is specific to neurons; costaining for glia with anti-GFAP showed no overlap with the Fox proteins, whereas staining with anti-NeuN gave a completely coincident pattern in both the hippocampus and the cortex (Fig. 2A and data not shown). As seen in the staining of cortical sections, both Fox-1 and Fox-2 are predominantly nuclear, as expected, but Fox-2 shows some additional cytoplasmic staining (Fig. 2C). Staining other regions of the brain gave similar results. Interestingly, there were some neurons in both the cerebellum and the olfactory bulb that expressed only Fox-1 or Fox-2, but not both (data not shown). In all experiments, these proteins were specific to neurons and were not seen in glia (Fig. 2A and 2C).
FIG. 2.
Fox-1 and Fox-2 are coexpressed in the nuclei of most neurons of the adult mouse brain but excluded from glia. (A) Immunostaining of Fox proteins in hippocampus. Fox-1 expression (red) overlaps completely with neuronal marker NeuN (green) but shows no overlap with glial marker GFAP (blue). Individual channels are shown in small panels on the right, and overlay images are shown in larger panels on the left. The boxed region in the left panel is shown in the right panels. (B) In the hippocampus, Fox-1 expression (red) also overlaps completely with Fox-2 expression (green). (C) High-power confocal microscopy in the cortex reveals that both Fox-1 (red) and Fox-2 (green) are expressed primarily in nuclei (blue). Some small nuclei (arrowheads), likely glia, contain no Fox labeling.
Fox-1 and Fox-2 activate splicing of neuronal exons through binding to downstream UGCAUG elements.
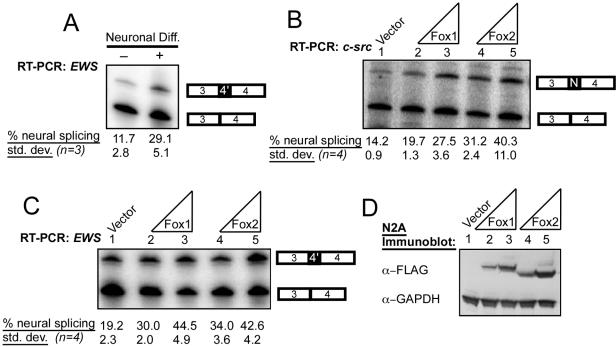
We previously observed that splicing of the c-src N1 exon increases about twofold in mouse neuroblastoma cells when they are induced to differentiate, correlating with the observed increase in Fox protein expression (Fig. 1C, lanes 6 to 9) (17). To test whether other exons were spliced in parallel with Fox protein expression, we tested these cells for splicing of the neuron-specific 4′ exon in the EWS gene, which has regulatory elements similar to those of c-src N1 (31). Both genes have UGCAUG elements in the downstream intron which are conserved between multiple mammalian species (36). As seen previously for c-src N1, induction of N2A cells with DMSO and low serum caused an increase in EWS exon 4′ splicing, in parallel with the change in Fox expression (Fig. 3A).
FIG. 3.
Fox proteins enhance neuron-specific splicing patterns in mouse N2A mRNAs. (A) EWS neuron-specific splicing is induced during neuronal differentiation of N2A cells. Cells were grown in standard media or differentiation media for 5 days, and RNA was harvested. EWS exon 4′ splicing was assayed by RT-PCR and quantified. The percent exon inclusion is indicated. std. dev., standard deviation. (B) Inclusion of the N1 exon in the endogenous c-src mRNA is enhanced by Fox overexpression. Each Fox plasmid (100 ng or 1 μg) was transfected into N2A cells, and total RNA was harvested after 60 h. RNA was assayed by RT-PCR and quantified. (C) Inclusion of the 4′ exon in the endogenous EWS mRNA is enhanced by Fox overexpression; similar to panel B, except assayed for EWS splicing by RT-PCR using an end-labeled primer in EWS exon 4. (D) Immunoblot confirming overexpression of Fox-1 and Fox-2 proteins in N2A cells. Whole-cell lysates (50 μg) were separated by 10% NuPage and subjected to immunoblotting with FLAG and GAPDH antibodies.
