Abstract
The Saccharomyces cerevisiae Trf4 and Trf5 proteins are members of a distinct family of eukaryotic DNA polymerase β-like nucleotidyltransferases, and a template-dependent DNA polymerase activity has been reported for Trf4. To define the nucleotidyltransferase activities associated with Trf4 and Tr5, we purified these proteins from yeast cells and show that whereas both proteins exhibit a robust poly(A) polymerase activity, neither of them shows any evidence of a DNA polymerase activity. The poly(A) polymerase activity, as determined for Trf4, is strictly Mn2+ dependent and highly ATP specific, incorporating AMP onto the free 3′-hydroxyl end of an RNA primer. Unlike the related poly(A) polymerases from other eukaryotes, which are located in the cytoplasm and regulate the stability and translation efficiency of specific mRNAs, the Trf4 and Trf5 proteins are nuclear, and a multiprotein complex associated with Trf4 has been recently shown to polyadenylate a variety of misfolded or inappropriately expressed RNAs which activate their degradation by the exosome. To account for the effects of Trf4/Trf5 proteins on the various aspects of DNA metabolism, including chromosome condensation, DNA replication, and sister chromatid cohesion, we suggest an additional and essential role for the Trf4 and Trf5 protein complexes in generating functional mRNA poly(A) tails in the nucleus.
The TRF4 gene of Saccharomyces cerevisiae was first identified in a genetic screen for mutations that are synthetically lethal with mutations in DNA topoisomerase I (20). TRF5 is a homolog of TRF4, and deletion of both genes is lethal, indicating that their functions are overlapping and essential for cell viability (5). A role for TRF4 in chromosome condensation is indicated from the observation that a top1 trf4 ts double mutant fails to establish and maintain chromosome condensation in rRNA genes at mitosis (4). Furthermore, the Trf4 protein interacts physically with the Smc1 and Smc2 proteins that bind chromosomes and cause condensation (4). TRF4 and TRF5 have also been suggested to have an essential role in the coordination between DNA replication and sister chromatid cohesion (25). In another study, the deletion of TRF4 alone was found to have no effect on sister chromatid cohesion in mitotic metaphase cells or on chromosome segregation during meiosis; in this study, however, the possibility that TRF5 compensates for the absence of TRF4 has not been excluded (18).
Trf4 and Trf5 are members of a distinct family of eukaryotic DNA polymerase (Pol) β-like nucleotidyltransferases (1), and a template-dependent DNA polymerase activity has been reported for Trf4 and named DNA polymerase σ (3, 25). More recently, the Trf4 and Trf5 proteins have been shown to interact with the C-terminal domain of Pol2, the catalytic subunit of Polɛ, and stimulation of Polɛ DNA synthetic activity was observed in the presence of Trf4 (7). In keeping with these observations, the Trf4/Trf5 proteins are located in the nucleus (12, 23).
The Trf4/Trf5-related proteins from other eukaryotes, however, such as Cid1 and Cid13 from Schizosaccharomyces pombe, have been shown to have a poly(A) polymerase (PAP) activity and they exhibit no DNA polymerase activity (19). For example, Cid1 readily extends a 15-nucleotide (nt)-long synthetic RNA substrate (A15) in the presence of ATP and Mg2+, but it is inactive on dT25/dA40 DNA polymerase substrate in the presence of all four deoxynucleoside triphosphates (dNTPs) and Mg2+ (19). Cid13 specifically targets suc22 mRNA that encodes a subunit of ribonucleotide reductase, and cid13 mutants have reduced dNTP pools and are sensitive to hydroxyurea, a ribonucleotide reductase inhibitor (21). The Cid1 and Cid13 proteins are located in the cytoplasm where they presumably affect the stability and activation of specific mRNAs (19, 21).
The GLD-2 and GLD-3 proteins control different aspects of germ line development in Caenorhabditis elegans. GLD-2 is a member of the Trf4/Trf5 family, and GLD-3 belongs to the bicaudal C family of RNA binding proteins, and both proteins are located in the cytoplasm of germ line and early embryonic cells (24). GLD-2 alone shows a very low poly(A) polymerase activity, but this activity is greatly stimulated by GLD-3, which by itself has no PAP activity. Thus, GLD-2 and GLD-3 represent a new type of cytoplasmic PAP in which GLD-2 provides the catalytic subunit and GLD-3 provides the RNA binding function (14, 24).
