Abstract
The xlnD gene from Pseudomonas alcaligenes NCIMB 9867 (strain P25X) was shown to encode 3-hydroxybenzoate 6-hydroxylase I, the enzyme that catalyzes the NADH-dependent conversion of 3-hydroxybenzoate to gentisate. Active recombinant XlnD was purified as a hexahistidine fusion protein from Escherichia coli, had an estimated molecular mass of 130 kDa, and is probably a trimeric protein with a subunit mass of 43 kDa. This is in contrast to the monomeric nature of the few 3-hydroxybenzoate 6-hydroxylases that have been characterized thus far. Like other 3-hydroxybenzoate 6-hydroxylases, XlnD could utilize either NADH or NADPH as the electron donor. P25X harbors a second 3-hydroxybenzoate 6-hydroxylase II that was strictly inducible by specific aromatic substrates. However, the degradation of 2,5-xylenol and 3,5-xylenol in strain P25X was found to be dependent on the xlnD-encoded 6-hydroxylase I and not the second, strictly inducible 6-hydroxylase II.
Oxygenases play crucial roles in microbial catabolic pathways as these enzymes initiate the degradation of aromatic compounds by either hydroxylating the aromatic ring in preparation for ring cleavage or catalyzing the ring fission reaction itself. One of the early steps in aromatic degradation involves the introduction of one or two hydroxyl groups on the aromatic ring; the introduction of single hydroxyl groups on the aromatic ring is generally catalyzed by monooxygenases (8). Dihydroxylated aromatic compounds are, in turn, substrates for ring cleavage dioxygenases which cleave open the aromatic ring by the incorporation of two atoms of oxygen. The products of the aromatic ring cleavage are then further metabolized to tricarboxylic acid cycle intermediates (7).
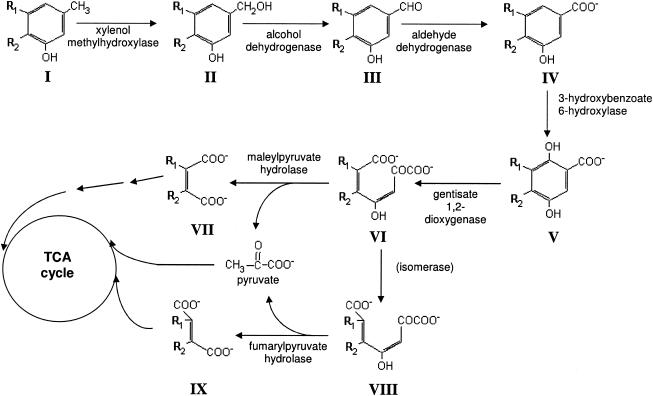
Pseudomonas alcaligenes NCIMB 9867 (strain P25X) is able to utilize 2,5-xylenol, 3,5-xylenol, and m-cresol as sole carbon and energy sources via the gentisate pathway (Fig. 1) (10, 11). Prior research with several P25X-derived catabolic mutants revealed the existence of isofunctional enzymes of the gentisate pathway—one set being constitutively expressed and the other set being strictly inducible by aromatic substrates such as 3-hydroxybenzoate and gentisate (20, 21). We recently cloned and characterized the constitutive copy of the gentisate 1,2-dioxygenase gene, designated xlnE, from strain P25X and showed that it is part of an operon of six genes, xlnEFGHID, that is transcribed from a σ70-type promoter, PxlnE, located 123 bp upstream of the xlnE start codon (29). Transcription from PxlnE was found to be inducible by specific aromatic substrates, in particular, 3-hydroxybenzoate, and its relatively high basal levels of expression in the absence of aromatic inducers is responsible for the apparent constitutive activity of one of the two isofunctional gentisate 1,2-dioxygenases, designated GDO-I, that was described earlier (20). The functions for the other genes in the xln operon have yet to be determined. Based on sequence similarity, the last gene in the operon, xlnD, was postulated to be 3-hydroxybenzoate 6-hydroxylase that catalyzes the conversion of 3-hydroxybenzoate to gentisate (29). The genetics and catalytic mechanism of hydroxylases that catalyze hydroxylation at a position ortho to an existing hydroxyl group of the aromatic ring has been widely characterized for 4-hydroxybenzoate hydroxylase (1, 4, 5, 19, 27). However, very little is known about the genetics and biochemistry of hydroxylases that catalyze the hydroxylation of the aromatic ring at a position para to an existing hydroxyl group, with 3-hydroxybenzoate 6-hydroxylase having been purified and characterized thus far only from Pseudomonas aeruginosa (6), Micrococcus sp. (22). Burkholderia (formerly Pseudomonas) cepacia (28) and Klebsiella pneumoniae (12, 25), from which the gene was recently cloned (16). Here, we show that xlnD from strain P25X does indeed encode for 3-hydroxybenzoate 6-hydroxylase I and present its biochemical and catalytic properties. This is, to our knowledge, the first detailed characterization of the P. alcaligenes-encoded 3-hydroxybenzoate 6-hydroxylase I, a crucial enzyme in the gentisate pathway of aromatic hydrocarbon degradation. From an xlnD knockout constructed from strain P25X, we offer evidence indicating that the degradation of 2,5-xylenol and 3,5-xylenol in strain P25X is dependent on the xlnD-encoded 3-hydroxybenzoate 6-hydroxylase I and not the second, strictly inducible 3-hydroxybenzoate 6-hydroxylase II.
