Abstract
Two important metal-responsive regulators, NikR and Fur, are involved in nickel and iron homeostasis and controlling gene expression in Helicobacter pylori. To date, they have been implicated in the regulation of sets of overlapping genes. We have attempted here dissection of the molecular mechanisms involved in transcriptional regulation of the NikR and Fur proteins, and we investigated protein-promoter interactions of the regulators with known target genes. We show that H. pylori NikR is a tetrameric protein and, through DNase I footprinting analysis, we have identified operators for NikR to which it binds with different affinities in a metal-responsive way. Mapping of the NikR binding site upstream of the urease promoter established a direct role for NikR as a positive regulator of transcription and, through scanning mutagenesis of this binding site, we have determined two subsites that are important for the binding of the protein to its target sequence. Furthermore, by alignment of the operators for NikR, we have shown that the H. pylori protein recognizes a sequence that is distinct from its well-studied orthologue in Escherichia coli. Moreover, we show that NikR and Fur can bind independently at distinct operators and also compete for overlapping operators in some coregulated gene promoters, adding another dimension to the previous suggested link between iron and nickel regulation. Finally, the importance of an interconnection between metal-responsive gene networks for homeostasis is discussed.
Virtually all organisms require iron for survival, and nickel acts as an important cofactor for many cellular enzymatic processes; however, these metals can be toxic in high concentrations. Consequently, microorganisms have evolved tightly regulated systems for both uptake and sequestration of these essential elements (21, 29). The ferric uptake regulator (Fur) is fundamental for maintaining iron homeostasis in many prokaryotes (14, 16). Similarly, the Ni-responsive repressor NikR has been shown in Escherichia coli to control transcription of the nikABCDE operon encoding a nickel permease (7, 8).
In Helicobacter pylori, an important human pathogen associated with stomach diseases, homologues of the NikR and Fur proteins have been shown to be involved in the regulation of overlapping gene circuits important for the survival of H. pylori in the host (6, 31). Both regulatory proteins have been reported to be involved in autoregulation through direct repression of their upstream promoter elements (9, 11), and mutants of each regulator show pleiotropic transcriptional responses in a variety of genes, some of which are in common (9, 13, 31). In H. pylori the Fur repressor has been well characterized at the molecular level, and its operators have been studied in a number of iron-responsive gene promoters (11, 12, 33). Analysis of its metal-binding affinity has revealed interesting insights, and it is known to control iron homeostasis through complex repression and derepression mechanisms at iron-repressed and iron-activated promoters. It has furthermore been implicated in acid resistance (4, 31) and nickel induction of the urease gene (30), and its target genes have been shown to respond to metal signals other than iron (3, 31), indicating that its regulatory role may expand outside that solely of iron metabolism.
In comparison, relatively little is currently known about the molecular mechanisms involved in NikR-mediated regulation in H. pylori. For example, it is not clear whether its role is solely as a nickel-responsive repressor of gene transcription as with the NikR protein of E. coli (7) or whether it may also activate gene transcription. The H. pylori NikR orthologue was originally identified as a regulator required for mediating nickel induction of the urease gene at the transcriptional level (34). In addition, the cis-acting nickel-responsive region in the urease promoter was identified. However, this element does not resemble the palindromic operator (GTATGA-N16-TCATAC) of the E. coli protein nor the supposed similar (GCATGA-N16-TCATGC) sequence in the H. pylori nikR promoter region, which has been implicated in its autoregulation (9). Contreras et al. (9) identified a series of differentially regulated genes in the NikR mutant, including nixA and hpn involved in nickel uptake and storage, respectively, verifying a role for NikR in the regulation of nickel metabolism in H. pylori. Also included in the list of genes deregulated in the NikR mutant are genes of the Fur regulon involved in iron transport, storage, and regulation, highlighting a link between nickel and iron metabolism and the regulatory networks controlled by NikR and Fur, respectively. These two regulators were recently proposed as a metal-responsive repressor cascade involved in controlling the acid adaptation (31). Despite these works, nothing is known of how NikR regulates gene expression, its direct target genes, and how its action could be interconnected to that exerted by Fur on genes regulated by both proteins. For example, the operator site to which NikR binds to control gene transcription has not been identified in H. pylori, as well as whether it binds directly to all or any of its target genes.
In the present study, we initiated molecular analysis of the NikR protein of H. pylori and, in particular, we used in vitro binding assays with purified NikR and Fur proteins and studied the protein-promoter interactions of the regulators with known target promoters. We identified binding sites for NikR in various promoters in the H. pylori genome, and we studied the binding of NikR and Fur in particular at the PnikR and PexbB promoters where the proteins bind to overlapping and flanking operators in each promoter, respectively. In addition, we attempted to dissect the NikR-binding site located in the cis element upstream of the urease gene implicated in nickel responsive induction and located two subsite determinants of binding which are compatible with NikR as metal-responsive tetrameric protein as established here.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The H. pylori strains used in the present study are all derivatives of the wild-type strain G27 (35). Two fur mutant strains have been described previously; the fur-null mutant G27Δfur and the G27(H99I) mutant expressing an iron-blind Fur protein (11). The nikR-null mutant strain G27/nikR::km was similarly constructed through allelic replacement of the nikR gene with a kanamycin cassette as recently described (22). H. pylori strains were recovered from −80°C stocks on Columbia agar plates containing 5% horse blood, 0.2% cycloheximide, and Dent's or Skirrow's antibiotic supplement under microaerophilic conditions (Oxoid) and routinely cultured in a 5% CO2 to 95% air atmosphere at 95% humidity. Liquid cultures were grown in modified brucella broth containing 5% fetal calf serum and Dent's or Skirrow's antibiotic supplement. E. coli strains DH5-α (15) and BL21(DE3) (27) were cultured in Luria-Bertani medium and when required ampicillin was added to a final concentration of 100 μg/ml.
Cloning and mutagenesis of the promoters.