Because of the published evidence indicating that the S. cerevisiae Trf4/Trf5 proteins are DNA polymerases and because of our interest in DNA polymerases and in translesion DNA synthesis, we sought to define the DNA polymerase activity of Trf proteins further. For this purpose, we purified the Trf4 and Trf5 proteins to near homogeneity from yeast cells but have found no evidence for a DNA polymerase activity associated with either of these proteins. Rather, both proteins have a robust poly(A) polymerase activity, which we characterize in some detail. We discuss the implications of our observations on TRF4/TRF5-encoded PAP activity in relation to the more recent studies that have been reported for these proteins. We also speculate on the possible connections of Trf4/Trf5 PAP activity with the phenotypic consequences of mutations in these genes.
MATERIALS AND METHODS
Proteins.
The S. cerevisiae TRF4 and TRF5 open reading frames (ORFs) were amplified by PCR, and each ORF was sequenced to confirm the integrity of the sequence. The TRF4 and TRF5 ORFs were then cloned in frame with the glutathione S-transferase (GST) gene in plasmid pBJ842. Plasmids pPOL71 and pPOL118 harbor fusions of TRF4 and TRF5 with GST, respectively, under the control of a galactose-inducible phosphoglycerate kinase promoter. The GST-Trf4 and GST-Trf5 proteins were overexpressed from plasmids pPol71 and pPol118 in the protease-deficient yeast strain BJ5464. Cells were grown at 30°C to stationary phase in complete synthetic medium lacking leucine to select for the plasmid. The culture was then diluted 10-fold in fresh medium lacking dextrose but containing 2% glycerol and 2% lactic acid, followed by overnight incubation. Expression of recombinant protein was induced by adding 2% galactose, followed by further propagation for 7 h at 30°C. Cells were harvested by centrifugation and disrupted by a bead beater in buffer A (40 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol [DTT], 0.01% Nonidet P-40 [NP-40], 10% glycerol, 5 mM EDTA) supplemented with 1 M NaCl and protease inhibitor mixture (Mini-Complete; Roche, Indianapolis, IN). After clarification of the crude extract by ultracentrifugation, it was loaded onto a glutathione-Sepharose column (Amersham Pharmacia Biotech). First, the column was washed with buffer A plus 1 M NaCl followed by washing with buffer A plus 100 mM NaCl. Native Trf4 and Trf5 proteins were eluted in buffer A plus 100 mM NaCl by PreScission protease, which cleaved in the spacer region between GST and Trf4 or Trf5 protein and left a 7-amino-acid leader peptide (GPGGDPH) attached to the N-terminal ends of the purified proteins. Purified Trf4 and Trf5 proteins were frozen in aliquots under liquid nitrogen and kept at −70°C. We also generated the catalytic mutation of TRF4, in which the active-site aspartate residues 236 and 238 were replaced by alanine, using the QuikChange site-directed mutagenesis kit (Stratagene Corporation, La Jolla, California) and mutagenic oligonucleotides. The mutant Trf4 DD236,238,AA protein was then purified in parallel with the wild-type Trf4 and Trf5 proteins.
The T4 DNA polymerase was purchased from Roche (Indianapolis, IN).
Poly(A) polymerase and DNA polymerase substrates.
Poly(A), poly(C), and poly(dA) polynucleotides and 12- to 18-nt-long poly(dT) were purchased from Amersham Pharmacia Biotech. The 40-nt-long RNA 5′-GUU UUC CCA GUC ACG ACG AUG CUC CGG UAC UCC AGU GUA G-3′ was synthesized by Oligos Etc., Inc. DNA oligonucleotides were synthesized by Midland Certified Reagent Co. (Midland, TX). The 29/75-nt DNA polymerase substrate was generated by annealing a 75-nt oligomer template, 5′-AGC AAG TCA CCA ATG TCT AAG AGT TCG TAT TAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′, to the 5′-32P-labeled oligonucleotide primer 5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TA-3′. The oligo(dT)/poly(dA) DNA polymerase substrate was generated by annealing a 3′-32P-labeled mixture of 12- to 18-nt-long oligo(dT) primers to poly(dA) template.
DNA polymerase assays.