Since an increase in Fox protein expression during neuroblastoma differentiation correlates with splicing of neuronal exons in both c-src and EWS, we next tested whether either protein expressed alone could induce splicing of these exons. The Fox proteins were transiently expressed in N2A cells (untreated for differentiation), and splicing of the c-src and EWS exons was assayed by RT-PCR (Fig. 3B and C). The expression of the Fox proteins was monitored by immunoblotting for the Flag epitope tag on each construct (Fig. 3D). N2A cells transfected with empty expression vector included the N1 exon in 14% of the c-src mRNA and included the 4′ exon in 19% of the EWS mRNA (Fig. 3B and C, lanes 1). Overexpression of either Fox-1 or Fox-2 approximately doubled the splicing of each exon (Fig. 3B and C, lanes 2 to 5); splicing of c-src N1 went up to 27 to 40%, and splicing of EWS 4′ went up to 43%. Note that these assays were performed on the endogenous c-src and EWS mRNAs. Since a large fraction of the cells (30 to 40%) do not take up DNA in these transfections, the induction of neural exon splicing by Fox proteins is likely to be larger than can be measured in this experiment (data not shown).
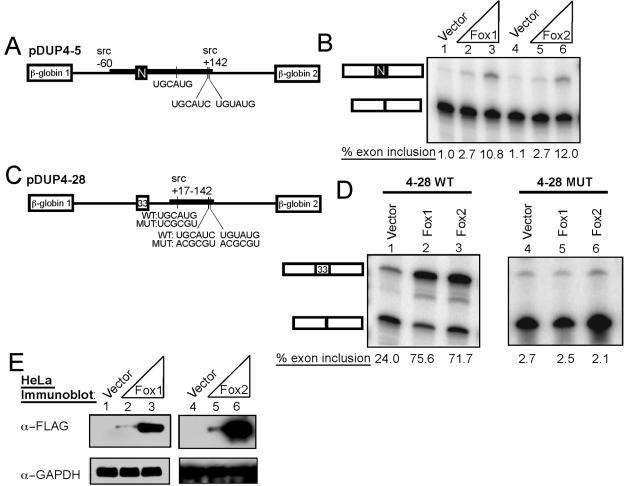
To examine the roles of particular RNA elements in the Fox protein activation of N1 splicing, we cotransfected N1 exon minigenes with the Fox expression plasmids (Fig. 4A). For this, we used HeLa cells, which have low levels of Fox-2 and no Fox-1 (Fig. 1C). As shown previously, a minigene carrying the N1 exon and its flanking intronic regulatory elements showed nearly complete (99%) skipping of the N1 exon in HeLa cells (pDUP4-5) (Fig. 4B, lane 1) (37). When the pDUP4-5 plasmid is coexpressed with either mFox-1 or mFox-2, the exon is activated 10- to 12-fold and is spliced into about 10 to 12% of the product mRNA (Fig. 4B, lanes 2 to 6). This level of N1 splicing in HeLa cells with Fox coexpression is similar to that seen with the reporter transfected into a mouse neuroblastoma cell line (37). This confirms that the N1 exon is strongly affected by either Fox protein and demonstrates that introducing a Fox protein into a nonneuronal cell line is sufficient to stimulate N1 inclusion.
FIG. 4.
Fox enhances splicing through the c-src intronic splicing enhancer. (A) A chimeric minigene, pDUP4-5, with β-globin exons 1 and 2 flanking the c-src N1 exon and its intronic regulatory regions. This is completely repressed in HeLa cells. (B) Fox expression enhances N1 splicing. Splicing reporter (100 ng) was cotransfected with empty expression vector (lane 1) or 100 ng (lanes 2 and 5) or 900 ng (lanes 3 and 6) of Fox expression plasmid. Lanes 2 and 3 show increasing Fox-1, and lanes 5 and 6 are increasing Fox-2. RNA was harvested after 48 h, assayed by RT-PCR, and quantified. (C) A minigene carrying β-globin exons 1 and 2 with a 33-nt generic internal exon composed of β-globin sequence. The wild-type (WT) (pDUP4-28) or mutant (MUT) (pDUP4-28Mut) c-src intronic enhancer sequence is inserted in the second intron (38). With the wild-type enhancer, the internal exon is partially included without Fox expression. The putative Fox sites are shown below, with the mutations made for disrupting enhancer activity. (D) Fox expression enhances splicing through sequences in the c-src enhancer. Cotransfections and analysis were as for panel B, except with pDUP4-28 and pDUP4-28Mut. (E) Immunoblot confirming expression of the Fox proteins in HeLa cells. Whole-cell lysates (50 μg) were separated by 10% NuPAGE, transferred to nitrocellulose, and probed with FLAG and GADPH antibodies.
