Abstract
Xeroderma pigmentosum is characterized by increased sensitivity of the affected individuals to sunlight and light-induced skin cancers and, in some cases, to neurological abnormalities. The disease is caused by a mutation in genes XPA through XPG and the XP variant (XPV) gene. The proteins encoded by the XPA, -B, -C, -D, -F, and -G genes are required for nucleotide excision repair, and the XPV gene encodes DNA polymerase eta, which carries out translesion DNA synthesis. In contrast, the mechanism by which the XPE gene product prevents sunlight-induced cancers is not known. The gene (XPE/DDB2) encodes the small subunit of a heterodimeric DNA binding protein with high affinity to UV-damaged DNA (UV-damaged DNA binding protein [UV-DDB]). The DDB2 protein exists in at least four forms in the cell: monomeric DDB2, DDB1-DDB2 heterodimer (UV-DDB), and as a protein associated with both the Cullin 4A (CUL4A) complex and the COP9 signalosome. To better define the role of DDB2 in the cellular response to DNA damage, we purified all four forms of DDB2 and analyzed their DNA binding properties and their effects on mammalian nucleotide excision repair. We find that DDB2 has an intrinsic damaged DNA binding activity and that under our assay conditions neither DDB2 nor complexes that contain DDB2 (UV-DDB, CUL4A, and COP9) participate in nucleotide excision repair carried out by the six-factor human excision nuclease.
UV-damaged DNA binding protein (UV-DDB) is a heterodimer of DDB1 (p127) plus DDB2 (p48) (23) that binds with high affinity to DNA damaged by UV and other physical and chemical agents (7) and is lacking in xeroderma pigmentosum group E (XP-E) patients (3) because of mutations in the DDB2 subunit (20, 33). Hence we will use XPE or DDB2 synonymously. Because of the high affinity of UV-DDB to UV-damaged DNA, in particular to UV-induced (6-4) photoproducts (37), the complex has been implicated in the damage recognition step of nucleotide excision repair (46). However, XP-E is the mildest form of xeroderma pigmentosum (4, 25), and in vivo studies with XP-E strains have raised some doubts about the role of XPE in excision repair (15, 20, 21, 24, 51). In vitro studies aimed at resolving the issue of the role of UV-DDB in excision repair have also resulted in some conflicting reports but on the whole support the view that UV-DDB does not directly participate in excision repair (39, 42). First, the human excision nuclease capable of excising Pyr<>Pyr and (6-4) photoproducts at relative rates comparable to the in vivo relative rates has been reconstituted in the absence of UV-DDB with six excision repair factors consisting of RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1 (30, 31). Second, addition of UV-DDB to the six-factor excision nuclease (22) or XP-E cell extracts (36) had no effect on the rate of excision of a (6-4) photoproduct or cisplatin-GpTpG diadduct, respectively, and at high concentration inhibited the excision of both adducts. However, a subsequent study found that the excision of Pyr<>Pyr by the six-factor excision nuclease was stimulated up to 17-fold by high concentrations of UV-DDB and that of (6-4) photoproducts was stimulated about 2-fold by low concentrations of UV-DDB (47) under conditions of low overall excision. Because the excision assay is very sensitive to reaction conditions, especially when the excision level is low, a systematic analysis of UV-DDB on excision of Pyr<>Pyr and (6-4) photoproducts was then carried out with the six-factor excision nuclease and it was found that UV-DDB did not stimulate the excision of either Pyr<>Pyr or (6-4) photoproducts under any circumstance and inhibited excision of the (6-4) photoproduct at relatively low concentrations and that of Pyr<>Pyr at much higher concentrations of UV-DDB (38).
In contrast to these generally negative data linking XPE to excision nuclease activity, a number of studies have indicated that UV-DDB is a multifunctional protein involved in several cellular processes, including transcription through association of DDB2 with E2F1 (12) and the transcriptional coactivators CBP/p300 and the STAGA complex (5, 27), cell cycle regulation (12, 14), and chromosome segregation (35). In addition, there appears to be a complex regulatory circuit between p53 and DDB2 (15, 18, 19), implicating UV-DDB in apoptosis. Finally, UV-DDB has been found to be associated with the Cullin 4A (CUL4A) complex (2, 32, 44) and the COP9 signalosome (10), which also includes the Cullin 4A complex that ubiquitinates proteins, including DDB2 and XPC, and targets them for degradation (2, 32, 50). Following UV damage, the UV-DDB/Cullin 4A complex is released from the COP9 signalosome and becomes tightly associated with chromatin (10). Collectively, these data strongly indicate that UV-DDB and its individual subunits perform multiple functions in the cell and that the XPE (DDB2) mutation may lead to UV-induced skin cancers, not necessarily by a defect in DNA repair but by defects in several other pathways that control cellular responses to DNA damage. Nevertheless, the recent description of several complexes that contain the XPE gene product and the fact that UV-DDB does have high affinity to damaged DNA raised the possibility that, if not UV-DDB, perhaps some of the other complexes may directly participate in the assembly of the human excision nuclease. Thus, we decided to isolate all of the complexes known to contain the XPE protein and test them for DNA binding and excision repair activities. We purified (i) the XPE (DDB2) protein free of other known interacting proteins, (ii) UV-DDB (DDB1 plus DDB2), (iii) the CUL4A complex, and (iv) the COP9 signalosome and analyzed them for damage-specific DNA binding activity and the effects of these various forms of XPE on the repair activity of the six-factor excision nuclease. Our data indicate that the DNA binding activity is intrinsic to DDB2, and in all heteromultimeric forms of XPE, the functional DNA binding entity is the DDB1-DDB2 complex. None of the four forms of XPE stimulates excision repair by the six-factor excision nuclease. Our data support the models that propose that XPE prevents cancer by regulating the cell cycle and the cellular response to DNA damage and apoptosis rather than by direct participation in the excision reaction itself.
MATERIALS AND METHODS
Expression and purification of proteins in insect cells.
DNA constructs for expression of DDB1 and DDB2 in insect cells, pBacPAK8-DDB1 and -DDB2, were obtained from Stuart Linn (34). DDB2 DNA was amplified by PCR with primers designed to incorporate the Flag epitope at the amino terminus and a His6 tag at the carboxyl terminus and was then subcloned into the p2Bac vector (Invitrogen); the DNA sequence was verified prior to use. The manufacturers' recommended procedures were used to establish virus stocks by cotransfecting Sf21 cells with pBacPAK8-DDB1 and BacPAK6 DNA (Clontech) or p2Bac-DDB2 and BaculoGold DNA (Pharmingen). Standard procedures were used for virus amplification and titer determination, and insect cells were cultured at 27°C in Grace's medium supplemented with 10% fetal bovine serum (FBS).