DNA manipulations were carried out routinely as described by Sambrook et al. (23). The promoter regions under study were amplified by PCR using G27 chromosomal DNA as a template and cloned into the pGemT vector (Promega). The nikR-exbB intergenic region was amplified with primers NKTB-F (5′-attcagggatccTCGGGTTGTCTAATTCGTC) and NKTB-R (5′-cggatgaattcCTCCTTGTCTATGATAAAAC) and cloned as a 407-bp BamHI/EcoRI fragment, generating plasmid pNKTB. The urease promoter region was amplified with the primers Ure-F (5′-attcaggatccCATAGTGGAGCATCAACTTGTC) and Ure-R (5′-attcagaattcGTCGTGGCCACCATTATCAC) and cloned as a 308-bp BamHI/EcoRI fragment, generating plasmid pGemUre. The pGemUre plasmid, consisting of the wild-type urease promoter was mutagenized with the QuikChange site-directed mutagenesis kit (Stratagene, Inc.), and primer pairs were designed according to the manufacturer's instructions for incorporation of ccc trinucleotide mutations (ure1-10, see Fig. 5C). This resulted in the generation of ten plasmids (pUreOP1-10) carrying mutant urease operators that were then radioactively labeled and used as probes for comparative affinity DNase I footprinting experiments. To estimate the relative affinities of the mutant promoters for NikR, the concentration of NikR for which there was a 50% reduction in the amount of radioactivity across the footprint was measured with a densitometer (Image Master VDS software; Pharmacia Biotech) and was taken to be the relative half-maximal binding concentration for that particular operator derivative. The comparative affinities could then be calculated with respect to the wild-type urease NikR operator.
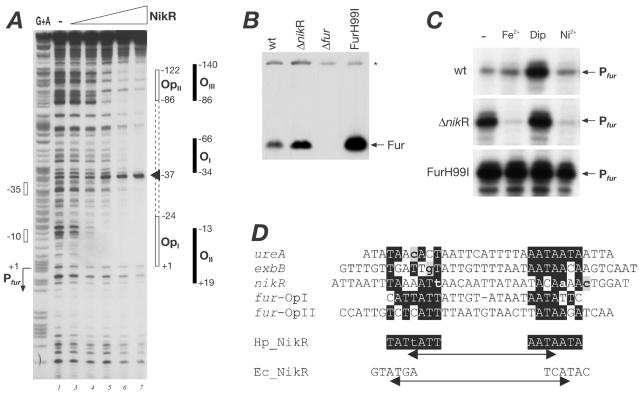
FIG. 5.
Identification of the NikR operator in the PureA promoter. (A) In vitro binding of rNikR to the urease promoter by DNase I footprinting analysis. The probe used consists of a 308-bp EcoRI-BamHI fragment containing the wild-type urease promoter region from pGemUre labeled at its BamHI site. The 5′-end-labeled probe was incubated with an increasing amount of purified NikR protein from lanes 1 to 6 corresponding to 0, 0.004, 0.011, 0.033, 0.10, and 0.30 μM concentrations of protein, respectively. The vertical open box on the right indicates the areas of DNase I protection resulting from binding of NikR; the numbers indicate the boundaries of the operator with respect to the +1 transcriptional start site of the PureA. The G+A lane is a G+A sequence reaction on the DNA probe used as size marker (19). The bent arrow and boxes to the left of the panel show the direction of transcription and position of the +1 and promoter elements of PureA. (B) Conservation and features of urease promoter sequence. Alignment of nucleotide sequences of the urease promoter region from the indicated five different strains of H. pylori (1, 18, 28, 34). The alignment was generated by CLUSTAL W, and the nonconserved nucleotides are highlighted in gray. The transcriptional start site (26) is shown by bent arrows, along with the “extended” −10 and −35 hexamers inferred, which are boxed. The proposed stem-loop structure that consists as the cis-acting NikR-mediated nickel-induction site (34) is indicated in underlined boldface. The NikR-operator as defined by DNase I footprinting in the present study is boxed. (C) Dissection of the NikR operator of the urease promoter. Scanning mutagenesis of the urease operator to determine nucleotides important for binding of NikR. The sequence changes and the fold decreases in affinity compared to the wild type of 10 ccc trinucleotide substitution (underlined in boldface) mutants of the urease operator are shown. By in vitro DNase I footprinting analysis with the purified NikR protein, the relative half-maximal binding was calculated for each mutant operator as the concentration at which 50% of the probe was estimated to be protected, and the relative fold changes in affinity were calculated accordingly. The wild-type operator sequence is shown, and the nucleotides that were determined important for binding are highlighted in gray and black.
Purification of NikR protein and generation of NikR antiserum.
The nikR gene was amplified from G27 chromosomal DNA with the primers NikR-UP (5′-atatatcatATGGATACACCCAATAAAGACG) and NikR-DO (5′-atatatggatccCTTTGTATTTTAAGACGCTATTC) and cloned as a 450-bp NdeI/BamHI fragment into the expression vector pET15b (Invitrogen), generating the pET15NikR plasmid. This plasmid was transformed into the E. coli BL21(DE3) strain, the expression of a recombinant NikR protein (His-NikR), containing a N-terminal histidine tag (His tag) was induced, and the protein was purified by Ni-NTA (QIAGEN) affinity chromatography under nondenaturing conditions according to the manufacturer's instructions. The His tag from 1 mg of the purified His-NikR protein was excised by digestion with 10 U of thrombin protease (Amersham-Pharmacia Biotech) at room temperature for 5 h, leaving only three amino acids on the N-terminal of the NikR protein (rNikR), and the excised His tag was subsequently removed by dialysis in phosphate-buffered saline (PBS) in 6,000- to 8,000-Da-MWCO tubing (Membrane Filtration Products, Inc.). Protein concentrations of the His-NikR and the rNikR-purified proteins were determined by the Bradford colorimetric method (Bio-Rad), and the proteins were stored in PBS at −80°C. For DNA-binding experiments, the rNikR protein was dialyzed overnight against buffer D (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol) containing 10% glycerol and then against buffer D containing 50% glycerol and subsequently stored at −20°C. To prepare anti-NikR antiserum, 20 μg of purified protein (His-NikR) was used to immunize 6-week-old CD1 female mice (Charles River Laboratories), and four mice were used. The protein was given intraperitoneally, together with complete Freund adjuvant for the first dose and incomplete Freund adjuvant for the second (day 21) and third (day 35) booster doses. Bleed-out samples were taken on day 49 and used in Western blot analysis by standard methods (23) at a dilution of 1:1,000. Generation of anti-Fur antisera and Western blot analysis has been previously described (11)
Gel filtration of the purified NikR protein.
The purified rNikR protein was dialyzed overnight in PBS alone or with the addition of 50 μM NiCl2. An aliquot of 50 to 100 μg of the purified rNikR protein was loaded onto a Superdex 200 HR 10/30 column (Amersham-Pharmacia Biotech) preequilibrated with PBS alone or with the addition of 50 μM NiCl2, and then the protein was eluted at 0.05 ml/min and collected in 500-μl fractions. The protein molecular weight standards, including albumin, ovalbumin, chymotrypsinogen A, and RNase A (LMW gel filtration calibration kit; Pharmacia Biotech) were applied to the column preequilibrated with PBS for column calibration, and the molecular weight of each protein standard (according to the manufacturer's specifications) was used to generate a standard curve to determine the molecular weight of the rNikR protein peaks.