The N1 exon is controlled by multiple regulatory proteins (5). Most notably, in HeLa cells, the exon is strongly repressed by PTB (1, 7, 9). To examine the enhancer activity of Fox protein in the absence of PTB repression and to confirm that the Fox stimulation required the enhancer sequence, we used a test exon that is activated by the N1 enhancer but is missing many of the other N1 regulatory sequences, including essential upstream PTB sites (pDup4-28) (Fig. 4C) (38). This enhancer contains PTB binding elements, but they are insufficient to mediate splicing repression without the two high-affinity PTB sites upstream of the alternative exon (1, 7). The enhancer fragment contains one UGCAUG element and two elements that match at five of six positions. This enhancer activates the test exon in the pDUP4-28 plasmid to 18 to 25% inclusion in HeLa cells (Fig. 4D, lane 1). Splicing enhancement is dependent on the three elements, as their mutation lowers test exon splicing to 2% (Fig. 4D, lane 4). When the enhancer-containing minigene is coexpressed with either Fox protein, test exon splicing is strongly stimulated to 70 to 75% inclusion (Fig. 4D, lanes 2 and 3). The construct with mutations in the UGCAUG-like elements did not respond to Fox expression (Fig. 4D, lanes 5 and 6). Thus, the stimulation of splicing by the Fox proteins is mediated by the enhancer sequences and is independent of splicing repression by PTB.
In previous work, it was shown that Fox-1 binds to the element GCAUG with high specificity (22, 46). To confirm that the Fox proteins were binding to this element in the context of the N1 exon enhancer, we performed site-specific labeling and cross-linking experiments. For this, we used HeLa nuclear extract, which contains low levels of Fox-2 and no Fox-1 but which is known to be active for splicing. The BS27 RNA was chosen for analysis because it lacks the upstream binding sites that are needed for repression of N1 splicing by PTB. Thus, PTB binds only weakly to the downstream enhancer region, and BS27 RNA is active for splicing in HeLa extract (7, 9). By reducing PTB binding to the enhancer, we hoped to increase Fox-2 binding, given its low level of expression in HeLa cells. To localize the observed protein binding to the enhancer element, BS27 RNA containing a single 32P label within the UGCAUG element of the N1 enhancer was synthesized (Fig. 5A). This was compared to a mutant BS27 RNA in which the UGCAUG was changed to UGACUG but which carried the label in the same position (Fig. 5A).
These RNAs were incubated in HeLa extract, irradiated with 254-nm UV light, and treated with RNase to leave a small oligonucleotide tag on the cross-linked proteins. For the wild-type RNA, a prominent set of cross-linked proteins was observed migrating at about 50 kDa on SDS-PAGE (Fig. 5B, lane 2). To confirm that these bands contained Fox-2 protein, the cross-linked proteins were immunoprecipitated with anti-Fox-2 antibodies. One 50-kDa protein and other minor bands were clearly precipitable with the Fox-2 antibodies (lane 4). The specificity of the immunoprecipitation was confirmed with preimmune serum and with Fox-2 antibodies preblocked with the antigenic peptide, neither of which reacted with any of the cross-linked proteins (Fig. 5B, lanes 3 and 5). The requirement for the UGCAUG element in the Fox-2 interaction was demonstrated by the mutant BS27 RNA. Cross-linking to this RNA yielded a different distribution of ∼50-kDa proteins, and none of these were brought down by the Fox-2 antibody (Fig. 5C). Thus, Fox-2 protein very specifically binds to the UGCAUG element within the context of the N1 enhancer.