The standard DNA polymerase reaction (10 μl) contained 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 20 mM NaCl, 20 mM KCl, 2 mM DTT, 2.5% glycerol, 200 μg/ml bovine serum albumin, and 100 μM of each dGTP, dATP, dTTP, and dCTP. As indicated in the figure legends, mutant and wild-type Trf4 and Trf5 proteins (10 nM) were mixed with 29/75-nt or oligo(dT)/poly(dA) primer/template DNA (20 nM). Assay mixtures were assembled on ice and incubated at 30°C for 10 min, and the assays were stopped by the addition of loading buffer (40 μl) containing 20 mM EDTA, 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were resolved on 15% polyacrylamide gels containing 8 M urea. Visualization of the results was done using a Molecular Dynamics STORM PhosphorImager and ImageQuant software.
Poly(A) polymerase assays.
Poly(A) polymerase assays were performed in 10-μl reaction buffer containing 25 mM Tris-HCl, pH 8.3, 0.5 mM MnCl2, 40 mM KCl, 0.5 mM DTT, 0.01% NP-40, 10% glycerol, 200 μg/ml bovine serum albumin, 50 μM [α-32P]ATP, 20 nM poly(A), and 10 nM of Trf4 or Trf5 protein. As indicated in the figure legends, in some particular experiments, we modified the components of the above reaction in the following ways: both 0.5 mM MnCl2 and 5 mM MgCl2 were added at the same time (see Fig. 2C); 5 mM MgCl2 replaced MnCl2 (see Fig. 3A); the reaction mixture contained 50 μM of either [α-32P]ATP, [α-32P]GTP, [α-32P]UTP, or [α-32P]CTP (see Fig. 3B); either poly(A), poly(C), or poly(dA) was used (see Fig. 3C), instead of poly(A), the 29-nt synthetic RNA was used in the presence of increasing concentrations of single α-32P-labeled nucleotide triphosphate (see Fig. 4). Assay mixtures were assembled on ice and incubated at 30°C for 10 min, and the assays were stopped by the addition of loading buffer (40 μl) containing 20 mM EDTA, 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were resolved on 15% polyacrylamide gels containing 8 M urea. Visualization of the results was done using a Molecular Dynamics STORM PhosphorImager and ImageQuant software.
FIG. 2.
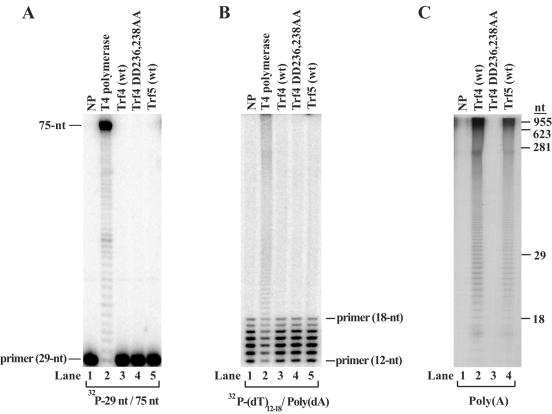
Trf4 and Trf5 exhibit a poly(A) polymerase activity but no DNA polymerase activity. (A) DNA polymerase assays on a 29/75-nt primer/template partial duplex DNA substrate. The 29-nt primer was 32P labeled at the 5′ end and annealed to a 75-nt template. Wild-type (wt) Trf4 (lane 3), Trf4 DD236,238AA (lane 4), and wild-type Trf5 (lane 5) (10 nM each) were incubated with the DNA substrate (20 nM) in the presence of each of the four dNTPs (100 μM). For positive and negative controls, parallel reactions were also carried out with T4 DNA polymerase (lane 2), and no added protein (NP) (lane 1). Reaction products were analyzed on a 15% polyacrylamide gel containing 8 M urea and analyzed by a PhosphorImager. (B) DNA polymerase assays on an oligo(dT)/poly(dA) DNA substrate. A mixture of 12- to 18-nt oligo(dT) primers was 5′ end labeled and annealed to poly(dA) template. DNA polymerase reactions were carried out as described for panel A. (C) Poly(A) polymerase assays on a poly(A) substrate. Trf4 (lane 2), Trf4 DD236,238AA (lane 3), and Trf5 (lane 4) (10 nM each) were incubated with poly(A) RNA and [α-32P]ATP (50 μM) in the presence of 5 mM Mg2+ and 0.5 mM Mn2+. A control reaction (lane 1) included no added protein (NP). Reaction products were resolved on a 15% polyacrylamide gel containing 8 M urea followed by PhosphorImager analyses of the incorporation of AMP into RNA tails. The sizes of the reaction products (in nucleotides) are indicated to the right of the gel.