RNA preparation, primer extension, and S1 nuclease protection analysis.
Total RNA was extracted from H. pylori and used in primer extension experiments as previously described (12). To ensure correct mapping of the promoter, a sequencing reaction was completed with a T7 sequencing kit (USB Corp.) using the same primer as in the primer extension reactions and the corresponding cloned promoter region. A radioactively labeled DNA probe for S1 nuclease mapping of the nikR promoter was prepared as follows: a 480-bp PCR product was amplified with primers Nik-6 (5′-ATCTTTCAACATTCCCACTG) and Nik-7 (5′-GCACTAATTCTGATCGAGAAG) and labeled at both extremities with T4 polynucleotide kinase (New England Biolabs, Inc.) and 4 pmol of [γ-32P]ATP (5,000 Ci/mmol; NEN). The labeled probe was then digested with SphI, and the 375-bp fragment, corresponding to the 5′ region of the nikR gene labeled at one extremity only, was purified by preparative polyacrylamide gel electrophoresis (PAGE). Briefly, the probe labeled at one extremity was extracted from a 6% polyacrylamide gel and eluted in 3 ml of elution buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 300 mM Na acetate [pH 5.2], 0.2% sodium dodecyl sulfate [SDS]) at 37°C overnight. The eluted probe was then extracted once with an equal volume of phenol-chloroform (1:1) and ethanol precipitated. Approximately 20,000 cpm of the labeled probe was coprecipitated with 20 μg of total RNA, resuspended in 20 μl of hybridization buffer (80% formamide, 60 mM Tris-HCl [pH 7.5], 400 mM NaCl, 0.4 mM EDTA), overlaid with 5 μl of paraffin oil and, after denaturation at 95°C for 2 min, incubated overnight at the calculated melting temperature of the probe. Subsequently, 180 μl of ice-cold S1 buffer (33 mM sodium acetate [pH 5.2], 5 mM ZnSO4, 250 mM NaCl) and 1 μl of S1 nuclease (150 U/μl; Invitrogen) were added, and the digestion was carried out for 30 min at 37°C. Samples were extracted with phenol-chloroform (1:1), ethanol precipitated, and resuspended in 5 μl of sequencing buffer. After denaturation at 100°C, samples were subjected to 6% urea-PAGE and autoradiographed. Quantification of the signals from extension products was performed by using the PhosphorImager and ImageQuant software (Molecular Dynamics).
Probe preparation and DNase I footprinting.
The pGemCl2 (11), pNKTB, and pGemUre containing the cloned fur, nikR-exbB, and urease promoter regions, respectively, or the pUreOP1-10 plasmids containing the mutant urease operators (Fig. 5C) were first linearized by digestion at a site at one extreme of the cloned fragment and then subjected to dephosphorylation using CIP (New England Biolabs, Inc.). Approximately 2 pmol of each plasmid was then 5′ end labeled using 5 pmol of [γ-32P]ATP with T4 polynucleotide kinase, digested at a site at the other extreme of the cloned fragment, and the promoter fragments, labeled at one extremity, were purified by preparative PAGE as described above. Approximately 20,000 cpm of the labeled probe was used in each reaction for the footprinting experiments carried out as previously described (12). The binding reactions were performed for 10 min at room temperature in 50 μl of binding buffer (50 mM Tris-HCl [pH 7.8], 50 mM KCl, 10 mM MgCl2, 0.01% NP-40, 10% glycerol) containing 1 μg of salmon sperm DNA as a nonspecific competitor and metals or metal chelators were added when appropriate to the final concentrations indicated.
RESULTS
To explore both the molecular properties of the NikR protein in recognition of specific genomic targets and the connection between its action and that of the Fur regulator, we selected gene promoters that appear to be coregulated by the two proteins (9) and studied protein-DNA interactions under different conditions. The genes selected for our studies include nikR and the upstream divergent predicted operon exbB-exbD-tonB coding for the membrane complex involved in iron uptake and the ureAB genes encoding the nickel-containing urease enzyme, which is a major colonization determinant of H. pylori (20). Furthermore, the fur gene involved in iron regulation in H. pylori (11-13, 32) was also selected for our studies since genes of the Fur regulon have been reported as deregulated in the H. pylori NikR mutant (9). Since the promoter regions of some of these genes (ureA and fur) have already been documented, we first carried out promoter mapping of the nikR and exbB divergent operons. A schematic representation of the promoters under study is shown in Fig. 1A.
FIG. 1.
Promoters used in the present study. (A) Diagrammatic representation of loci used in the present study. (B) Mapping of the promoters under study. The two panels show primer extension of PexbB promoter (primer Ton-2, 5′-CTCCTTGTCTATGATAAAAC) and S1 mapping of the PnikR promoter. In both cases, the radioactively labeled primer or DNA probe was hybridized to 15 μg of total RNA extracted from logarithmically grown H. pylori G27 and either reverse transcribed (primer extension) or digested with S1 nuclease (S1 mapping), and the elongated products or RNA-protected bands are separated on an 8 M urea-polyacrylamide gel (lanes G27). For correct mapping of the elongated primer products, a sequencing reaction on the cloned promoter region, with the same primer as for the extension, was run in parallel on the gel (lanes G, A, T, and C). Instead, for correct mapping of the RNA-protected S1 products a G+A sequence reaction (19) on the radioactively labeled DNA probe was run in parallel on the gel (lane G+A). Bands representing the initiation of transcription of each promoter are highlighted with arrows and labeled accordingly. (C) Nucleotide sequences of the promoters mapped in the present study PexbB, and PnikR (Fig. 1B) and previous studies PureA (26) and Pfur (11). Alignment of the promoter sequences with respect to their transcriptional start sites (+1, in boldface) is shown. Putative −10 and −35 hexamers are highlighted in boldface, and the conservation with the E. coli σ70 consensus is underlined.
Mapping the PexbB and PnikR promoters.
The nikR-exbB intergenic region was cloned and primer extension was performed with multiple primers complementary to their transcripts. A clear elongation product was generated by using an exbB-specific primer on total RNA from the G27 strain, and the resultant primer extension reaction is shown in Fig. 1B. This band maps to 71 bp upstream of the ATG translational start site of the exbB gene, and the corresponding upstream nucleotide sequence shows high conservation with known promoter elements (Fig. 1C). We were unable to obtain reproducible results by primer extension with nikR-specific primers; therefore, we set up an S1 nuclease protection assay with a radioactively labeled DNA probe for mapping of the nikR promoter. S1 nuclease digestion after hybridization of the DNA probe with total RNA from the G27 strain resulted in two major and a faint minor RNA-protected bands (Fig. 1B) mapping to 55, 62, and 70 nucleotides, respectively, upstream of the ATG start site of the nikR gene. We conclude from this analysis that the +1 initiation of transcription probably corresponds to the higher band at 70 nucleotides. Although the nucleotide sequence upstream shows a perfect TATAAT −10 hexamer, very low conservation is found with the −35 hexamer (Fig. 1C).