For DDB1 purification, 2.5 ×108 Sf21 cells were grown in a 250-ml suspension culture and infected with recombinant baculovirus at multiplicity of infection (MOI) of 10. After 48 h, cells were harvested by centrifugation and washed with phosphate-buffered saline and DDB1 was purified using modifications of a published procedure (34). Briefly, cells were lysed by sonication and DDB1 was purified by sequential chromatography on P11 phosphocellulose (Whatman), DEAE-Sepharose (GE Healthcare), and Superdex 200 10/300GL (GE Healthcare) columns. DDB1-containing fractions were identified after resolution by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% polyacrylamide gels containing sodium dodecyl sulfate followed by Coomassie blue staining, and fractions from the last column were stored at −80°C in PDG buffer (50 mM phosphate, 1 mM dithiothreitol, 10% [vol/vol] glycerol). The protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories).
To purify Flag-DDB2-His, 2 × 108 High Five cells were cultured in 150-mm dishes (2 × 107 cells/dish) and infected with recombinant baculovirus at an MOI of 10. After 48 h, cells were harvested by scraping and centrifugation, washed with phosphate-buffered saline, resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM β-glycerophosphate, 10% [vol/vol] glycerol, 1% Tween 20, 0.1% NP-40, 1 mM Na3VO4, 1 mM NaF), incubated on ice for 30 min, and lysed by sonication. Lysates were clarified by centrifugation at 16,000 × g for 30 min at 4°C, and the supernatant was incubated under constant rotation (Labquake device) overnight at 4°C with 150 μl anti-Flag (M2) antibody-affinity resin (Sigma). Beads and bound proteins were collected by centrifugation in a microcentrifuge and washed with Tris-buffered saline (TBS) containing 1 M NaCl. Proteins were eluted with TBS containing 150 mM NaCl, Flag peptide at 100 μg/ml, and 10% (vol/vol) glycerol and stored in small aliquots at −80°C. Protein-containing fractions were identified by Western blot analysis using anti-Flag (M2) antibodies, protein concentration was determined by the Bio-Rad protein assay, and the purity of eluted proteins was determined by SDS-PAGE followed by staining with Coomassie blue. Recombinant UV-DDB was affinity purified as described for Flag-DDB2 after coinfection of 2.5 × 108 Sf21 cells grown in a 250-ml suspension culture with the two baculovirus expression vectors, each at an MOI of 5. The DDB2 expressed in insect cells has both Flag and His6 tags, but for simplicity, we refer to this recombinant protein as Flag-DDB2.
Expression and purification of proteins in mammalian cells.
DNA constructs for mammalian expression of DDB2 and CUL4A were obtained from Yue Xiong (14). DDB2 and CUL4A gene sequences were amplified by PCR with primers designed to incorporate the Flag epitope at the amino terminus and were then subcloned into the pcDNA3 vector (Invitrogen); DNA sequences were verified prior to use. Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco's modified Eagle medium, supplemented with 10% FBS and 1% penicillin-streptomycin, and transfected with plasmid DNA using the calcium phosphate precipitation method; typically 180 μg DNA was used to transfect 2 × 107 cells in 150-mm dishes. After 48 h, cells were harvested, washed with phosphate-buffered saline, and incubated in lysis buffer for 30 min at 4°C. Lysates were clarified by centrifugation at 16,000 × g for 30 min at 4°C, and Flag-tagged proteins with their associated polypeptides were recovered as described above for Flag-DDB2, except beads were washed with TBS containing 150 mM NaCl. Protein-containing fractions were identified by Western blot analysis using anti-Flag (M2) antibodies, and the composition of complexes was determined by SDS-PAGE followed by staining with silver or Coomassie blue. Total protein concentrations were determined by the Bio-Rad protein assay, and the concentrations of DDB1 and DDB2 within the complexes were determined by scanning stained gels and comparing band intensities to the intensities of known amounts of bovine serum albumin resolved in the same gels after correcting for relative molecular weights.
Cell extracts and purified repair factors.
Chinese hamster ovary (CHO) AA8 cells were grown in 10-liter suspension cultures (Eagle's minimal essential medium supplemented with 10% FBS) and harvested while in log phase. Cell extracts (CFE) were prepared by the method of Manley (26) with modifications as described previously (40); dialyzed against 25 mM HEPES, pH 7.9, 100 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM dithiothreitol, and 12.5% (vol/vol) glycerol; and stored in small aliquots at −80°C. The six essential repair factors XPA, RPA, XPC-HR23B, TFIIH, XPG, and XPF-ERCC1 were purified and stored as described previously (40).
DNA substrates.
Internally radiolabeled DNA substrates containing a single, centrally located (6-4) photoproduct were prepared by annealing and ligating 4 or 6 oligomers as described previously (40). We used 136- and 50-bp duplexes for the excision and electrophoretic mobility shift assays, respectively; the sequence of the 50-bp substrate corresponds to nucleotides (nt) 44 to 93 of the 136-bp substrate. For electrophoretic mobility shift assays, we also used an undamaged 50-bp duplex (38).
Electrophoretic mobility shift assays.
A total of 2.5 to 5 fmol of the 50-bp duplex was incubated with the indicated amounts of proteins in 15- to 30-μl reaction mixtures as described previously (38). After a 30-min incubation at 30°C, glycerol was added to ∼8%, and reaction mixtures were resolved in 5% polyacrylamide gels at room temperature with a constant current of 25 mA. DNA binding was visualized by autoradiography and quantified using the Storm 860 system and ImageQuant 5.2 software (GE Healthcare). Binding was expressed as a percentage of radiolabel in the bound DNA relative to the total radiolabel in bound and free DNA.
Excision assays.