FIG. 1.
Gentisate pathway for the degradation of 2,5-xylenol, 3,5-xylenol and m-cresol by P. alcaligenes P25X. Compounds (when R1 = H and R2 = H): I, m-cresol; II, 3-hydroxy-benzylalcohol; III, 3-hydroxy-benzaldehyde; IV, 3-hydroxybenzoate; V, gentisate; VI, maleylpyruvate; VII, maleate; VIII, fumarylpyruvate; IX, fumarate. Compounds (when R1 = H and R2 = CH3): I, 2,5-xylenol; II, 3-hydroxy-4-methylbenzylalcohol; III, 3-hydroxy-4-methylbenzaldehyde; IV, 3-hydroxy-4-methylbenzoate; V, 4-methylgentisate; VI, 5-methylmaleylpyruvate; VII, citraconate. Compounds (when R1 = CH3 and R2 = H): I, 3,5-xylenol; II, 3-hydroxy-5-methylbenzylalcohol; III, 3-hydroxy-5-methylbenzaldehyde; IV, 3-hydroxy-5-methylbenzoate; V, 3-methylgentisate; VI, 6-methylmaleylpyruvate; VII, citraconate.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
Escherichia coli strain DH5α [ϕ80d lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169] (23) was used as the host for cloning experiments. E. coli BL21(DE3)/pLysS [F− ompT hsdSB (rB− mB−) dcm gal λ(DE3) pLysS (Cmr)] was purchased from Novagen and used as the host for the overexpression of genes cloned into the pET28a and pET30a expression vectors. E. coli S17-1 (thi pro hsdR hsdM+recA Strr RP4 tra) (24) was used as the host to mobilize pRK415 derivatives into P. alcaligenes.
E. coli strains were grown at 37°C in either Luria broth or agar (18), whereas Pseudomonas alcaligenes P25X and its derivatives were grown at 32°C in minimal media containing 20 mM sodium lactate as the sole carbon source (9). Aromatic hydrocarbons used as inducers such as 3-hydroxybenzoate, 3-hydroxy-4-methylbenzoate, and gentisate, were introduced into cultures at a final concentration of 2.5 mM. To assess growth on aromatic substrates, P25X wild-type and its derivatives were incubated at 32°C for 4 days in minimal agar containing 2.5 mM concentration of the following aromatic compounds as the sole carbon source: 2,5-xylenol, 3,5-xylenol, 3-hydroxybenzoate, 3-hydroxy-4-methylbenzoate, or gentisate. Growth of P25X cells was also assessed in liquid minimal media containing the same aromatic compounds by measuring the optical density at 600 nm (OD600) every 6 h for 96 h.
When required, antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg ml−1; kanamycin (Km), 60 μg ml−1; chloramphenicol (Cm), 34 μg ml−1; streptomycin (Sm), 100 μg ml−1; spectinomycin (Sp), 100 μg ml−1; and tetracycline (Tc), 30 μg ml−1.
DNA manipulations, sequencing, and sequence data analysis.
DNA manipulations and other molecular biology techniques were carried out by using established protocols (23). Plasmid DNA was isolated by using the Wizard SV Miniprep kit (Promega). DNA fragments and PCR products were purified by using the GFX purification kit from Amersham Biosciences, Sweden. DNA sequencing was carried out by using the BigDye terminator cycle sequencing ready reaction kit and an ABI Prism 377 DNA sequencer (Applied Biosystems). DNA sequences were compiled and analyzed by using the Lasergene sequence analysis software (DNASTAR). Comparison of nucleotide and amino acid sequence data was carried out by using BLAST at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/BLAST). The nucleotide sequence for the P. alcaligenes xln operon has been deposited in GenBank under accession no. AF173167.