We conclude that mapping of the promoters within the nikR-exbB intergenic region identified two single divergently oriented promoters, which we call PnikR and PexbB. We have previously reported the mapping of the PureA promoter (26) and the Pfur (11, 12). All genes appear to be transcribed from single promoters that are recognized by the H. pylori vegetative sigma factor (Fig. 1C).
NikR is a tetrameric protein.
In order to investigate the molecular properties of the NikR orthologue of H. pylori (HP1338 in reference 28), a recombinant form of the protein was expressed in the E. coli and purified by Ni affinity chromatography (Fig. 2A). The E. coli NikR protein has been shown to be tetrameric in structure independently of the nickel-bound status of the protein (24). To verify the oligomeric state of the protein, we performed gel filtration-size exclusion chromatography on a Superdex 200 HR 10/30 column. The protein was found to elute with a major peak at an elution volume (Ve) of 13.63 ml, and two minor peaks at Ve of 11.94 and 10.96 ml, respectively (Fig. 2B). On calibration of the column with standard molecular mass markers, the major peak was calculated to correspond to an estimated molecular mass of 64 kDa, and the minor peaks were estimated as 130 and 198 kDa, respectively. Since the NikR monomer has a predicted molecular mass of 17 kDa, the result from gel filtration is compatible with a predominant tetrameric state of the protein in solution. The two minor peaks of 130 and 198 kDa may correspond to interactions between two or three tetramers, respectively, although the origin of these peaks was not further investigated. The overnight dialysis of the protein in the presence of NiCl2 gave rise to a similar elution profile with a maximum peak eluting with an expected molecular mass of 64 kDa (data not shown); however, the overnight dialysis with nickel resulted in its partial precipitation. This suggests that NikR is present as a tetrameric protein in solution and, as with the E. coli orthologue, that Ni binding has no major effect on the oligomerization status of the protein.
FIG. 2.
NikR is a tetrameric protein. (A) SDS-PAGE of the nickel affinity purified protein preparation of the NikR protein before and after removal of the histidine tag. Lane 1, the purified protein with the histidine tag (His-NikR); lane 2, His-NikR after thrombin digestion and removal of the tag (rNikR); lane M, protein size standards, with molecular masses shown to the left. The arrows indicate the migration of the proteins. The protein was estimated to be 99% pure. (B) Elution profile of the purified rNikR protein from a Superdex 200 HR 10/30 column gel filtration column. The molecular masses corresponding to the peaks of maximal absorbance were calculated after calibration of the peaks with molecular mass standards (67, 43, 25, and 13.7 kDa with elution volumes of 13.52, 14.53, 16.31, and 17 ml, respectively).
Metal-dependent binding of NikR.
In a previous study with the NikR mutant of H. pylori, Contreras et al. (9) reported that NikR represses its own transcription and that of the exbBD-tonB genes located divergently upstream of the nikR gene itself. Furthermore, they showed that in gel shift assays the purified NikR protein was able to bind and retard a probe consisting of the upstream intergenic region, and they also suggested that a 28-bp palindrome within this region may represent the NikR operator. To identify the operator(s) of NikR within the nikR-exbB intergenic region, we performed DNase I footprinting analysis with the purified recombinant protein and a radioactively labeled probe (Fig. 3). We carried out multiple in vitro binding experiments in parallel with the addition of 100 μM NiCl2, 100 μM MnCl2, or 100 μM EDTA to each buffer to either provide or chelate divalent metal ions in each reaction in order to determine the metal dependency of the binding activity of the protein. In the presence of nickel, two areas of protection were observed on addition of 33 nM concentrations of the tetrameric NikR protein (panel I, lane 3). One region of protection spans from positions +1 to −37 with respect to the PexbB transcriptional start site, and the other region spanned from positions +10 to −27 with respect to the PnikR transcriptional start site. It is worth noting that at higher concentrations of NikR protein a hypersensitive band appears in the middle of the protected region overlapping the PnikR promoter at approximately −6. Protection is only partially visible at these operators at the highest protein concentration (300 nM) in the presence of 100 μM EDTA (panel II), indicating the chelator greatly reduced the affinity of the protein for these operators. Interestingly, in the presence of manganese salts protection at the operators is visible on addition of 100 nM concentrations of protein (panel III).
FIG.3.
In vitro metal-dependent binding of NikR and Fur proteins to the PnikR and PexbB promoters. DNase I footprinting with the NikR (A) or Fur (untagged and purified as described in reference 12) (B) proteins on a probe of the nikR-exbB intergenic region and in binding buffer containing different metals (100 μM NiCl2, 100 μM MnCl2) or metal chelators (100 μM EDTA, 100 μM dipyridyl) as indicated. The probe used consists of a 407-bp EcoRI-BamHI fragment containing the nikR-exbB intergenic region labeled at its BamHI site. The 5′-end-labeled probe was incubated with increasing amounts of purified protein: from lanes 1 to 5 corresponding to 0, 0.033, 0.1, 0.3, and 1 μM purified NikR tetrameric protein, respectively, in panel A and from lanes 1 to 5 corresponding to 0, 0.055, 0.165, 0.5, and 1.5 μM purified Fur dimer, respectively, in panel B. The vertical open and filled boxes on the right of each panel indicate the areas of DNase I protection resulting from binding of NikR and Fur proteins, respectively, and the numbers indicate the boundaries of the operators with respect to the +1 transcriptional start site of the PexbB and PnikR respective promoters as indicated. The arrowhead indicates a band of hypersensitivity that appears at high concentrations of rNikR protein on the PnikR promoter at approximately −6 with respect to transcriptional +1. The G+A lane is a G+A sequence reaction on the DNA probe used as size marker (19). The bent arrows to the left of the panels show the direction of transcription and position of the +1 of the PexbB and PnikR respective promoters within the probe. The converging arrows mark the position of a previously proposed NikR binding site (7, 9).
In summary, we identified two operators for the NikR protein within the nikR-exbB intergenic region, which overlap with the PnikR and PexbB promoters, respectively. The specificity of binding was not altered by the presence or absence of metal ions; however, the affinity of binding was seen to be altered and dependent on metal ions, and in vitro NikR bound more efficiently in the presence of Ni2+ ions than Mn2+ ions. These results are compatible with nickel-responsive DNA-binding activity. Furthermore, the palindromic sequence within this regulatory region (Fig. 3A and Fig. 4A), which has been hypothesized as the operator of NikR (7, 9), was not protected in DNase I assays with the NikR protein.