FIG. 3.
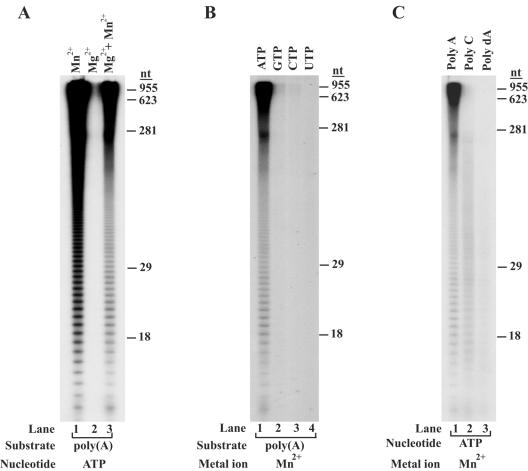
Characterization of the poly(A) polymerase activity of Trf4. (A) Mg2+ versus Mn2+ dependence. Trf4 (10 nM) was incubated with poly(A) RNA and [α-32P]ATP (50 μM) in the presence of either Mn2+ (0.5 mM; lane 1), Mg2+ (5 mM; lane 2), or both Mn2+ and Mg2+ ions (0.5 mM and 5 mM, respectively; lane 3). (B) Nucleotide substrate dependence. Trf4 (10 nM) was incubated with poly(A) RNA in the presence of Mn2+ (0.5 mM) and with either [α-32P]ATP (lane 1), [α-32P]GTP (lane 2), [α-32P]CTP (lane 3), or [α-32P]UTP (lane 4) (50 μM each). (C) Polynucleotide substrate dependence. Trf4 (10 nM) was incubated in the presence of [α-32P]ATP (50 μM) and 0.5 mM Mn2+ with either poly(A) (lane 1), poly(C) (lane 2), or poly(dA) (lane 3). The sizes of the reaction products (in nucleotides) are indicated to the right of the gels in panels A, B, and C.
FIG. 4.
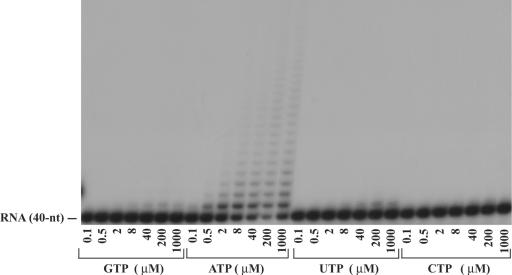
Nucleotide incorporation into the 3′ end of a synthetic RNA by Trf4. Trf4 (10 nM) was incubated with a synthetic 40-mer RNA substrate (20 nM) and increasing concentration of a single nucleotide for 5 min at 30°C. Reaction products were resolved on a 15% polyacrylamide gel containing 8 M urea followed by PhosphorImager analysis.
RESULTS
Purification of wild-type and active-site mutant Trf4 and Trf5 proteins.
S. cerevisiae Trf4 and Trf5 proteins belong to the Pol β-nucleotidyltransferase superfamily, and a DNA polymerase activity has been reported for Trf4 (25). This finding was congruent with genetic studies indicating an involvement of TRF4 and TRF5 in many DNA metabolic events, including chromosome condensation, DNA replication, and sister chromatid cohesion. Also, the fact that Trf4 and Trf5 are nuclear proteins was in accord with these observations.