Construction of pXlnD::SmSp and a P25X xlnD knockout mutant strain, G50.
The xlnD gene on pTOPOxln, which comprised the 5.4-kb xln operon and 2.0 kb of upstream sequences cloned into the pCR-TOPO-XL cloning vector (Invitrogen), was disrupted through in vitro transposition with a custom streptomycin-spectinomycin resistance (Sm/Spr) transposon constructed by using the EZ::TN pMOD MCS transposon construction vector kit (Epicenter Technologies) as had been previously described for xlnE (29). DNA sequencing of one of the transposon mutants obtained, pXlnD::SmSp, showed that the custom Sm/Spr transposon had inserted into xlnD at nucleotide 8086. pXlnD::SmSp was then introduced into strain P25X, which is naturally competent toward its own homologous DNA (15), by natural transformation and selected on LB agar containing Sm and Sp. To allow for the uptake of pXlnD::SmSp into P25X cells, a 10-ml overnight culture of P25X cells was harvested and resuspended in 0.1 ml of LB broth. Approximately 0.1 μg of DNA was mixed with the resuspended cells which were then spotted onto a prewarmed LB agar plate and left overnight at 32°C. The cells were later harvested from the plate using a sterile inoculating loop and resuspended in 1 ml of LB broth. Aliquots of 0.1 ml were then plated out on the selection plates. Transformation frequencies of ca. 5 × 10−7 transformants per bacterial recipient were obtained. The resulting xlnD knockout mutant, designated strain G50, is a result of gene replacement through a double crossover, and this was confirmed by PCR amplification of xlnD from the genome of G50 followed by sequence analysis.
Complementation of xlnD in G50.
To complement the xlnD::SmSp lesion in strain G50, intact xlnD was provided in trans on the broad-host-range plasmid, pRK415 (14). xlnD was PCR amplified from the genome of P25X wild-type by using Pfu DNA polymerase (Stratagene) and the primers xlnDKpnF (5′-ATC TGG GTA CCG ACG ACG ACG AC-3′) and xlnDKpnR (5′-CTT TAG GTA CCG AGC CTA AGA CTA-3′), which incorporated KpnI restriction sites at the 5′ ends of the primers. The amplified product was digested with KpnI and cloned into the KpnI site of pRK415, resulting in the recombinant plasmid designated pRK415xlnD. Transcription of xlnD in pRK415xlnD is driven by the lac promoter of the pRK415 vector and its expression was confirmed by carrying out enzyme assays in the E. coli DH5α host. pRK415xlnD was mobilized into P. alcaligenes strain G50 by conjugation with E. coli S17-1 carrying the recombinant plasmid, and transconjugants were selected on minimal media plates containing 3-hydroxybenzoate and supplemented with Tc, Sm, and Sp.
Construction of recombinant plasmids for overexpression of xlnD in E. coli.
To overexpress xlnD in E. coli, xlnD was cloned into the expression vectors pET28a and pET30a (Novagen). xlnD was PCR amplified from P25X wild-type genomic DNA by using Pfu DNA polymerase (Stratagene) and primers XlnD-SN-F (5′-AGG AGC TCC CAT GGT GAT GCA TAA TAA TAT CTT GAT-3′) and XlnD-S-R (5′-ACA AGA GCT CCG GCG ATG GTG TCA-3′), both of which contain SacI restriction sites (underlined) at their 5′ ends. The XlnD-SN-F primer also contains an NcoI site (italicized) immediately following the SacI site and the ATG start codon of xlnD, which is indicated in boldface. The XlnD-S-R primer was designed to anneal 64 nucleotides (nt) downstream of the xlnD stop codon. The amplified product obtained was digested with SacI and cloned into pUC18, resulting in the recombinant plasmid pUC18xlnD, which was subsequently sequenced to ensure that no mutations were incorporated into xlnD during PCR amplification. The xlnD fragment in pUC18xlnD was then excised with NcoI and SacI digestions and cloned into NcoI- and SacI-digested pET28a and pET30a, resulting in the recombinant plasmids designated pET28xlnD and pET30xlnD, respectively. Since the NcoI site in pET28a is upstream of the hexahistidine (His6) coding sequence, recombinant pET28xlnD would express xlnD from the T7 promoter of pET28a but without the His6 tag. For pET30xlnD, xlnD is cloned downstream and in-frame with the His6 coding sequence and would thus result in the expression of an N-terminal His6-XlnD fusion protein. Both pET28xlnD and pET30xlnD constructs were subsequently transformed into E. coli BL21(DE3)/pLysS for overexpression.