FIG. 4.
Binding of the NikR and Fur proteins at the overlapping and flanking operators of the PnikR or PexbB target promoters, respectively. (A) Summary of the key regulatory elements of the nikR-exbB locus that have been localized in the present study, including a schematic representation of the intergenic region and underneath the double-stranded nucleotide sequence spanning from the ATG translational starts (in boldface) of nikR-exbB genes, with determinants highlighted. The bent arrows indicate the direction of transcription and position of the +1 of the PexbB and PnikR promoters, and the −10 and −35 promoter elements are highlighted in black, respectively. The open boxes represent NikR binding sites or protected nucleotides resulting from the DNase I footprinting assays, and the triangles mark the boundaries of the NikR operators. The hatched gray boxes represent the Fur binding sites or protected nucleotides resulting from the DNase I footprinting assays, and the asterisks mark the boundaries of the Fur operators. The converging arrows highlight the positioning of the palindromic sequence (underlined) that has been proposed until now as the NikR operator within this region (7, 9). (B) Competition or independent binding of NikR and Fur at the operators at the overlapping and flanking operators of the PnikR or PexbB target promoters, respectively. All footprinting assays were performed with the addition of NiCl2 to the binding buffer. An equivalent amount of 5′-end-labeled probe was incubated without any protein (−); with increasing amounts of NikR (panel I) and Fur (panel II), independently; or with fixed relative maximal binding (RMB) concentration of Fur and increasing amounts of NikR (panel III); or with a fixed RMB concentration of NikR and increasing amounts of Fur (panel IV) as indicated above each lane. The RMB concentration for each protein is the minimal concentration tested that allows complete binding at both operators and was determined for each protein independently from panels I and II. The triangles and asterisks indicate the boundaries of the NikR and Fur operators, respectively. The arrowhead indicates a band of hypersensitivity that appears at high concentrations of NikR protein on the PnikR promoter.
Metal-dependent binding of Fur to the PexbB and PnikR promoters.
The TonB-ExbBD complex is an energy-transducing membrane complex, which provides energy across the cell envelope for high-affinity iron-uptake, and the genes in this operon are usually iron and Fur regulated (5). To determine whether Fur also binds to an operator(s) in the nikR-exbB intergenic region, we performed DNase I footprinting with purified Fur protein (12) and the radioactively labeled nikR-exbB probe. Again, multiple in vitro binding experiments were performed in parallel with the addition of 100 μM dipyridyl, 100 μM MnCl2, or 100 μM NiCl2 to each buffer. The results in Fig. 3 show that Fur binds specifically and with different affinities to two binding sites, resulting in two regions of protection within the probe. In the presence of the iron chelator dipyridyl (panel I), addition of 500 nM Fur dimer results in a region of protection spanning −37 to −70 of the PexbB promoter (lane 4) and at the next higher concentration of Fur (1.5 μM, lane 5) results in another region of protection spanning +3 to −30 of the PnikR promoter. In the presence of divalent metal ions (panel II and III), protection at PexbB and PnikR occurs on addition of 165 nM Fur (lanes 3) and 500 nM Fur dimer (lanes 4), respectively, suggesting that under these conditions Fur exhibits at least threefold-higher affinity for these operators, respectively. In conclusion, Fur binds in a metal-dependent manner to two operators within the nikR-exbB intergenic region, and Ni2+ or Mn2+ ions result in similar in vitro binding affinities of the protein. Fur binds with higher affinity to an operator upstream and proximal to the PexbB promoter and to a second operator with lower affinity overlapping the PnikR promoter.
Competition of Fur and NikR for PnikR and PexbB.
In the footprinting experiments described above, we identified and localized operators for NikR and Fur at the PexbB and PnikR promoters, and the relative positioning and corresponding nucleotide sequence are represented in Fig. 4A. Both proteins result in footprints in which approximately 35 contiguous nucleotides are protected due to binding. Whereas Fur and NikR bind to overlapping operators at the PnikR promoter and cause protection over a similar 30 nucleotides spanning +3 to −27 (Fur, +3 to −30; and NikR, +10 to −27), their operators at PexbB are distinct. Binding of Fur and NikR at the PexbB promoter occur in flanking regions (Fur, −35 to −70; and NikR, +1 to −37) with only two overlapping nucleotides resulting from the tandem location of their operators.
In order to analyze the possible interactions (either negative, i.e., competition, or positive, i.e., cooperativity) of these repressors at each promoter, we performed in vitro competition binding experiments. We set up binding reactions using individually the minimum concentration of each protein that resulted in complete protection at both of its operators and then added the second protein in increasing concentrations in order to observe the resulting pattern of binding in footprinting experiments. In Fig. 4B, subpanels I and II represent each of the Fur and NikR footprints when bound individually to the nikR-exbB radioactive probe and to facilitate recognition of the respective operators we have highlighted the boundaries of each with different symbols. Subpanel III shows the competition experiment in which the Fur dimer is constant at the 500 nM concentration which allows complete binding of both operators (lane 2), and the NikR protein was then added in increasing concentrations (lanes 3 to 6). As can be seen, binding at the PexbB promoter occurs at 33 nM concentrations of NikR tetramer in the presence of the Fur protein (subpanel III, lane 3) and does not interfere with the binding of Fur since its operator protection occurs at both flanking sites on the probe (lanes 4 to 6). In contrast, at the PnikR promoter, whereas in the absence of NikR the footprint is clearly due to the Fur protein (lane 2, borders indicated with an asterisk) at concentrations of ≥300 nM of NikR a pattern of protection of the NikR protein is evident at the PnikR promoter (lanes 5 and 6, indicated with “Δ”). This indicates an outcompetition by NikR of the Fur protein for occupancy of the overlapping region of the probe. Similarly, in the presence of 100 nM, NikR binding was established at both operators (subpanel IV). The addition of Fur in increasing concentrations results in protection of the PexbB operator at 165 nM (subpanel IV, lane 3) and did not affect NikR binding, whereas at concentrations of ≥250 nM it out-competed NikR for occupancy of the PnikR operator (subpanel IV, lanes 4 and 5).
These experiments demonstrate that the NikR and Fur protein compete with each other for occupancy of the PnikR operator but can bind simultaneously and independently of each other at the PexbB promoter.
NikR binds with high affinity to the urease promoter.