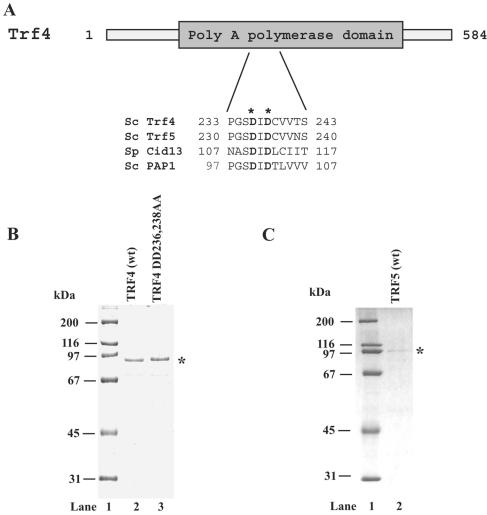
To facilitate the purification of Trf4 and Trf5 proteins, the TRF4 and TRF5 cDNAs were fused downstream of the GST gene and expressed in a protease-deficient S. cerevisiae strain. GST-Trf4 and GST-Trf5 proteins were purified on glutathione beads, followed by elution of native Trf4 and Trf5 with PreScission protease, which removed the GST tag. For a control, we generated a cDNA encoding a mutant Trf4 in which the aspartate residues 236 and 238 have been replaced by alanine. These residues correspond to the conserved amino acids shown to be essential for catalysis in many Pol β-family members (Fig. 1A). The mutant Trf4 DD236,238,AA was purified in parallel with the wild-type proteins. On denaturing polyacrylamide gels, both the wild-type and mutant Trf4 proteins migrate at ∼90 kDa, while the Trf5 protein migrates at ∼97 kDa. The fact that the predicted molecular masses of Trf4 and Trf5 proteins are 66 kDa and 73 kDa, respectively, suggests that the proteins possess features that confer a lower electrophoretic mobility on denaturing polyacrylamide gels. Coomassie blue staining of the gels revealed that the wild-type and mutant Trf4 proteins were purified to >95% homogeneity, whereas Trf5, which was expressed at a lower level in yeast cells than the Trf4 protein, was ∼85% pure. Nevertheless, because of the parallel purifications, the Trf4 DD236,237AA-containing sample could serve as a control in enzymatic assays for both Trf4 and Trf5.
FIG. 1.
Purity of wild-type Trf4 and Trf5 proteins and of active-site mutant Trf4 protein. (A) Schematic representation of Trf4. The conserved sequence motif characteristic for the Pol β family is shown as a gray box. A highly conserved region known to be involved in metal binding and catalysis was aligned for the S. cerevisiae (Sc) Trf4 and Trf5, S. pombe (Sp) Cid13, and the S. cerevisiae canonical PAP enzyme. The conserved catalytic aspartates at positions 236 and 238 in Trf4, which correspond to the metal-binding aspartates at positions 100 and 102 in the canonical PAP are indicated by asterisks. (B) Purified Trf4 and mutant Trf4 DD236,238AA, in which aspartates at positions 236 and 238 have been changed to alanines, were analyzed on a 10% denaturing polyacrylamide gel and stained with Coomassie blue. The position of the purified proteins is indicated by an asterisk. Lane 1, molecular mass standards; lane 2, 0.5 μg of wild-type (wt) Trf4 protein; lane 3, 0.5 μg of Trf4 DD236,238AA protein. (C) Purified Trf5 protein (150 ng). The position of purified Trf5 is indicated by an asterisk.
Trf4 and Trf5 have no DNA polymerase activity.
To test whether Trf4 and Trf5 possess DNA polymerase activity, first we used a DNA substrate containing a 75-nt template oligomer primed with a 5′-32P-labeled 29-nt oligomer and incubated the purified proteins with the DNA substrate in the presence of all four deoxynucleotides (Fig. 2A). Whereas the control reaction with T4 polymerase resulted in the extension of the primer up to the end of the template (Fig. 2A, lane 2), Trf4 and Trf5 did not exhibit any DNA polymerase activity (Fig. 2A, lanes 3 and 5). Because in the original study, the DNA polymerase activity of Trf4 was examined on an oligo(dT)/poly(dA) DNA substrate (25), we considered the possibility that Trf4 had some specificity for this substrate. Therefore, we examined the DNA polymerase activity using oligo(dT)/poly(dA) DNA, but again, in contrast to the robust DNA polymerase activity of the control T4 DNA polymerase, neither Trf4 nor Trf5 exhibited any detectable DNA polymerase activity (Fig. 2B, compare lane 2 to lanes 3 and 4).