Assays with CHO cell extracts and the repair system reconstituted with purified factors were conducted as described previously (38). Briefly, the indicated amounts of DDB proteins or storage buffer (PDG or TBS) were preincubated for 10 to 15 min at 30°C with 15 fmol of 136-bp duplex DNA in 17 to 20 μl of reaction buffer, CHO cell extract (75 μg) or the six-factor excision nuclease was added, and the 25-μl reaction mixtures were incubated at 30°C for 90 min. For kinetic analyses with cell extracts, aliquots were removed at the indicated time points. Reactions were terminated with proteinase K digestion followed by extraction with phenol and phenol-chloroform-isoamyl alcohol. Deproteinized DNA was precipitated with ethanol, resuspended in formamide-dye mixture, resolved in 10% denaturing (7.7 M urea) polyacrylamide gels, visualized by autoradiography, and quantified using the Storm 860 system and ImageQuant 5.2 software (GE Healthcare). Excision levels for each reaction were determined as a percentage of radiolabel in the 20-to 35-nt region of the gel relative to the total radiolabel in the substrate migrating at 136 nt plus the excision products in that lane.
RESULTS
Purification of XPE (DDB2)-containing protein complexes.
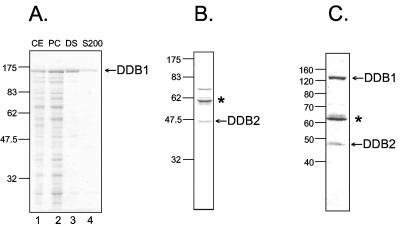
To investigate the effect of XPE on excision repair, we purified the protein in isolation and in the XPE complexes that have been identified to date. To purify DDB1 and DDB2 individually and in complex, we used the insect cell/baculovirus expression system. Insect Sf21 or High Five cells were infected with vectors expressing DDB1, Flag-DDB2, or a combination of the two vectors. DDB1 was purified by conventional chromatography as described previously (34); DDB2 and the DDB1-DDB2 complex were purified by affinity chromatography using anti-Flag (M2) antibody resin (Fig. 1). DDB1 was highly pure; DDB2 and the complex were of high purity as well but contained a contaminant that was identified as the heat shock protein HSP70, a protein known to bind to Flag resin nonspecifically and with high affinity.
FIG. 1.
Purification of DDB1, DDB2, and UV-DDB. Insect cells were infected with baculoviruses expressing either subunit alone or coinfected with baculoviruses expressing DDB1 and DDB2. Purified proteins were separated by 10% SDS-PAGE and stained with Coomassie blue. The numbers to the left of each panel indicate the positions of molecular weight standards in thousands). Asterisks in panels B and C indicate the position of HSP70 (identified by mass spectrometry), which is known to bind with relatively high affinity to the Flag resin used in the purification of the proteins in these panels. (A) Purification of DDB1 by conventional chromatography (33). Lane 1, cell extract (CE, 10 μg); lane 2, phosphocellulose (PC) fraction (9 μg); lane 3, DEAE-Sepharose fraction (1.3 μg); lane 4, Superdex 200 (S200) fraction (1 μg). (B) Flag-DDB2 (1 μg) purified with anti-Flag M2 affinity resin. (C) UV-DDB (0.6 μg) purified with anti-Flag M2 affinity resin from insect cells that were coinfected with baculoviruses expressing DDB1 and Flag-DDB2.
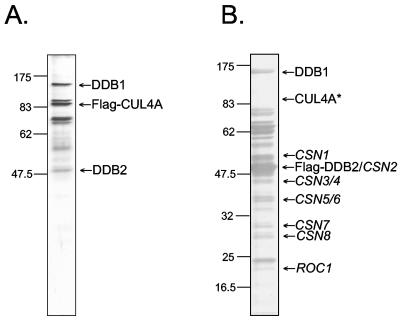
To obtain the CUL4A and COP9 signalosome forms of XPE, we transfected HEK293T cells with vectors expressing Flag-CUL4A or Flag-DDB2, respectively, and purified the complexes by Flag affinity chromatography. When the isolated complexes were analyzed by SDS-PAGE, the protein profiles of isolated complexes were quite similar to those published previously (2, 10, 14) for these complexes (Fig. 2). The CUL4A complex contained CUL4A, DDB1, and DDB2 with an approximately threefold molar excess of DDB1 to DDB2 (Fig. 2A) as reported previously (14). The COP9 complex purified by DDB2 affinity chromatography exhibited a polypeptide profile consistent with that of COP9 (10). In this particular preparation (Fig. 2B), there was an approximately 10-fold molar excess of DDB2 relative to DDB1 and the CUL4A band was too faint to be seen but was readily detectable by Western blotting. In addition to these complexes, we purified UV-DDB to near homogeneity from HeLa cells by multistep conventional chromatography (23) and used this preparation as a reference bona fide UV-DDB protein in our assays.
FIG. 2.
Isolation of DDB2 in CUL4A and COP9 complexes. HEK293T cells were transfected with vectors expressing either Flag-CUL4A or Flag-DDB2, and protein complexes were isolated by affinity chromatography on anti-Flag (M2) antibody resin, separated by 10% SDS-PAGE, and visualized by silver staining. The numbers to the left of each panel indicate the positions of the molecular size markers (in kilodaltons). (A) The CUL4A complex (1.1 μg). The identities of the bands designated DDB1, Flag-CUL4A, and DDB2 were confirmed by Western blotting with the appropriate antibodies. (B) The COP9 complex (2.5 μg). To the right of the panel, the identities of COP9 signalosome subunits are assigned according to the band pattern of COP9 previously purified by this procedure (10). The DDB1 and DDB2 bands were verified by Western blotting. The CUL4A band indicated by an asterisk is not apparent in this photographic reproduction, but its presence at the indicated position was demonstrated by Western blotting.
DNA binding activity of XPE and XPE-containing multiprotein complexes.