The urease promoter has been previously mapped (26), and deletion and mutation analysis has identified a cis-acting region upstream of the promoter elements that, although not required for basal transcription of PureA (10), is responsible for NikR-mediated nickel-responsive induction of the promoter (34). In order to understand the exact nature of this region, we investigated whether it may contain a NikR binding site and the DNase I footprinting of a radioactively labeled probe containing the PureA promoter region is shown in Fig. 5A. On addition of 11 nM concentrations of NikR tetramer, protection of nucleotides spanning −56 to −87 with respect to the PureA transcriptional start site occurs, indicating that NikR binds with high affinity to an operator in the urease promoter. In Fig. 5B we aligned the urease promoter sequences of five strains of H. pylori and reported the position of the regulatory elements therein. The protected nucleotides corresponding to the ureA NikR-operator correlate very closely to the cis-acting site from −48 to −68 pinpointed by van Vliet et al. (34) and which in turn corresponds to a region of high conservation within the 5 strains (Fig. 5B). However, comparing the sequence of the urease operator and the two previously defined operators overlapping the PnikR and PexbB promoters (above) showed no obvious similarities to the palindromic operator (GTATGA-N16-TCATAC) of the E. coli NikR protein.
In order to define the NikR operator sequences required for high-affinity binding of the protein to its operator, we performed scanning mutagenesis of the nucleotides of the urease operator. The urease operator was chosen since NikR shows the highest affinity to this operator of the ones tested thus far. The mutant operators numbered 1 to 10 were constructed with ccc trinucleotide substitutions spanning the operator as shown in Fig. 5C, and the fold-affinity changes of binding of NikR to the mutant operators were assessed in parallel footprinting experiments performed with the wild-type urease probe. NikR was added in twofold increasing amounts, the concentration at which 50% of the probe was estimated to be protected was taken as the relative half-maximal binding for that operator derivative (data not shown), and the relative fold changes in affinity were calculated accordingly (Fig. 5C). Two of the mutant operators OP-Ure3 and OP-Ure8 showed drastic reductions in affinity for NikR of more than 256- and 128-fold, respectively, and another three mutations—OP-Ure2, OP-Ure4, and OP-Ure9—showed moderate reductions of 32-, 16-, and 8-fold, respectively (Fig. 5C). At the remaining operator mutants OP-Ure1, OP-Ure5, OP-Ure6, OP-Ure7, and OP-Ure10 no significant changes in affinity were detectable.
These experiments indicate that there are two subsites that are important for the binding of the H. pylori NikR protein within a maximum of 26 nucleotides of operator with at least nine nucleotides in the center which have no role in the affinity of the binding per se. The two subsites in the urease promoter, although generally rich in AT, do not show a strong palindromic pattern to their sequence, and the right-hand subsite is more conserved among the five strains (Fig. 5B).
Identification of NikR operators at the fur promoter and regulation of Pfur transcription by metal-responsive regulators and metals.
We have shown above that NikR and Fur can bind and compete for occupancy of specific operators over the PnikR promoter. We have previously shown that Fur binds to its own promoter and to the promoter of the pfr gene, and both of these genes have been reported as being deregulated in the NikR mutant (9). In order to identify possible operators for NikR within these promoters, we performed footprinting on radioactively labeled probes containing the promoters of the pfr and fur genes. No protection was observed on binding of NikR with the pfr probe (data not shown), and the resulting footprint on the fur promoter probe is shown in Fig. 6A. On addition of 100 nM NikR protein protection is detectable at two regions of the fur probe which we call OpI and OpII spanning from −1 to −27 and from −86 to −122 with respect to the Pfur promoter (lane 4). At higher concentrations of protein, the protection expands between the operators and results in the appearance of a hypersensitive band at −37 of the Pfur promoter. Previous studies have identified the location of Fur operators, OI to OIII, at the fur promoter and are indicated to the right of Fig. 6A, and the OpI and OpII NikR operators overlap the OII and OIII Fur operators, respectively. It is worth noting also that Fur did not result in protection of the PureA probe in similar footprinting experiments (data not shown). We conclude that the NikR protein interacts in vitro with the Pfur promoter, and therefore fur may represent a member of the NikR regulon. Furthermore, while the urease promoter appears to be a direct target only of NikR, the pfr promoter is only bound by Fur (12).
FIG. 6.
Binding and regulation of the metal-responsive regulators to the fur promoter. (A) Identification of NikR operators in the fur promoter by DNase I footprinting of NikR on a radioactively labeled Pfur probe consisting of a 220-bp BamHI-PstI fragment from plasmid pGemCl2 labeled at its BamHI site. An equivalent amount of 5′-end-labeled probe was incubated with increasing amounts of NikR, with 0, 0.011, 0.033, 0.10, 0.30, 0.9, and 2.5 μM purified protein corresponding to lanes 1 to 7, respectively. The vertical open bars on the right indicate the areas of DNase I protection resulting from binding of NikR; the filled boxes represent the positions of footprints due to the Fur protein previously reported (9, 11), and the numbers indicate the boundaries of the operators with respect to the +1 transcriptional start site of the respective promoters as indicated. Lane G+A is a G+A sequence reaction on the DNA probe used as a size marker (19). The open bars and the bent arrow on the left indicate the position of the Pfur promoter. (B) Western blot showing derepression of the Fur protein in mutants of the metal-responsive regulators NikR and Fur. Total protein extracts of the G27 wild-type strain (wt), a NikR-null mutant (ΔnikR), a Fur-null mutant (Δfur), and the mutant G27(H99I) expressing the iron-blind protein (FurH99I) collected at logarithmic phase were transferred after SDS-PAGE onto nitrocellulose and stained with anti-Fur antiserum. The band corresponding to the Fur protein is highlighted with an arrow, and the asterisk denotes a cross-reacting band which serves as a control. (C) Quantitative primer extension of the Pfur promoter showing transcriptional regulation in response to metals and metal-responsive regulators. Total RNA was extracted from cells of the G27 wild type (wt), the NikR-null mutant (ΔnikR), and G27(H99I) expressing the FurH99I iron-blind protein, which were grown to logarithmic phase (−) and then treated for 20 min. with 1 mM FeCl2 (Fe2+), 100 μM 2,2′-dipyridiyl (Dip), or 5 mM NiCl2 (Ni2+). Primer extension was performed on 10 μg of total RNA from each preparation with the Fur-R2 primer (11), and the elongation product corresponding to the Pfur transcript is indicated. (D) Alignment of the NikR operators with respect to the determinants of the urease operator. Variations in the nucleotide sequences between the available sequences are shown in lowercase gray-shaded letters. Conserved bases and the proposed consensus operator are shaded in black. The E. coli NikR consensus is reported underneath.