To demonstrate that the UGCAUG hexanucleotide alone was sufficient for Fox activation of splicing, we tested several minigenes carrying short synthetic enhancers containing this element (Fig. 6). As shown previously, a fragment carrying three copies of the element separated by 10-nucleotide spacer sequences from β-globin intron 1 activates the test exon weakly in HeLa cells but is more potent in neuronal cell lines. The background enhancer activity in HeLa cells is presumably due to a low level of endogenous Fox-2 (Fig. 1C). If the pDUP4-108 construct is coexpressed with either Fox-1 or Fox-2, splicing is stimulated from 7 to 9% to 24 to 35% (Fig. 6A). A similar fragment with the spacer sequences reversed (pDUP4-108Rev) was also activated by Fox proteins (Fig. 6B). In contrast, mutating 2 nucleotides within each element to UGACUG in the pDUP4-108Mut reporter eliminated splicing enhancement altogether, indicating that enhancement is indeed dependent on the UGCAUG element (Fig. 6C). A fragment carrying only one element did not enhance splicing, with or without expressed Fox protein. Similarly, a fragment carrying three directly abutted hexanucleotide elements without spacers showed much weaker activity than three elements spaced by 10 nucleotides (data not shown). These experiments indicate that the number and spacing of the elements are important for their activity.
Loss of Fox proteins leads to loss of UGCAUG enhancer function.
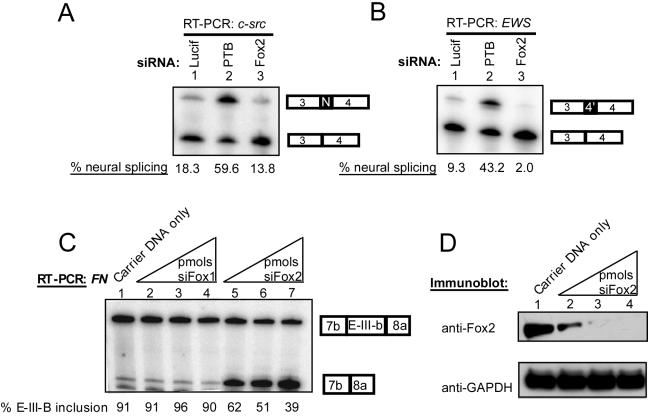
In addition to assessing the effect of Fox protein overexpression, we examined whether splicing was altered when the Fox proteins were eliminated using RNA interference (RNAi). Several siRNAs were tested in HeLa cells using transfected Fox cDNAs as targets (data not shown and reference 13). siRNAs that led to the specific reduction in Fox-1 or Fox-2 were identified (data not shown). HeLa cells exhibit efficient RNAi but contain no Fox-1 protein and very little Fox-2 protein. To assess whether this small amount of Fox-2 was responsible for the low enhancer activity seen in HeLa cells, we used the pDUP4-183 reporter, which is enhanced for basal exon inclusion by the addition of a binding site for ASF/SF2 within the alternative exon (Fig. 6D) (37, 41a). The combination of this exonic splicing enhancer with the triple UGCAUG yields about 30% exon inclusion (Fig. 6D, lane 1). Cotransfection with an siRNA against Fox-1 had no effect on splicing (Fig. 6D, lane 2). However, the siRNA against Fox-2 reduced exon inclusion to about 8% (lane 3). Thus, the small amount of Fox-2 in HeLa cells drives the basal activity of the UGCAUG enhancer.
Measuring alterations in endogenous c-src splicing by RNAi presented technical difficulties. Neuronal cell lines that exhibited high levels of N1 inclusion proved inefficient for siRNA transfection (data not shown). Conversely, efficient RNAi-mediated knockdown of Fox-2 was observed in N2A cells, but these cells exhibit only low levels of N1 inclusion, making it difficult to observe a large loss of splicing for N1. The N2A cells showed a consistent reduction of N1 inclusion, dropping from 18% with a control siRNA to 13% with Fox-2 siRNA (Fig. 7A, lanes 1 and 3). The low splicing of N1 in these cells is in part due to PTB repression; when PTB was targeted with an siRNA, N1 exon splicing increased to 59% (Fig. 7A, lane 2). Consistent with its near absence in N2A cells, a Fox-1 siRNA showed no effect on c-src splicing (data not shown). EWS 4′ splicing paralleled the regulation of c-src N1 (Fig. 7B). In this case, the siRNA against PTB increased exon 4′ inclusion from 9% to 43% and the siRNA against Fox-2 decreased inclusion to 2% (Fig. 7B, lanes 1 to 3). Thus, both the c-src and EWS neuron-specific exons share regulation by the repressor protein, PTB, and the enhancer protein, Fox-2.
FIG. 7.