Preparation of E. coli cell extracts.
E. coli BL21(DE3)/pLysS cells carrying either pET28xlnD or pET30xlnD were grown overnight in 10 ml of LB broth containing Km. A 2.5-ml aliquot of the overnight culture was inoculated into fresh 250 ml of LB broth and grown with shaking at 37°C until an optical density at 600 nm (OD600) value of ca. 0.6. The cultures were then induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM and allowed to grow with shaking for another 4 h. The cells were harvested by centrifugation (7,000 × g, 5 min, 4°C) and resuspended in 50 ml of 50 mM MOPS (morpholinepropanesulfonic acid; pH 7.4) containing 10% glycerol. Cell lysis was carried out by passing the resuspended cell solution through a French press Basic Z model (Constant Systems, Ltd., United Kingdom) operating at a pressure of 15,000 lb/in2. Cell extracts were clarified by centrifugation (100,000 × g, 1 h, 4°C) and used in the enzyme assays as well as for protein purification.
Preparation of P. alcaligenes cell extracts.
A 5-ml overnight LB culture of strain P25X or its derivatives was inoculated into 250 ml of minimal medium containing 20 mM lactate. The culture was allowed to grow with shaking at 32°C until the OD600 reached 0.5 to 0.6. Aromatic hydrocarbons used as inducers, i.e., 3-hydroxybenzoate, 3-hydroxy-4-methylbenzoate, and gentisate, were introduced into the cultures at a final concentration of 2.5 mM, and growth was continued for further 6 h. The cells were then harvested by centrifugation (7,000 × g, 5 min, 4°C) and resuspended in 50 mM MOPS (pH 7.4) containing 10% glycerol. Cells were lysed by passing through a French press operating at a pressure of 15,000 lb/in2, and the resulting cell extract clarified by centrifugation at 100,000 × g for 1 h at 4°C prior to carrying out enzyme assays.
Assay for 3-hydroxybenzoate 6-hydroxylase and determination of its kinetic properties.
The 3-hydroxybenzoate 6-hydroxylase activity was determined by measuring the decrease in absorbance at 340 nm due to the substrate-dependent oxidation of NADH, the molar extinction coefficient of which was taken as 6,200 M−1 cm−1 (16, 28). Assays were carried out in 100 mM potassium phosphate buffer (pH 7.4) at 23°C in a UV-2550 spectrophotometer (Shimadzu, Japan). The sample cuvette contained 100 mM potassium phosphate buffer, 0.2 μmol of substrate, and 0.5 μmol of NADH. The problem of high endogenous NADH oxidase activity, present in crude cell extracts of P. alcaligenes and the E. coli host but not in the purified 6-hydroxylase, was overcome by using the reference blank cuvette containing all assay components except the substrate to compensate for NADH oxidase, similar to the spectrophotometric assay used to detect salicylate 5-hydroxylase activity (30). The assay was initiated by the addition of ultracentrifuged extracts to both sample and reference cuvettes. One unit of enzyme activity is defined as the conversion of 1 μmol of NADH to NAD+ per min at 23°C. Specific activities are expressed as units per mg of protein. Protein concentration was determined by the method of Bradford (3) with bovine serum albumin as the standard.
A series of substrate solutions (3-hydroxybenzoate or 3-hydroxy-4-methylbenzoate) with different concentrations, ranging from 4 to 1,600 μM, were prepared for the determination of Km values of the purified 3-hydroxybenzoate 6-hydroxylase. Spectrophotometric assays were carried out with the same amount of enzyme against every substrate concentration in 100 mM phosphate buffer (pH 7.4) to obtain the initial velocity of the purified enzyme. Lineweaver-Burk plots against different substrate concentrations were used to determine the Km values.
Protein purification.
All procedures were carried out at 4°C. Crude cell extracts were applied to a HiTrap chelating HP column (Amersham Biosciences, Sweden) containing 5 ml of chelating Sepharose that was previously loaded with 5 ml of 0.1 M NiSO4 solution and equilibrated with 25 ml of binding buffer (0.02 M KH2PO4, 0.5 M KCl [pH 7.4]). The column containing the crude cell extracts was washed with 25 to 50 ml of binding buffer, and the protein was eluted with 10 to 25 ml of elution buffer (0.02 M KH2PO4, 0.5 M KCl, 0.5 M imidazole [pH 7.4]). The desired fractions were desalted and concentrated by using Centricon-30 (Millipore).