In order to understand the physiological relevance of the protein-promoter interactions at Pfur we investigated whether the expression of the Fur protein was altered in H. pylori NikR and Fur mutants. To do this, we performed Western blot analysis to compare the amount of Fur in the wild type to that in isogenic NikR and Fur mutants, including a mutant strain expressing an iron-blind Fur protein (FurH99I), previously generated for autoregulatory studies (11). As expected (Fig. 6B), while no Fur protein was detected in the Fur-null mutant (Δfur), high levels of expression are detected in the FurH99I mutant. Interestingly, in the NikR mutant (ΔnikR) the amount of Fur increases with respect to the wild-type strain, though not to the same levels as in the FurH99I mutant. This suggests that NikR represses expression of the fur gene, although to a lesser extent compared to the autoregulatory effect of Fur.
We also measured the transcription from the Pfur promoter in the wild type and NikR and FurH99I mutants in response to treatment with iron, iron chelation, and nickel. Quantitative primer extension on total RNA from untreated cells (−) of the three strains confirmed the results in the Western blot that the transcription is derepressed in the NikR and FurH99I mutants (Fig. 6C). In the wild-type strain addition of chelator derepressed the promoter; however, the addition of iron or nickel did not significantly change the amount of transcript (top panel). In the NikR mutant the promoter is repressed on addition of iron and nickel and derepressed on iron chelation (middle panel), and finally in the FurH99I mutant the promoter no longer responds to iron or iron chelator but is slightly repressed by the addition of nickel (bottom panel). Taken together, these results suggest that Fur can repress its own promoter, Pfur, in response to iron and nickel and that the NikR protein represses also the Pfur promoter, although to a lesser extent, in response to nickel.
In light of these results we can align the operator sequences of the in vitro binding sites of NikR determined in the present study and, on the basis of the functional data from the scanning mutagenesis of the high-affinity urease operator, we propose a consensus NikR operator consisting of 2 AT-rich inverted repeats (Fig. 6D).
DISCUSSION
In this study, we initiated the in vitro characterization of the H. pylori NikR protein and also investigated its interplay with the other metal responsive regulator protein of H. pylori Fur. We identified a number of distinct operators for NikR and for Fur in promoters of coregulated target genes at which they may compete for occupancy or bind independently. Collectively, the in vitro experiments fit a relatively simple model of regulation whereby the metal responsive regulator proteins sense Ni2+ and Fe2+ directly to effect affinity for promoter elements of metal-regulated genes.
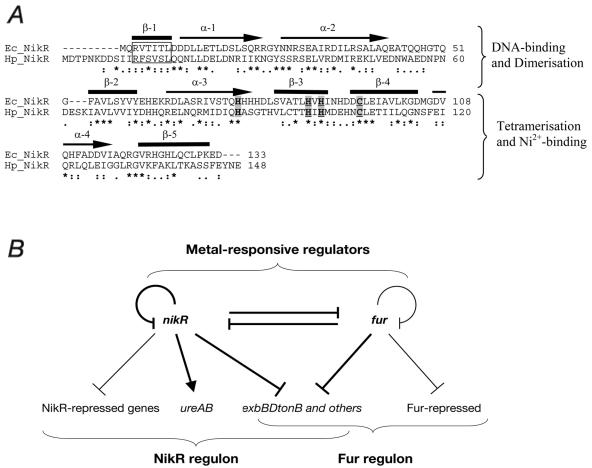
The nikR gene of H. pylori is a homologue of the nickel-responsive repressor NikR of E. coli, although they only share 28% identity, the predicted secondary structure of the proteins are similar and they share 58% similarity (Fig. 7A). The results reported here highlight a number of differences between it and the E. coli NikR protein. With respect to DNA recognition, we demonstrated that NikR of H. pylori binds to an inverted repeat with two distally located subsites, although to an operator different from that which has been considered its binding site up to now. The NikR homologue of E. coli has been shown to bind to an operator consisting of two 5′-GTATGA-3′ half sites related by dyad symmetry and separated by 16 bp (7). A related sequence was identified in the H. pylori nikR promoter region 5′-GCATGA-3′ exhibiting perfect dyad symmetry and similarly separated by 16 bp (7), and site-directed mutagenesis of one of the hexamers was reported to result in deregulation of nikR-lacZ promoter fusion in the E. coli background (9). In the footprinting experiments presented in the present study, we demonstrated that NikR protein does not bind to this palindromic sequence within the nikR-exbB intergenic region but instead to two distinct operators overlapping the PnikR and PexbB elements (Fig. 3A). The deregulation observed by Contreras et al. (9) may have been a result of the destruction of a stem-loop structure in the RNA. Indeed, the PnikR promoter proved very difficult to map correctly and this may also be due to complex secondary structures in the 5′ end of the mRNA.
FIG. 7.
(A) Alignment of the amino acid sequences of the H. pylori (Hp) and E. coli (Ec) NikR proteins with secondary structure elements highlighted and Ni2+-binding amino acids shaded as elucidated from the crystal structure of E. coli NikR (24). The N terminus The β-1 strand of the RHH structure responsible for DNA recognition is boxed. (B) Model encompassing the direct interplay of the metal-responsive regulators of H. pylori elucidated in footprinting experiments in the present study (thick lines) and thin lines indicate the interplay reported in previous studies (11, 12, 33) and possibly others. According to gene organization deduced from the genome sequence, the exbB, exbD, and tonB genes appear to constitute an operon.
We identified a number of distinct operators for the protein at promoters of genes that have been reported to be NikR-regulated (9, 31, 34). It was surprising to us that there was no obvious conserved “NikR-box” sequence within these operators that were bound by the protein. It would appear that compared to the perfect palindromic sequence of the E. coli NikR operator, the H. pylori NikR tolerates sequence variations within the subsites to a significant degree. Through scanning mutagenesis of the highest-affinity operator, we show that two subsites determine the NikR operator and this is consistent with the tetrameric oligomerization status of the protein in solution. The functional unit for most ribbon-helix-helix (RHH) members is a tetramer in which the antiparallel sheets of each dimer recognizes a subsite of the operator (Arc, Mnt, NikR) (17). From alignment of the NikR operators, a consensus sequence consisting of an inverted repeat can be proposed consisting of an AT-rich palindrome (Fig. 6B). This consensus shows remarkable differences both in sequence and distance of the subsites compared to the E. coli one. It is well documented that sequence specificity in this family of RHH regulators arises from direct interactions between side chains of the N-terminal beta-strands and the edges of the bases in the major groove (24). The N terminus of the NikR protein differs greatly from the E. coli homologous protein in that it contains an additional nine amino acid residues and, more importantly, there are only two of six identical residues in the beta-ribbon (Fig. 7A). This difference in amino acid residues may account for the different sequence specificity of the two proteins. Furthermore, the C-terminal region, responsible for tetramerization, may dictate the distance between the subsites (Fig. 6B).