RNAi of Fox-2 affects splicing of endogenous mRNAs in N2A cells. (A) Splicing of the c-src N1 exon after siRNA treatment. Cells were transfected with 1 μg of pUC plasmid and 20 pmols of the siRNA indicated at the top. After 60 h, RNA was harvested and assayed by RT-PCR. (B) Splicing of EWS 4′ exon after siRNA treatment. Similar to the conditions in panel A but assayed for EWS exon 4′. (C) Splicing of fibronectin EIIIB after siRNA treatment. The same procedure as in panel A, except with 5, 10, or 20 pmol of siRNA and with fibronectin primers. Note that an siRNA directed against Fox-1 had no effect (lanes 2 to 4), but the Fox-2 siRNA strongly increased skipping of fibronectin EIIIB (lanes 5 to 7; products labeled to the right). (D) Immunoblot of N2A whole-cell lysates after siRNA treatment. Carrier pUC DNA and siRNA were cotransfected, and whole-cell lysates were harvested after 60 h. Protein (50 μg) was separated by 10% NuPAGE, transferred to nitrocellulose, and probed with Fox-2 NT and GAPDH antibodies. The protein in lane 1 is from cells transfected with pUC plasmid only. Lanes 2 to 4 contain 1 μg pUC and 5 pmol, 10 pmol, or 20 pmol of Fox-2 siRNA, respectively. An immunoblot for Fox-1 showed no detectible expression in this experiment. GADPH is shown as a loading control.
To measure a stronger effect from the Fox siRNA on an endogenous transcript, we needed an exon that exhibited a higher starting level of exon inclusion. For this, we used the fibronectin EIIIB exon. Although not a neuron-specific exon, EIIIB is known to be dependent on UGCAUG enhancer elements, and overexpression of Fox-1 was shown to increase its inclusion in minigene transcripts (20, 22, 29). N2A cells show high levels of EIIIB inclusion (91%) (Fig. 7C, lane 1). N2A cells make little Fox-1 protein, and an siRNA targeting the Fox-1 transcript had little effect on EIIIB splicing (lanes 2 to 4). In contrast, transfection of the Fox-2 siRNA led to a large decrease in EIIIB splicing (39% exon inclusion) (lanes 5 to 7). The progressive loss of Fox-2 protein with higher doses of Fox-2 siRNA correlates with the loss of EIIIB splicing (Fig. 7C and D). Thus, the splicing of UGCAUG-dependent exons is very sensitive to the loss of endogenous Fox protein.
The loss of Fox-2 affected both endogenous N1 exon splicing and the activity of the triple UGCAUG enhancer. To examine whether the loss of endogenous Fox-2 protein specifically affected the activity of the src N1 enhancer, we cotransfected siRNAs with the pDUP4-28 minigene into N2A cells in which the N1 enhancer was active. Treatment with Fox-2 siRNA reduced enhancer activity by 50% (data not shown). Taken together, these data indicate that an endogenous Fox protein is needed for activity of the c-src intronic splicing enhancer. In N2A cells, the active family member is Fox-2.
DISCUSSION
Many neuronally regulated exons are controlled by intronic splicing enhancers containing the element UGCAUG (4, 18, 24). We show that mammalian Fox-1 and its homologue Fox-2 are specifically expressed in neurons and that both proteins activate neuron-specific exons through UGCAUG-dependent enhancers. Recently, it was demonstrated that Fox proteins could also enhance splicing of a neuron-specific exon from the nonmuscle myosin heavy chain pre-mRNA (39). Thus, the Fox proteins are likely key regulators of splicing in the nervous system.
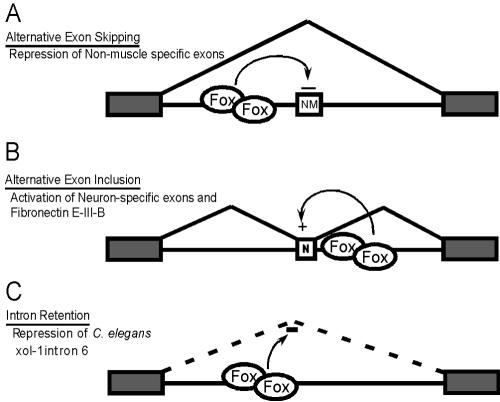
In addition to neurons, the Fox proteins also play roles in other tissues. Fibronectin exon EIIIB, whose inclusion is not neuron specific, is also dependent upon Fox proteins. The proteins are also expressed in heart and muscle (22). In muscle cells, Fox-1 was shown to act as a splicing repressor, inhibiting the splicing of exons in actinin and ATP synthase transcripts. These exons are normally skipped in muscle but used elsewhere (Fig. 8A). Interestingly, the GCAUG regulatory elements needed for this repression are upstream of the skipped exons. So far, all of the UGCAUG enhancer elements known to activate splicing are found downstream of the regulated exon (36) (Fig. 8B). Thus, the Fox proteins can apparently act either positively or negatively, depending on where they bind relative to the affected exon. Similar location dependence is seen in the SR proteins that activate splicing when bound in exons but inhibit splicing when bound to intronic elements (14, 23).