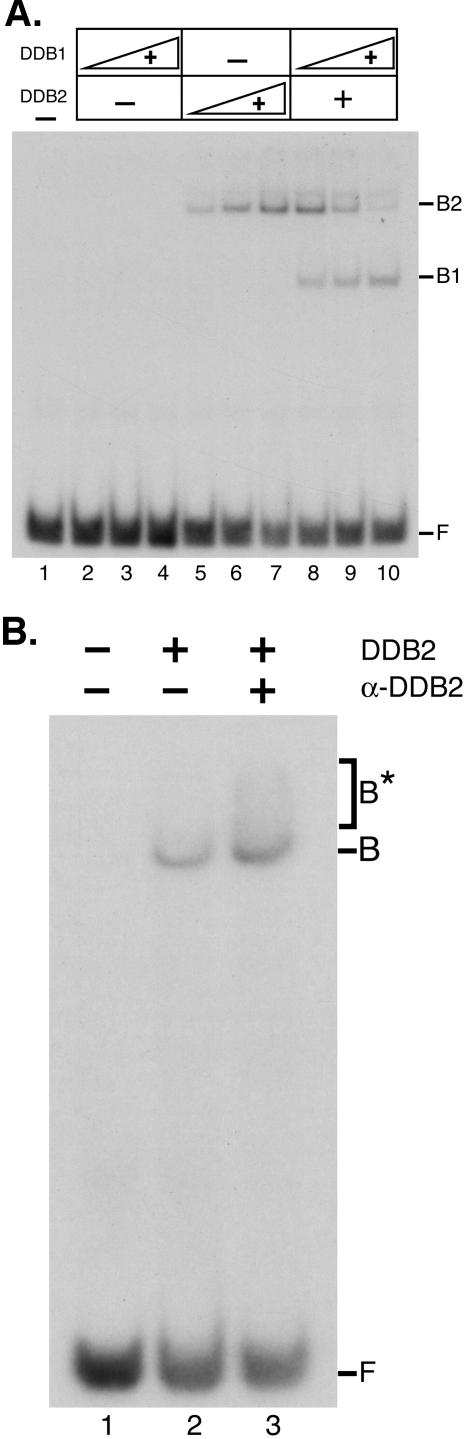
Although UV-DDB has been investigated extensively, there is no consensus about whether one or the other subunit, or the heterodimer only, has intrinsic DNA binding capacity. It has been suggested that the large subunit (37), the large subunit activated in a catalytic manner (“hit-and-run”) by the small subunit (16), or only the DDB1-DDB2 complex (34) possesses damage-specific DNA binding activity. The availability of the individual subunits in reasonable quantities and purity enabled us to address this question in a systematic manner. First, we tested recombinant DDB1 and DDB2 for binding activity. To our surprise, as shown in Fig. 3A, DDB2 bound an oligonucleotide containing a (6-4) photoproduct, but DDB1, in the concentration range used in this experiment, did not. Interestingly, when increasing amounts of DDB1 were added to DDB2 the DNA-protein band observed with DDB2 alone (B2) gradually disappeared in parallel with the appearance of a faster-migrating band (B1) with an electrophoretic mobility identical to that of UV-DDB (data not shown). Because of this relatively unusual observation of slower migration of a DNA-protein complex compared to a complex of larger mass, we considered the possibility of a contaminant in the DDB2 preparation that might give rise to this slower-migrating species in the electrophoretic mobility shift assay. To address this question, we performed an antibody supershift assay. As seen in Fig. 3B, the band assigned to the DDB2-DNA complex is supershifted by antibodies specific to DDB2. Thus, in conjunction with data in Fig. 3A, this establishes DDB2 as the DNA binding subunit of UV-DDB. The slower migration of DDB2-DNA complexes may result from binding by a DDB2 homodimer (17), or it may be caused by the high pI, 9.6, of DDB2 compared to UV-DDB, which also contains DDB1 with a low pI, 5.1.
FIG. 3.
DNA binding activity of UV-DDB is intrinsic to DDB2. Recombinant DDB1 and Flag-DDB2 purified from insect cells were used in electrophoretic mobility shift assays with a 50-bp duplex containing a (6-4) photoproduct in the middle (0.17 nM). (A) Binding of DDB1, DDB2, and the combination of the two subunits. Lane 1 contains DNA only; lanes 2 to 4 contain 30, 60, and 120 nM DDB1, respectively. Lanes 5 to 7 contain 1, 2, and 4 nM DDB2, respectively. Lanes 8 to 10 contain 4 nM DDB2 and increasing concentrations of DDB1 (30, 60, and 120 nM). F, free DNA; B1, UV-DDB-DNA complex; B2, DDB2-DNA complex. (B) Identification of DDB2 as the DNA binding protein in the recombinant DDB2 preparation by antibody supershift. Lane 1, DNA; lane 2, DNA plus 4 nM DDB2; lane 3, DNA plus 4 nM DDB2 plus anti-DDB2 antibody (α-DDB2). F, free DNA; B, DNA-protein complex; B*, DNA-protein-antibody complex.
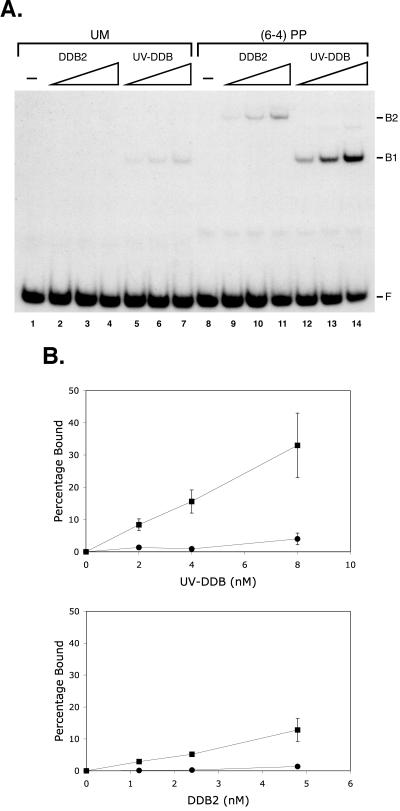
To determine if DDB2, like UV-DDB, has preference for UV-damaged DNA, we performed electrophoretic mobility shift experiments with DDB2 and UV-DDB using either an undamaged oligonucleotide or an oligonucleotide containing a (6-4) photoproduct (Fig. 4A). Both DDB2 and UV-DDB bind undamaged DNA weakly and bind damaged DNA with much higher affinity. We find that UV-DDB bound damaged DNA about twofold more efficiently than DDB2, and, importantly, we find that DDB2 and UV-DDB bound to the (6-4) photoproduct with comparable damage discrimination (Fig. 4B). Thus, we conclude that the damaged DNA binding property of UV-DDB is conferred by the DDB2 subunit.
FIG. 4.
Specificity of DDB2 binding to damaged DNA. Recombinant DDB2 and UV-DDB isolated from insect cells infected with the appropriate baculoviruses were mixed with 0.4 nM 50-bp duplexes without (unmodified [UM]) or with a (6-4) photoproduct [(6-4PP)], and DNA-protein complexes were visualized by autoradiography following a gel mobility shift assay. (A) Autoradiograph of a representative gel. The DDB2 concentrations in lanes 2 to 4 and 9 to 11 were 1.2, 2.4, and 4.8 nM; the UV-DDB concentrations in lanes 5 to 7 and 12 to 14 were 2, 4, and 8 nM. Because of the low concentration of the DDB2 stock, we were unable to use it at higher concentrations in this series of experiments. F, free DNA; B1, UV-DDB-DNA complex; B2, DDB2-DNA complex. (B) Binding isotherms of DDB2 and UV-DDB. Circles and squares represent UM and (6-4) photoproduct-containing DNA, respectively. Bars indicate standard deviations for two independent experiments.