In this report by studying the binding of purified NikR and Fur to various target promoters in vitro, we have begun to define the overlapping regulons of these metal-responsive regulators, which brings important implications into the transcriptional regulation in H. pylori. We have expanded the Fur regulon to include novel operators in the promoter regions of PnikR and PexbB, and Fig. 7B shows a model of the direct links we defined here (and in previous studies) in relation to the regulation that has been previously reported. The binding of NikR either independently or in competition with Fur to PnikR and PexbB may result in occlusion of the RNA polymerase or inhibition of open complex formation and repression of these two promoters and their downstream genes. The TonB-ExbBD complex is involved in high-affinity iron uptake and is usually iron regulated (5). Contreras et al. (9) have demonstrated that these genes are derepressed in the NikR mutant. In the present study, we identified operators for both the NikR and the Fur protein overlapping the PexbB promoter. It would appear from our competition studies that NikR and Fur can bind to adjacent operators at the PexbB promoter independently of each other in vitro, whereas they can outcompete each other for the overlapping operators at PnikR. Indeed, the PexbB promoter is derepressed in the Fur mutant, and no significant differences in the PnikR transcript could be detected in a Fur− background (I. Delany and V. Scarlato, unpublished results). Fur autoregulates its own promoter in response to iron by binding at three operators, each playing a distinct role in its regulation (11), and once again here we show that NikR also may bind to the Pfur promoter and repress it in response to nickel (Fig. 6). This is in agreement with data from van Vliet et al. (31) in which through Northern blot analysis the Fur transcript was highly derepressed in the nikR mutant of H. pylori. Furthermore, two promoters are bound and directly regulated exclusively by NikR or Fur and these are the urease and pfr promoters, respectively (9, 12, 13, 34). Moreover, we demonstrated that NikR binds to a high-affinity operator upstream of the urease promoter that overlaps with the previously identified cis-acting region of this promoter which is responsible for NikR-mediated nickel-induction of the promoter (34). This introduces a second regulatory function for this protein other than repression, that NikR may act directly as a nickel responsive activator of the PureA promoter; however, it is also possible that NikR may recruit the actual activator protein to the urease promoter. The RHH family consists mainly of transcriptional repressors; however, two of the protein AlgZ and TraY have also been shown to be involved in upregulation or activation of promoters (2, 25).
In conclusion, we clarified certain distinctions between the E. coli and H. pylori orthologues of NikR. Whereas NikR of E. coli is known only as a specific repressor of the nickel permease operon with a very defined operator, H. pylori NikR appears to have many target genes at which it binds to operators with different affinities. Furthermore, NikR is now directly implicated in the upregulation of the urease promoter. NikR and Fur regulatory circuits seem to be intricately intertwined since each protein may bind separately or in competition with each other for the occupancy of overlapping but distinct operators on some coregulated genes (Fig. 7B). Furthermore, not only do these proteins autoregulate their own promoters, they also may directly bind to each other's promoters and, in doing so, indirectly regulate the expression of the genes of each other's regulons. Finally, there seems to be an exquisite equilibrium in the action of these two regulators highlighting the importance of metal homeostasis within H. pylori.
Acknowledgments
We thank Giorgio Corsi for artwork and Catherine Mallia for manuscript editing.
This study was supported by Chiron and partially by a grant from the Italian Ministry of Education, University, and Research and by the University of Bologna to V.S.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baynham, P. J., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. [DOI] [PubMed] [Google Scholar]
- 3.Bereswill, S., S. Greiner, A. H. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J., B. Waidner, A. H. Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 6.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 7.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high-affinity nickel uptake in bacteria: Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 8.Chivers, P. T., and R. T. Sauer. 2002. NikR repressor: high-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chem. Biol. 9:1141-1148. [DOI] [PubMed] [Google Scholar]
- 9.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947-963. [DOI] [PubMed] [Google Scholar]
- 10.Davies, B. J., N. de Vries, S. G. Rijpkema, A. H. van Vliet, and C. W. Penn. 2002. Transcriptional and mutational analysis of the Helicobacter pylori urease promoter. FEMS Microbiol. Lett. 213:27-32. [DOI] [PubMed] [Google Scholar]
- 11.Delany, I., G. Spohn, A. B. Pacheco, R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2002. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46:1107-1122. [DOI] [PubMed] [Google Scholar]
- 12.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and iron-repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 13.Ernst, F. D., S. Bereswill, B. Waidner, J. Stoof, U. Mader, J. G. Kusters, E. J. Kuipers, M. Kist, A. H. van Vliet, and G. Homuth. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiol. 151:533-546. [DOI] [PubMed] [Google Scholar]
- 14.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 17.Knight, K. L., and R. T. Sauer. 1989. DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal residues. Proc. Natl. Acad. Sci. USA 86:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labigne, A., V. Cussac, V., and P. Courcoux. 1991. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol. 173:1920-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulrooney, S. B., and R. P. Hausinger. 2003. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 27:239-261. [DOI] [PubMed] [Google Scholar]
- 22.Pflock, M., S. Kennard, I. Delany, V., Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 76:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Schreiter, E. R., M. D. Sintchak, Y. Guo, P. T. Chivers, R. T. Sauer, and C. L. Drennan. 2003. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10:794-799. [DOI] [PubMed] [Google Scholar]
- 25.Silverman, P. M., and A. Sholl. 1996. Effect of traY amber mutations on F-plasmid traY promoter activity in vivo. J. Bacteriol. 178:5787-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 28.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 29.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 30.van Vliet, A. H., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Vliet, A. H., E. J. Kuipers, J. Stoof, S. W. Poppelaars, and J. G. Kusters. 2004. Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade. Infect. Immun. 72:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Vliet, A. H., J. Stoof, R. Vlasblom, S. A. Wainwright, N. J. Hughes, D. J. Kelly, S. Bereswill, J. J. Bijlsma, T. Hoogenboezem, C. M. Vandenbroucke-Grauls, M. Kist, E. J. Kuipers, and J. G. Kusters. 2002. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237-244. [DOI] [PubMed] [Google Scholar]
- 33.van Vliet, A. H., J. Stoof, S. W. Poppelaars, S. Bereswill, G. Homuth, M. Kist, E. J. Kuipers, and J. G. Kusters. 2003. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J. Biol. Chem. 278:9052-9057. [DOI] [PubMed] [Google Scholar]
- 34.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang, Z., S. Censini, P. F. Babeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]