PCNA, along with replication factor C (RFC) and replication protein A (RPA), greatly stimulates the DNA synthetic activity of many DNA polymerases. To examine the possibility that PCNA, RFC, and RPA activate DNA synthesis by Trf4 and Trf5, we used the assay conditions and DNA substrate we have employed previously for studies on PCNA-dependent stimulation of translesion synthesis of DNA polymerases (8-11). The Trf4 or Trf5 protein was incubated in the presence of PCNA, RFC, and RPA, all four deoxynucleotides, and a DNA substrate, in which the 75-nt template was bound to biotin at both the ends, and following its annealing to the 5′-32P-labeled oligonucleotide primer, both of these biotins were coupled to streptavidin to prevent the sliding of PCNA from the ends after it has been loaded onto the DNA by RFC. Trf4 and Trf5, however, did not exhibit any DNA polymerase activity even in the presence of PCNA, RFC, and RPA, whereas a robust stimulation of yeast Polη was observed by these protein factors in the control reaction (data not shown).
Trf4 and Trf5 exhibit a robust poly(A) polymerase activity.
To examine whether Trf4 and Trf5 have a poly(A) polymerase activity, we incubated these proteins with single-stranded poly(A) RNA and [α-32P]ATP. Since a particular poly(A) polymerase may require either Mg2+ or Mn2+ for its activity, first we carried out the reactions in the presence of both metal ions. As shown in Fig. 2C, Trf4 as well as Trf5 incorporated AMP into poly(A) during the incubation. Since a similar reaction with the control Trf4 DD236,238AA protein did not result in any AMP incorporation (Fig. 2C, lane 3), this confirmed that the poly(A) polymerase activity was intrinsic to both the Trf4 and Trf5 proteins. This experiment not only establishes a poly(A) polymerase activity for the Trf4 and Trf5 proteins but also indicates that the lack of DNA polymerase activity with either of these proteins cannot be attributed to the inactivity of our protein samples.
Characterization of poly(A) polymerase activity of Trf4.
To explore the enzymatic properties of Trf4 further, we tested it for metal ion, nucleotide substrate, and RNA substrate dependence. As shown in Fig. 3A, in the presence of ATP and poly(A), Trf4 required Mn2+ for its poly(A) polymerase activity. Mn2+ could not be replaced by Mg2+, as there was no detectable activity when Mg2+ was used as the sole metal ion (Fig. 3A, compare lanes 1 to 3).
A high degree of specificity for ATP over the other nucleotides is a characteristic feature of all the previously characterized poly(A) polymerases. In accordance with this, we observed that Trf4 also shows a high degree of selectivity for AMP incorporation into poly(A) RNA, and GTP, CTP, and UTP were incorporated very poorly (Fig. 3B).
To test whether Trf4 can extend only a preexisting poly(A) tail or whether it can initiate AMP incorporation into other polynucleotides as well, we examined the activity of Trf4 using either poly(C) or poly(dA) polynucleotide substrate. As shown in Fig. 3C, Trf4 preferentially extended the poly(A) substrate, but it incorporated AMP into the poly(C) substrate also, although not as well (Fig. 3C, compare lanes 1 and 2). In contrast, Trf4 was very inefficient on the poly(dA) substrate.
To gain more insight into the specificity of Trf4 for its nucleotide substrate, we examined its poly(A) polymerase activity in the presence of various concentrations of single nucleotides. Trf4 was incubated with a synthetic 40-mer 5′-32P-labeled single-stranded RNA in the presence of increasing concentrations of a single ribonucleotide, GTP, ATP, UTP, or CTP. As shown in Fig. 4, although Trf4 could use all four nucleotides, GTP, UTP, and CTP were incorporated very poorly, and at most, one residue was incorporated. In contrast, with ATP, incorporation of up to ∼20 AMP residues could be detected. In addition, in our assay conditions, incorporation of AMP was detected at as low as 0.5 μM ATP concentration, while GMP, UMP, and CMP incorporation required much higher concentrations. Thus, Trf4 exhibits a high degree of selectivity for ATP. This enzymatic property of Trf4, taken together with the fact that ATP is the most abundant nucleotide in vivo, indicates that Trf4 functions solely as a poly(A) polymerase.
DISCUSSION
Although Trf4 has been reported to have a DNA polymerase activity, we have found no evidence for such an activity with the Trf4 or Trf5 protein, either alone or in the presence of PCNA, RFC, and RPA. Since the purified Trf4 and Trf5 proteins both exhibit a robust poly(A) polymerase activity, their lack of DNA polymerase activity cannot be ascribed to our protein preparations being rendered inactive during the purification protocol; instead, our observations provide strong evidence that the DNA polymerase activity observed with Trf4 previously was not intrinsic to the protein. This conclusion is also supported by the fact that the active-site Trf4 mutant protein in which the aspartate 236 and 238 residues had been changed to alanines still exhibited a considerable level of DNA polymerase activity (25).