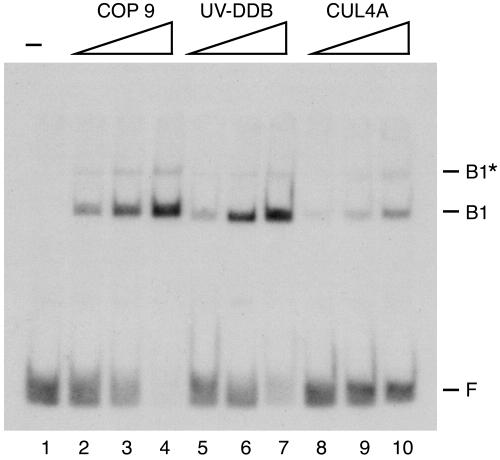
Having found that DDB2 is the damaged DNA binding subunit that confers upon UV-DDB its unique DNA binding properties, we wished to find out if the CUL4A and COP9 complexes, which contain several other polypeptides in addition to DDB2, make DNA-protein complexes with electrophoretic mobilities different from those of DDB2 and UV-DDB. The results, shown in Fig. 5, reveal that these higher-order complexes containing DDB1 and DDB2 make DNA-protein complexes with mobilities identical to that of UV-DDB. The different intensities of the retarded bands observed with the three forms are due to different amounts of DDB2 present in each complex and do not necessarily indicate differential affinities of UV-DDB, CUL4A, and COP9 to damaged DNA. These data suggest that the UV-DDB heterodimer within CUL4A and COP9 is in dynamic equilibrium with other polypeptides in these complexes that readily dissociate from DDB1-DDB2 during formation of DNA-protein complexes. It is also conceivable that the other polypeptides in CUL4A and COP9 are present in the initial DNA-protein complex but that these polypeptides dissociate as the complex enters the band shift assay gel. Further work is needed to distinguish between these possibilities, but the data suggest that CUL4A and COP9 do not make stable complexes with damaged DNA even though they are likely to be recruited to DNA by UV-DDB or DDB2.
FIG. 5.
DNA binding with multiprotein complexes of DDB2. The three indicated complexes known to contain DDB2 were used in an electrophoretic mobility shift assay. The reaction mixtures contained a 50-bp duplex with a (6-4) photoproduct (0.1 nM) and COP9 (0.2, 0.4, and 0.8 nM), UV-DDB (0.1, 0.2, and 0.4 nM), and CUL4A (1.1, 2.2, and 4.4 nM) as indicated. These concentrations are expressed relative to the limiting subunit in each complex. UV-DDB contains stoichiometric amounts of DDB1 and DDB2, whereas CUL4A, purified by CUL4A immunoaffinity resin, contains about 3-fold more DDB1 than DDB2, and COP9, purified by DDB2 immunoaffinity resin, contains about 10-fold more DDB2 than DDB1. F, free DNA; B1, UV-DDB-DNA complex; B1*, two UV-DDB-DNA complexes per DNA molecule.
Effect of various DDB2 (XPE) complexes on excision repair.
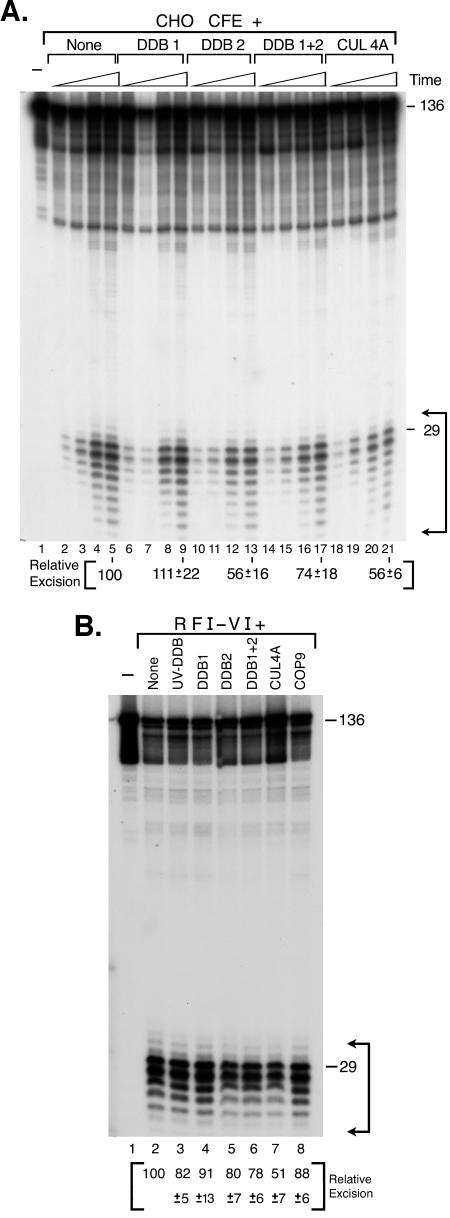
All the previous in vitro work addressing the potential role of XPE in excision repair was carried out with UV-DDB. Although some data have suggested a stimulatory role (47, 48), the overwhelming evidence is that UV-DDB does not stimulate excision at physiological concentrations and is inhibitory at high concentrations (21, 36, 38). It was, however, conceivable that XPE (DDB2) alone or XPE within the CUL4A or COP9 complexes might have different effects on the repair reaction. Therefore, we carried out excision reactions in the presence of all complexes known to contain XPE, including XPE (DDB2) alone, DDB1 plus DDB2, UV-DDB (purified from HeLa cells), CUL4A, and COP9. We used either CHO cell extract that is known to be free of DDB2 (16) or the reconstituted six-factor excision nuclease for these experiments. The results of the excision assays are shown in Fig. 6. With CHO cell extract, DDB1 had no effect on excision and amounts of DDB2, DDB1 plus DDB2, UV-DDB, and CUL4A sufficient to bind approximately 25% of the substrate in gel mobility shift assays resulted in 25 to 50% inhibition of the excision reaction (Fig. 6A). Because CHO extracts contain DDB1, it is possible that the inhibition by DDB2 is by the UV-DDB that forms upon addition of DDB2 to the extract (16). Importantly, neither of the UV-DDB subunits nor UV-DDB or CUL4A stimulates excision of (6-4) photoproducts. Similar results were obtained when the subunits and higher-order complexes of UV-DDB were added to the six-factor human excision nuclease. Again DDB1 had no statistically significant effect on the reaction, and all forms of DDB2 inhibited the reaction (Fig. 6B). Inhibition appeared to be most severe in the presence of the CUL4A complex. Clearly, further work is needed to explain the significance of this effect. However, collectively the data unambiguously show that neither XPE alone nor known complexes of XPE stimulate the excision reaction, suggesting that the pathogenesis of xeroderma pigmentosum in XP-E patients is not related to a defect in nucleotide excision repair.