FIG. 8.
Fox proteins bind to introns to influence the splicing of adjacent exons by multiple mechanisms. (A) Fox promotes exon skipping. In muscle cells, Fox proteins bind upstream of a non-muscle-specific exon (NM) to promote skipping (22). (B) Fox promotes exon inclusion. As described here, Fox proteins bind downstream of a neuron-specific exon (N) or the fibronectin EIIIB exon and activate splicing. (C) Fox promotes intron retention. Intron 6 of the C. elegans Xol-1 transcript is spliced poorly in the presence of C. elegans Fox-1 (43). The protein may bind to this intron to decrease its splicing efficiency.
In addition to the location of the Fox protein binding site, the particular Fox isoform expressed is likely to affect its activity. In mammals, Fox-1 and Fox-2 are members of an extended family of proteins arising from multiple genes and complex alternative splicing. Recent work demonstrated that the expression of certain spliced isoforms is specific to particular tissues and that their enhancer activities are variable (39). It will thus be important to characterize which forms are expressed in which cell types and to determine whether particular isoforms are needed for the regulation of specific transcripts.
The Fox proteins have been identified in other contexts. In C. elegans, the original Fox gene is required for proper assessment of the X-to-autosome ratio for dosage compensation during sexual development (19, 33, 40, 43). Interestingly, this worm protein controls a downstream transcript, Xol-1, which exhibits alternative splicing patterns. This worm system may show an effect on splicing different from that seen in mammals. The sixth intron of Xol-1 is more efficiently spliced in the absence of Fox-1, which may cause its retention (Fig. 8C) (43). This intron contains two copies of the pentamer GCAUG, but whether this is the target sequence for the worm protein has not been reported. The RRM of the worm Fox protein is 75% identical to the mouse homologues, and its auxiliary domains have similarities to mammalian splice variants. Worm Fox-1 also functions in the adult, where it is expressed in some neurons and other cells. Thus, Fox-1 may function in the adult worm in a manner similar to its role in mammalian cells. Conversely, there may be additional embryonic functions for the mammalian protein.
The mammalian Fox proteins have also been identified in other studies. Fox-1 was identified in yeast two-hybrid screens as interacting with ataxin-2 and was called A2bp1 (42). Trinucleotide expansions in the ataxin-2 gene cause spinocerebellar ataxia type 2 (21). Interestingly, ataxin-2 also has homologues in worms and other distantly related species and is also thought to be an RNA binding protein (10, 25). A2bp1 has been reported to localize to the Golgi apparatus (26, 42). We see predominantly the nuclear localization expected for a splicing regulator. However, we cannot rule out the possibility that the cytoplasmic population is Golgi associated. It also appears that different isoforms of the protein show differential localization (39).
Interestingly, human mutations in A2bp1 were identified in a limited number of patients with an inherited epilepsy and mental retardation disorder (3). The Fox1/A2bp1 gene also lies within an autism susceptibility locus on chromosome 16 (2). Given its function described here, these mutations in Fox-1 will lead to changes in the neuronal regulation of splicing. Thus, it will be very interesting to test for changes in the splicing of UGCAUG-dependent exons in multiple forms of neurological disease, including ataxias, epilepsy, and autism. The recent implication of splicing defects in other neurological disorders, such as frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17) and myotonic dystrophy, provides an interesting precedent for this idea (15, 16).
Studies of Fox-2 (RBM9) identified it as a gene upregulated by androgens and as a repressor of tamoxifen activation of the estrogen receptor (28, 41). There are splice variants of Fox-2 that, judging from EST frequency data, appear specific to breast, ovary, and other estrogen-sensitive tissues (data not shown). Thus, hormone signaling may regulate alternative splicing through changes in Fox transcription or isoform ratios. Analysis of downstream Fox targets in these tissues is likely to uncover another important role for Fox proteins.