The poly(A) polymerase activity of Trf proteins, as characterized here for Trf4, shows a strong dependence upon Mn2+, is inactive in the presence of Mg2+, and shows a high degree of specificity for ATP, incorporating AMP onto the free 3′-hydroxyl end of an RNA primer. In many respects, the Trf proteins resemble the eukaryotic canonical poly(A) polymerase, which like the S. cerevisiae Trf proteins, is nuclear and carries out the polyadenylation of mRNAs in the nucleus (6). Attachment of poly(A) tails facilitates the export of mRNAs from the nucleus, increases the stability of mRNAs in the cytoplasm, and increases the efficiency of translation initiation (6).
The crystal structures of canonical yeast and bovine poly(A) polymerases have indicated that the polymerase is a U-shaped molecule comprised of three domains: the N-terminal catalytic domain, which is structurally similar to the palm domain of DNA polymerases (2, 16, 17) and harbors the three conserved acidic residues that coordinate the two metal ions required for catalysis; the central domain, which like the fingers domain of DNA polymerases, orients the incoming ATP; and a C-terminal domain, which contains an RNA binding domain (2, 16, 17). Thus, a canonical poly(A) polymerase can bind the RNA substrate at the 3′ end via its C terminus and carry out poly(A) addition onto the mRNAs.
In contrast to the tripartite domain organization of canonical PAP, the GLD-2 protein of C. elegans has been suggested to be a bipartite domain PAP, where GLD-2 contributes the catalytic N-terminal and the central domains, and GLD-3 contributes the RNA binding domain (14, 24). In a number of studies that have been published very recently, the yeast Trf4 protein has also been suggested to similarly provide the catalytic subunit of a poly(A) polymerase complex in which the Air1 and Air2 proteins constitute the RNA binding subunits (15, 22, 26). Briefly, in all these studies, a poly(A) polymerase activity was observed with the tandem affinity-purified (TAP)-Trf4 multisubunit protein complex which contained the Air1 and Air2 proteins, in addition to other proteins, whereas no poly(A) polymerase activity was observed with the purified TAP-Trf5 multiprotein complex. Overall, these studies have indicated that Trf4 alone has no polyadenylation activity; this Trf4 activity is expressed only together with the Air1 or the Air2 protein, and simultaneous addition of both these proteins leads to a higher level of poly(A) addition activity than with either protein alone. These observations have led to the suggestion that Trf4 is the catalytic subunit of a multiprotein complex in which the Air1 and Air2 proteins confer poly(A) addition activity to the Trf4 subunit by providing the RNA binding domain (15, 22, 26).
By contrast to the above noted observations for the Trf4 and Trf5 proteins, we show here that both the proteins possess a robust poly(A) polymerase activity and they require no additional proteins for its expression. From our observations, we infer that like the canonical PAP, Trf4 and Trf5 are three-domain proteins and they contain, in addition to the N-terminal catalytic and the central ATP binding domains, an RNA binding domain as well.
A role for the Trf4 complex, containing Trf4, Air1, Air2, and a putative RNA helicase Mtr4, has been indicated in the polyadenylation of small nuclear RNAs, small nucleolar RNAs, pre-rRNA, and rRNA. The addition of poly(A) tails onto these RNAs activates processive 3′ degradation by the exosome (15). The Trf4 complex-mediated polyadenylation is responsible for exosomal degradation of PolII transcripts that result from the adventitious transcription of silent intergenic regions in the genome (26), and the polyadenylation activity of the Trf4 complex also stimulates the degradation of unmodified tRNAs (13, 22). Thus, Trf4-mediated polyadenylation promotes the exosomal degradation of a variety of RNAs that are incorrectly folded or that result from inappropriate expression of genetic information. It remains unclear, however, how any of these activities could account for the roles of Trf4 and Trf5 proteins in the various aspects of DNA metabolism, including chromosome condensation, DNA replication, and sister chromatid cohesion. One possibility is that Trf4 and Trf5 play an additional and essential role in generating functional mRNA poly(A) tails in the nucleus. Such a role for the Trf proteins would then be in addition to the role of canonical PAP, and in this role the Trf protein complexes could either act alone or in conjunction with the canonical PAP.