FIG. 6.
Effect of DDB proteins on mammalian excision nuclease activity. A 136-bp duplex with a (6-4) photoproduct (0.8 nM) was incubated either with CHO cell extract (CFE) or reconstituted human excision nuclease (RFI-VI) in the presence of the indicated proteins, the products were separated in sequencing gels, and the levels of excision were quantified. (A) Excision kinetics with CHO cell extract. Reaction mixtures contained 75 μg of extract and, where indicated, DDB1 (175 nM), DDB2 (2.4 nM), DDB1 plus DDB2 (2.4 nM DDB2 but 175 nM DDB1), or CUL4A (2 nM DDB2 but 7 nM DDB1) and were incubated at 30°C for 15, 30, 60, and 90 min. Excision levels in the absence of additions (lanes 2 to 5) were 1.8%, 4.9%, 12.1%, and 21.3%, respectively. Excision levels at 90 min in the presence of the added factors are expressed relative to the 90-min control reaction (lane 5). The values are from three experiments, and standard errors are given. (B) Effect on reconstituted human excision nuclease. UV-DDB purified from HeLa cells (0.2 nM), DDB1 (175 nM), DDB2 (2.4 nM), DDB1 plus DDB2 (2.4 nM DDB2 but 175 nM DDB1), CUL4A (2 nM DDB2 but 7 nM DDB1), and COP9 (0.24 nM DDB1 but 3 nM DDB2) were added to the reconstituted human excision nuclease as indicated and incubated at 30°C for 90 min. Excision in the absence of added UV-DDB-related factors (lane 2) was 56.2% ± 2.3%, and the other values are expressed as percentages of this value. The values are averages of two experiments with standard deviations as indicated.
DISCUSSION
XPE is a DNA binding protein with high affinity for certain DNA lesions such as the (6-4) photoproduct. XPE is also a component of UV-DDB, CUL4A, and COP9 signalosome complexes that participate in protein modification such as sumoylation (50) and ubiquitylation (28, 45, 50), cell cycle regulation by direct interaction with E2F1 (12, 43), and apoptosis through a complex regulatory circuit that controls the level of p53 in the cell (18, 19). Models for the physiological role of XPE in the cellular response to DNA damage fall into two broad categories. In one model, XPE is mainly a DNA damage recognition protein that directly participates in damage recognition and removal (46). Our results (38; this work) suggest that XPE does not directly participate in excision repair. In an alternative model, XPE is primarily involved in cell cycle regulation (12), DNA damage checkpoint control (2, 14), and apoptosis (18, 19), with no or minimal role in DNA repair.
Evidence for a putative repair function of XPE.
The following observations have been used as evidence for the model that posits XPE as a key protein in the damage recognition step of nucleotide excision repair. (i) XP-E cell lines are moderately more sensitive to UV than wild-type controls (4). (ii) XP-E strains have been reported to have 40 to 60% unscheduled DNA synthesis (UDS) relative to wild-type controls (24, 25, 51). (iii) UV-DDB binds to UV-irradiated DNA (3, 7) and in particular to (6-4) photoproducts (37) with high affinity and specificity. In apparent accord with these findings, it has been reported that XP-E cells are defective in Pyr<>Pyr excision but have nearly normal (6-4) photoproduct excision (15) as determined by immunochemical methods. [However, as noted below, another study using the same methodology found that the repair kinetics of Pyr<>Pyr were identical in XP-E and wild-type strains and that the initial rate of (6-4) photoproduct removal in XP-E was about 75% of the wild-type rate (21).] (iv) The XPE gene is induced by UV in a p53-dependent manner (15) and p53−/− cells, which cannot up-regulate DDB2, and rodent cells that do not express DDB2 are sensitive to UV and defective in global genomic repair (9). (v) Using micropore irradiation/immunostaining techniques, it was found that DDB2 (presumably in the form of UV-DDB) was the first known protein to accumulate at sites of UV damage and that this accumulation was independent of XPA and XPC (47) but that translocation of XPC to these foci was dependent on XPE (8, 49). (vi) XPE, either in the form of UV-DDB or CUL4A complex, is involved in a number of UV-induced sumoylation and ubiquitylation reactions of proteins including XPE itself and XPC (28, 45, 50), and it has been reported that the ubiquitylation of XPC by UV-DDB improves its DNA repair activity (45). (vii) It has been reported that UV-DDB can stimulate the in vitro excision of Pyr<>Pyr marginally (45, 48) and of (6-4) photoproducts more extensively under special circumstances (47, 48).
Evidence for cell cycle and apoptosis regulatory functions of XPE.
An equally compelling set of reports and functional considerations suggest that the primary role of XPE is in the control of cell cycle/DNA damage checkpoints and apoptosis. (i) A carefully controlled study comparing the UV sensitivity of XP-E strains found that under identical experimental conditions there was no difference in the UV sensitivity of XP-E and wild-type strains and no difference in the level of UDS in the two cell types (20). (ii) XP-E primary fibroblasts excise Pyr<>Pyr at normal rates and (6-4) photoproducts at near-normal rates as measured by immunochemical methods (21). (iii) Rapid accumulation of UV-DDB in UV foci (47, 49) before XPC and XPA does not establish a cause-and-effect relationship because CHO cells that do not express DDB2 excise Pyr<>Pyr at a rate similar to that in human cells and (6-4) photoproducts at about 75% of the rate of human cells (29), casting some doubt about the functional relevance of such foci. (iv) XPE as a component of UV-DDB regulates the activity of the cell cycle-specific transcription factor E2F1 (12, 43). (v) XPE as a component of CUL4A and COP9 complexes is involved in ubiquitylation and degradation of a number of proteins, including DDB2 (2, 28, 32) and CDT1 (14). These reactions are of potential significance in the DNA damage checkpoint response. (vi) In all XP-E strains tested, p53 is down-regulated 3- to 10-fold and as a consequence XP-E cells exhibit defective UV-induced apoptosis (18). (vii) The six-factor human excision nuclease as well as human and CHO cell extracts that do not contain DDB2 excise (6-4) photoproducts (22, 38), Pyr<>Pyr (38), and the cisplatin-1,3-d(GpTpG) diadduct (36) at rates indistinguishable from those in in vitro systems containing UV-DDB under a variety of experimental conditions.