The mechanism of splicing activation by the Fox proteins and their interaction with other splicing regulators are also interesting directions for future studies. Fox-regulated exons often show repression by PTB, and PTB binding elements are often found adjacent to Fox sites. It will be interesting to examine whether these proteins bind in a common regulatory complex or perhaps antagonize each other's binding.
The roles of the conserved N- and C-terminal Fox domains are also unknown. Understanding their interactions with the general splicing machinery will be important in determining the mechanism of splicing regulation. Most intronic regulatory elements studied to date cluster near 5′ or 3′ splice sites and often overlap with them (5). The neuronal exons studied here have a UGCAUG element within 100 nucleotides of the alternative 5′ splice site. Many other neuron-specific exons have at least one conserved UGCAUG in this region (6). However, fibronectin exon EIIIB and calcitonin/CGRP carry important UGCAUG elements at a distance of several hundred nucleotides from the affected splice sites (18, 29). Splice site proximity and enhancer location relative to the exon are additional features that must be understood to predict Fox-regulated exons and whether Fox will be a repressor or enhancer.
UGCAUG is among the most precisely conserved splicing regulatory elements. The presence of these elements in groups of commonly regulated exons is a clue to the biological role of regulation by Fox proteins (36). This biological role is likely conserved across diverse species. Genetic analyses in the worm, fish, and mouse will yield information on their common function in the lives of these different organisms.
Supplementary Material
Acknowledgments
We thank Dan Geschwind, in whose laboratory the immunostaining was performed, and John Winkelmann and Donald McDonnell for providing Fox cDNAs. Thanks are given to members of the Black laboratory for advice, discussions, and manuscript review. We are grateful to Brent Graveley and Jiuyong Xie for comments on the manuscript.
J.G.U. was supported by a USPHS National Research Service Award (GM07185). J.D.D. was supported by a Howard Hughes Medical Institute predoctoral fellowship. This work was supported by NIH grant RO1 GM49662 to D.L.B., who is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amir-Ahmady, B., P. L. Boutz, V. Markovtsov, M. L. Phillips, and D. L. Black. 2005. Exon repression by polypyrimidine tract binding protein. RNA 11:699-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnby, G., A. Abbott, N. Sykes, A. Morris, D. E. Weeks, R. Mott, J. Lamb, A. J. Bailey, and A. P. Monaco. 2005. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am. J. Hum. Genet. 76:950-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhalla, K., H. A. Phillips, J. Crawford, O. L. McKenzie, J. C. Mulley, H. Eyre, A. E. Gardner, G. Kremmidiotis, and D. F. Callen. 2004. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J. Hum. Genet. 49:308-311. [DOI] [PubMed] [Google Scholar]
- 4.Black, D. L. 1992. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell 69:795-807. [DOI] [PubMed] [Google Scholar]
- 5.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 6.Brudno, M., M. S. Gelfand, S. Spengler, M. Zorn, I. Dubchak, and J. G. Conboy. 2001. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 29:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, R. C., and D. L. Black. 1997. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 17:4667-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, M. Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, M. Y., J. G. Underwood, J. Nikolic, M. H. Luu, and D. L. Black. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5:949-957. [DOI] [PubMed] [Google Scholar]
- 10.Ciosk, R., M. DePalma, and J. R. Priess. 2004. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131:4831-4841. [DOI] [PubMed] [Google Scholar]
- 11.Deguillien, M., S. C. Huang, M. Moriniere, N. Dreumont, E. J. Benz, Jr., and F. Baklouti. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98:3809-3816. [DOI] [PubMed] [Google Scholar]
- 12.Del Gatto, F., A. Plet, M. C. Gesnel, C. Fort, and R. Breathnach. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 14.el Ibrahim, C., T. D. Schaal, K. J. Hertel, R. Reed, and T. Maniatis. 2005. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. USA 102:5002-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 16.Grabowski, P. J., and D. L. Black. 2001. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65:289-308. [DOI] [PubMed] [Google Scholar]