Acknowledgments
We thank Dianne Johnson for technical assistance in the cloning of the TRF4 and TRF5 genes.
This work was supported by National Institutes of Health Sciences grant CA107650, the Wellcome Trust International Senior Research fellowship, the Hungarian Science Foundation grant (OTKA T043354), and the Marie Curie International Reintegration grant.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bard, J., A. M. Zhelkovsky, S. Helmling, T. N. Earnest, C. L. Moore, and A. Bohm. 2000. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science 289:1346-1349. [DOI] [PubMed] [Google Scholar]
- 3.Burgers, P. M., E. V. Koonin, E. Bruford, L. Blanco, K. C. Burtis, M. F. Christman, W. C. Copeland, E. C. Friedberg, F. Hanoka, D. C. Hinkle, C. W. Lawrence, M. Nakanishi, H. Ohmori, L. Prakash, S. Prakash, R. Claude-Agnes, A. Sugino, T. Todo, Z. Wang, J.-C. Weill, and R. Woodgate. 2001. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276:43487-43490. [DOI] [PubMed] [Google Scholar]
- 4.Castano, I. B., P. M. Brzoska, B. U. Sadoff, H. Chen, and M. F. Christman. 1996. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 10:2564-2576. [DOI] [PubMed] [Google Scholar]
- 5.Castano, I. B., S. Heath-Pagliuso, B. U. Sadoff, J. Fitzhugh, and M. F. Christman. 1996. A novel family of TRF (DNA topisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 24:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, S., C. M. Li, D. L. Levy, J. Brown, P. M. Snow, and J. L. Campbell. 2003. Saccharomyces cerevisiae DNA polymerase ɛ and polymerase σ interact physically and functionally, suggesting a role for polymerase ɛ in sister chromatid cohesion. Mol. Cell. Biol. 23:2733-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haracska, L., R. E. Johnson, I. Unk, B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21:7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haracska, L., C. M. Kondratick, I. Unk, S. Prakash, and L. Prakash. 2001. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8:407-415. [DOI] [PubMed] [Google Scholar]
- 11.Haracska, L., I. Unk, R. E. Johnson, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2002. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huh, W.-K., J. V. Falvo, L. C. Gerke, A. D. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature (London) 425:686-691. [DOI] [PubMed] [Google Scholar]
- 13.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebush, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAmet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller, W., and G. Martin. 2002. Reviving the message. Nature (London) 419:267-268. [DOI] [PubMed] [Google Scholar]
- 15.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 16.Martin, G., W. Keller, and S. Doublie. 2000. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19:4193-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, G., A. Moglich, W. Keller, and S. Doublie. 2004. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 341:911-925. [DOI] [PubMed] [Google Scholar]
- 18.Petronczki, M., B. Chwalla, M. F. Siomos, S. Yokobayashi, W. Helmhart, A. M. Deutschbauer, R. W. Davis, Y. Watanabe, and K. Nasmyth. 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-α-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 117:3547-3559. [DOI] [PubMed] [Google Scholar]
- 19.Read, R. L., R. G. Martinho, S.-W. Wang, A. M. Carr, and C. J. Norbury. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99:12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadoff, B. U., S. Heath-Pagliuso, I. B. Castano, Y. Zhu, F. S. Kieff, and M. F. Christman. 1995. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 141:465-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitoh, S., A. Chabes, W. H. McDonald, L. Thelander, J. R. Yates III, and P. Russell. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109:563-573. [DOI] [PubMed] [Google Scholar]
- 22.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedelin, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:986-997. [First published 19 April 2005; 10.1371/journal.pbio.0030189.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walowsky, C., D. J. Fitzhugh, I. B. Castano, J. Y. Ju, N. A. Levin, and M. F. Christman. 1999. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J. Biol. Chem. 274:7302-7308. [DOI] [PubMed] [Google Scholar]
- 24.Wang, L., C. R. Eckmann, L. C. Kadyk, M. Wickens, and J. Kimble. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature (London) 419:312-316. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Z., I. B. Castano, A. de las Penas, C. Adams, and M. F. Christman. 2000. Pol κ: a DNA polymerase required for sister chromatic cohesion. Science 289:774-779. [DOI] [PubMed] [Google Scholar]
- 26.Wyers, F., M. Rougemaille, G. Badis, J.-C. Rouselle, M.-E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]