An exhaustive critique of all of these seemingly contradictory reports is outside the scope of this discussion. Undoubtedly, the recent generation of Xpe−/− mice (19, 52) will be invaluable in defining the role of XPE in the cellular response to DNA damage. Studies with fibroblasts from these mice and with the mice themselves have already revealed some important facts. First, the Xpe−/− fibroblasts are either more resistant than wild-type fibroblasts to UV-induced killing (19) or are at least as resistant as the wild-type controls (52). Second, mouse embryo fibroblasts from Xpe+/+ and Xpe−/− animals remove Pyr<>Pyr at identical rates (52). Third, Xpe−/− cells exhibit reduced basal and inducible levels of p53 and are more resistant to UV-induced apoptosis relative to Xpe+/+ cells (19). Finally, Xpe−/− mice developed spontaneous malignant tumors in internal organs at a high rate (52), perhaps because of disrupted cell cycle regulation and diminished apoptosis. This is in contrast to Xpa−/− mice (6), which are totally defective in excision repair and are extremely susceptible to UV-induced skin cancers but do not show elevated levels of spontaneous tumors. These results strongly support the suggestion that the primary role of XPE in preventing skin cancer is to promote apoptosis (18, 19), and any involvement in DNA repair, which may or may not exist, is of secondary significance. This model, however, does not address the issue of high-affinity binding of UV-DDB to DNA.
XPE and models for repair of UV damage.
The conclusion that XPE is mainly a cell cycle and checkpoint protein brings back into focus the question we wished to address in this paper: the effect of binding of XPE to UV damage on repair rates. The (6-4) photoproduct is recognized with moderate selectivity by the three excision repair factors RPA, XPA, and XPC (38, 39), but Pyr<>Pyr is not (38), and in fact, it has been reported that Pyr<>Pyr is excluded by XPC (13), meaning that in this study undamaged DNA was bound with higher affinity by XPC than DNA containing a Pyr<>Pyr. UV-DDB only poorly discriminates between Pyr<>Pyr and undamaged DNA (37, 38). Yet, despite these findings and the extensive data summarized above indicating normal or near-normal Pyr<>Pyr excision both in vivo and in vitro by mammalian systems in the absence of UV-DDB, a rather commonly advanced model for repair of UV photoproducts is as follows (1, 11, 15, 45, 46). Both (6-4) photoproducts and Pyr<>Pyr in the template strand of transcribed sequences block RNA polymerase, which with the aid of CSA and CSB recruits XPA, RPA, TFIIH, XPG, and XPF-ERCC1 to carry out excision without the involvement of XPC or XPE. When the photoproducts are in nontranscribed sequences, XPC recognizes the (6-4) photoproducts and recruits the other five basal repair factors to execute excision. In the case of Pyr<>Pyr, according to the model, the damage is recognized by UV-DDB, which recruits XPC to the damage site, and UV-DDB either dissociates from the site or is destroyed—leaving XPC bound to Pyr<>Pyr, where it recruits the remaining basal factors of the excision nuclease. This model cannot explain the efficient recognition and removal of Pyr<>Pyr in the absence of UV-DDB in a number of in vivo and in vitro rodent and human excision repair systems summarized above. We considered the possibility that complexes containing DDB2 other than UV-DDB may in fact play the facilitator role for XPC (15, 45) or XPA (48), but we failed to observe any stimulatory effect of either CUL4A or COP9 complexes containing XPE (this work). Moreover, with both rodent and human cell-free systems we observe efficient repair of Pyr<>Pyr (31, 38, 41) and of (6-4) photoproducts (38; this work) in the absence of XPE in its various forms. We conclude that the thermodynamic recognition model that posits recognition of damage by a given repair protein followed by recruitment of the remaining repair factors is not compatible with the known properties of human excision repair factors and that the high specificity combined with the moderate rate of the human excision nuclease can be accomplished only by cooperative recognition of damage by RPA, XPA, and XPC coupled with kinetic proofreading afforded by the helix-unwinding activity of TFIIH (38, 39) without the need for additional damage specificity factors.
Acknowledgments
This work was supported by National Institutes of Health grant GM32833 (to A.S.) and a training grant from TUBITAK, The Scientific and Technical Research Council of Turkey (to G.K.).
We thank Y. Xiong (University of North Carolina, Chapel Hill) for CUL4A antibodies and mammalian expression vectors containing DDB1, DDB2, and CUL4A genes; P. Raychaudhuri (University of Illinois, Chicago) for antibodies to DDB1 and DDB2; and S. Linn (University of California, Berkeley) for insect expression vectors containing DDB1 and DDB2.