- 17.Hall, M. P., S. Huang, and D. L. Black. 2004. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 15:774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedjran, F., J. M. Yeakley, G. S. Huh, R. O. Hynes, and M. G. Rosenfeld. 1997. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl. Acad. Sci. USA 94:12343-12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgkin, J., J. D. Zellan, and D. G. Albertson. 1994. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development 120:3681-3689. [DOI] [PubMed] [Google Scholar]
- 20.Huh, G. S., and R. O. Hynes. 1994. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 8:1561-1574. [DOI] [PubMed] [Google Scholar]
- 21.Imbert, G., F. Saudou, G. Yvert, D. Devys, Y. Trottier, J. M. Garnier, C. Weber, J. L. Mandel, G. Cancel, N. Abbas, A. Durr, O. Didierjean, G. Stevanin, Y. Agid, and A. Brice. 1996. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 14:285-291. [DOI] [PubMed] [Google Scholar]
- 22.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 24.Kawamoto, S. 1996. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem. 271:17613-17616. [PubMed] [Google Scholar]
- 25.Kiehl, T. R., H. Shibata, and S. M. Pulst. 2000. The ortholog of human ataxin-2 is essential for early embryonic patterning in C. elegans. J. Mol. Neurosci. 15:231-241. [DOI] [PubMed] [Google Scholar]
- 26.Kiehl, T. R., H. Shibata, T. Vo, D. P. Huynh, and S. M. Pulst. 2001. Identification and expression of a mouse ortholog of A2BP1. Mamm. Genome 12:595-601. [DOI] [PubMed] [Google Scholar]
- 27.Kubo, Y. 1991. Comparison of initial stages of muscle differentiation in rat and mouse myoblastic and mouse mesodermal stem cell lines. J. Physiol. 442:743-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman, A. P., D. L. Friedlich, G. Harmison, B. W. Howell, C. L. Jordan, S. M. Breedlove, and K. H. Fischbeck. 2001. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-2 and fox-1. Biochem. Biophys. Res. Commun. 282:499-506. [DOI] [PubMed] [Google Scholar]
- 29.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markovtsov, V., J. M. Nikolic, J. A. Goldman, C. W. Turck, M. Y. Chou, and D. L. Black. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7463-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melot, T., L. Dauphinot, N. Sevenet, F. Radvanyi, and O. Delattre. 2001. Characterization of a new brain-specific isoform of the EWS oncoprotein. Eur. J. Biochem. 268:3483-3489. [DOI] [PubMed] [Google Scholar]
- 32.Meneely, P. M. 1997. The chromosomal signal for sex determination in Caenorhabditis elegans. Bioessays 19:945-948. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, B. J. 2000. Sex in the wormcounting and compensating X-chromosome dose. Trends Genet. 16:247-253. [DOI] [PubMed] [Google Scholar]
- 34.Min, H., R. C. Chan, and D. L. Black. 1995. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9:2659-2671. [DOI] [PubMed] [Google Scholar]
- 35.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 36.Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak, and J. G. Conboy. 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33:714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modafferi, E. F., and D. L. Black. 1999. Combinatorial control of a neuron-specific exon. RNA 5:687-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33:2078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicoll, M., C. C. Akerib, and B. J. Meyer. 1997. X-chromosome-counting mechanisms that determine nematode sex. Nature 388:200-204. [DOI] [PubMed] [Google Scholar]
- 41.Norris, J. D., D. Fan, A. Sherk, and D. P. McDonnell. 2002. A negative coregulator for the human ER. Mol. Endocrinol. 16:459-468. [DOI] [PubMed] [Google Scholar]
- 41a.Rooke, N., V. Markovtsov, E. Cagavi, and D. L. Black. 2003. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata, H., D. P. Huynh, and S. M. Pulst. 2000. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum. Mol. Genet. 9:1303-1313. [DOI] [PubMed] [Google Scholar]
- 43.Skipper, M., C. A. Milne, and J. Hodgkin. 1999. Genetic and molecular analysis of fox-1, a numerator element involved in Caenorhabditis elegans primary sex determination. Genetics 151:617-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamm, S., S. Ben-Ari, I. Rafalska, Y. Tang, Z. Zhang, D. Toiber, T. A. Thanaraj, and H. Soreq. 2005. Function of alternative splicing. Gene 344:1-20. [DOI] [PubMed] [Google Scholar]
- 45.Underwood, J. G., and D. L. Black. Unpublished observations.
- 46.Winkelmann, J. C., W. Chen, Z. L. Chu, and R. I. Blough. 2001. Presented at the Cold Spring Harbor Meeting on Eukaryotic RNA Processing, Cold Spring Harbor, N.Y., August, 2001.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.