REFERENCES
- 1.Adimoolam, S., and J. M. Ford. 2003. p53 and regulation of DNA damage recognition during nucleotide excision repair. DNA Repair (Amsterdam) 2:947-954. [DOI] [PubMed] [Google Scholar]
- 2.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 3.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242:564-567. [DOI] [PubMed] [Google Scholar]
- 4.Cleaver, J. E., and K. H. Kraemer. 1989. Xeroderma pigmentosum, p. 2949-2971. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic basis of inherited disease, vol. 2. McGraw-Hill, New York, N.Y. [Google Scholar]
- 5.Datta, A., S. Bagchi, A. Nag, P. Shiyanov, G. R. Adami, T. Yoon, and P. Raychaudhuri. 2001. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res. 486:89-97. [DOI] [PubMed] [Google Scholar]
- 6.deVries, A., C. T. M. van Oostrom, F. M. A. Hofhuis, P. M. Dortant, R. J. W. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. A. Capel, H. van Steeg, and S. J. Verbeek. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377:169-173. [DOI] [PubMed] [Google Scholar]
- 7.Feldberg, R. S., and L. Grossman. 1976. A DNA binding protein from human placenta specific for ultraviolet damaged DNA. Biochemistry 15:2402-2408. [DOI] [PubMed] [Google Scholar]
- 8.Fitch, M. E., S. Nakajima, A. Yasui, and J. M. Ford. 2003. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 278:46906-46910. [DOI] [PubMed] [Google Scholar]
- 9.Ford, J. M., and P. C. Hanawalt. 1995. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc. Natl. Acad. Sci. USA 92:8876-8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357-367. [DOI] [PubMed] [Google Scholar]
- 11.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 21:8949-8956. [DOI] [PubMed] [Google Scholar]
- 12.Hayes, S., P. Shiyanov, X. Chen, and P. Raychaudhuri. 1998. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol. 18:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hey, T., G. Lipps, K. Sugasawa, S. Iwai, F. Hanaoka, and G. Krauss. 2002. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry 41:6583-6587. [DOI] [PubMed] [Google Scholar]
- 14.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6:1003-1009. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang, B. J., S. Toering, U. Francke, and G. Chu. 1998. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 18:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoki, T., S. Yamagami, Y. Inoki, T. Tsuru, T. Hamamoto, Y. Kagawa, T. Mori, and H. Endo. 2004. Human DDB2 splicing variants are dominant negative inhibitors of UV-damaged DNA repair. Biochem. Biophys. Res. Commun. 314:1036-1043. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, T., C. O'Shea, and S. Linn. 2003. Impaired regulation of tumor suppressor p53 caused by mutations in the xeroderma pigmentosum DDB2 gene: mutual regulatory interactions between p48DDB2 and p53. Mol. Cell. Biol. 23:7540-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, T., D. Cado, R. Kamide, and S. Linn. 2004. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc. Natl. Acad. Sci. USA 101:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh, T., S. Linn, T. Ono, and M. Yamaizumi. 2000. Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. J. Investig. Dermatol. 114:1022-1029. [DOI] [PubMed] [Google Scholar]
- 21.Itoh, T., T. Mori, H. Ohkubo, and M. Yamaizumi. 1999. A newly identified patient with clinical xeroderma pigmentosum phenotype has a non-sense mutation in the DDB2 gene and incomplete repair in (6-4) photoproducts. J. Investig. Dermatol. 113:215-257. [DOI] [PubMed] [Google Scholar]
- 22.Kazantsev, A., D. Mu, A. F. Nichols, X. Zhao, S. Linn, and A. Sancar. 1996. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc. Natl. Acad. Sci. USA 93:5014-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney, S., G. J. Chang, and S. Linn. 1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268:21293-21300. [PubMed] [Google Scholar]
- 24.Keeney, S., A. P. M. Eker, T. Brody, W. Vermeulen, D. Bootsma, J. H. J. Hoeijmakers, and S. Linn. 1994. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc. Natl. Acad. Sci. USA 91:4053-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer, K. H., E. A. de Weerd-Kastelein, J. H. Robbins, W. Keijzer, S. F. Barrett, R. A. Petinga, and D. Bootsma. 1975. Five complementation groups in xeroderma pigmentosum. Mutat. Res. 33:327-340. [DOI] [PubMed] [Google Scholar]
- 26.Manley, J. L., A. Fire, A. Cano, P. A. Sharp, and M. L. Gefter. 1980. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. USA 77:3855-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda, N., K. Azuma, M. Saijo, S.-i. Iemura, Y. Hioki, T. Natsume, T. Chiba, K. Tanaka, and K. Tanaka. 2005. DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair (Amsterdam) 4:537-545. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, D. L., C. A. Haipek, and J. M. Clarkson. 1985. (6-4) photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat. Res. 143:109-112. [DOI] [PubMed] [Google Scholar]
- 30.Mu, D., C.-H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 31.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 32.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb− phenotype. J. Biol. Chem. 271:24317-24320. [DOI] [PubMed] [Google Scholar]
- 34.Nichols, A. F., T. Itoh, J. A. Graham, W. Liu, M. Yamaizumi, and S. Linn. 2000. Human damage-specific DNA-binding protein p48. J. Biol. Chem. 275:21422-21428. [DOI] [PubMed] [Google Scholar]
- 35.Obuse, C., H. Yang, N. Nozaki, S. Goto, T. Okazaki, and K. Yoda. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9:105-120. [DOI] [PubMed] [Google Scholar]
- 36.Rapić-Otrin, V., I. Kuraoka, T. Nardo, M. McLenigan, A. P. M. Eker, M. Stefanini, A. S. Levine, and R. D. Wood. 1998. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol. 18:3182-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reardon, J. T., A. F. Nichols, S. Keeney, C. A. Smith, J.-S. Taylor, S. Linn, and A. Sancar. 1993. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J. Biol. Chem. 268:21301-21308. [PubMed] [Google Scholar]
- 38.Reardon, J. T., and A. Sancar. 2003. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 17:2539-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reardon, J. T., and A. Sancar. 2005. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 79:183-235. [DOI] [PubMed] [Google Scholar]
- 40.Reardon, J. T., and A. Sancar. Purification and characterization of E. coli and human nucleotide excision repair enzyme systems. Methods Enzymol., in press. [DOI] [PubMed]
- 41.Reardon, J. T., L. H. Thompson, and A. Sancar. 1993. Excision repair in man and the molecular basis of xeroderma pigmentosum syndrome. Cold Spring Harbor Symp. Quant. Biol. 58:605-617. [DOI] [PubMed] [Google Scholar]
- 42.Sancar, A., L. A. Lindsey-Boltz, K. Ünsal-Kaçmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 43.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 45.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387-400. [DOI] [PubMed] [Google Scholar]
- 46.Tang, J., and G. Chu. 2002. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amsterdam) 1:601-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakasugi, M., A. Kawashima, H. Morioka, S. Linn, A. Sancar, T. Mori, O. Nikaido, and T. Matsunaga. 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277:1637-1640. [DOI] [PubMed] [Google Scholar]
- 48.Wakasugi, M., M. Shimizu, H. Morioka, S. Linn, O. Nikaido, and T. Matsunaga. 2001. Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J. Biol. Chem. 276:15434-15440. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Q.-E., Q. Zhu, G. Wani, J. Chen, and A. A. Wani. 2004. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 25:1033-1043. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Q.-E., Q. Zhu, G. Wani, M. A. El-Mahdy, J. Li, and A. A. Wani. 2005. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 33:4023-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaizumi, M., and T. Sugano. 1994. U.V.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene 9:2775-2784. [PubMed] [Google Scholar]
- 52.Yoon, T., A. Chakrabortty, R. Franks, T. Vali, H. Kiyokawa, and P. Raychaudhuri. 2005. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene 24:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]