PAGE.
Both native polyacrylamide gel electrophoresis (PAGE) and sodium dodecyl sulfate (SDS)-PAGE were performed using MiniProtean II (Bio-Rad) and 5% acrylamide for the stacking gel and 10% acrylamide for the separating gel. The Precision Plus Protein standards with defined molecular masses from Bio-Rad were used as markers for SDS-PAGE, whereas HMW Native from Amersham Biosciences, Sweden, was used as a marker for native PAGE.
Isoelectric focusing.
Ampholine PAGE plates (pH, 3.5 to 9.5) (Amersham Biosciences) were used to determine the pI of purified recombinant 3-hydroxybenzoate 6-hydroxylase I. Gels were run on Multiphor II electrophoresis unit (Amersham Biosciences) connected to the Multistep III thermostatic circulator (Amersham Biosciences) for 1,500 V for 2 h at 4°C.
Gel filtration.
Gel filtration of purified recombinant XlnD was carried out by using Superdex-200 column chromatography (Amersham Biosciences) with thyroglobulin (Mr, 669,000), ferritin (Mr, 440,000), catalase (Mr, 232,000), adolase (Mr, 158,000), chymotrypsinogen A (Mr, 25,000), and RNase A (Mr, 13,700) as molecular weight standards. The column was equilibrated and eluted with 0.15 M phosphate buffer (pH 7.0) at a flow rate of 0.1 ml/min.
RESULTS
Sequence analysis.
The xlnD-encoded protein was found to belong to a family of proteins that have been annotated mainly as either salicylate hydroxylase or 2-polyprenyl 6-methoxyphenol hydroxylase (PMH). With the exception of MhbM from Klebsiella pneumoniae M5a1 (16), SalA from Acinetobacter sp. strain ADP1 (13), and NahW from Pseudomonas stutzeri AN10 (2), the functionality of other XlnD homologs has yet to be ascertained. MhbM was recently reported to be a functional 3-hydroxybenzoate 6-hydroxylase from K. pneumoniae (16), whereas SalA from Acinetobacter sp. strain ADP1 and NahW from P. stutzeri AN10 have been shown to be salicylate 1-hydroxylase that catalyzes the conversion of salicylate into catechol (2, 13). PMH is an enzyme that is part of the ubiquinone biosynthetic pathway and is encoded by ubiH in E. coli (17). PMH catalyzes the conversion of 2-polyprenyl 6-methoxyphenol to 2-polyprenyl 6-methoxy-1, 4-benzoquinol, a hydroxylation reaction that occurs para to an existing hydroxyl group, similar to the reaction catalyzed by 3-hydroxybenzoate 6-hydroxylase.
Overexpression of XlnD in E. coli.
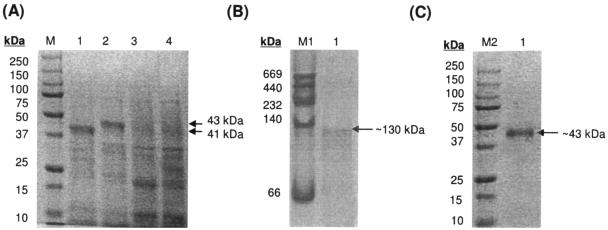
The xlnD reading frame was cloned under the transcriptional regulation of the IPTG-inducible T7 promoter in the expression vectors pET28a and pET30a as detailed in Materials and Methods. The pET28xlnD clone was constructed such that the protein is expressed in its native state, whereas the expression of the pET30xlnD clone would result in an N-terminal His6 fusion protein. Cell extracts of IPTG-induced E. coli BL21(DE3)/pLysS harboring these constructs were separated on SDS-PAGE (Fig. 2A) and an overexpressed protein band of ∼41 kDa could be seen for extracts from the pET28xlnD recombinant, whereas a slightly larger band of ∼43 kDa was observed for the pET30xlnD recombinant due to the presence of the N-terminal His6 tag, corresponding to their respective expected sizes.
FIG. 2.
(A) Extracts of E. coli BL21(DE3)/pLysS overexpressing XlnD separated by SDS-10% PAGE. E. coli BL21(DE3)/pLysS cells harboring the following plasmids were harvested and lysed 4 h after IPTG induction: pET28xlnD (lane 1), pET30xlnD (lane 2), pET28a (lane 3), and pET30a (lane 4). Lane M contains protein standards with molecular masses as indicated. Note the overexpressed protein bands of approximately 41 kDa in lane 1 and 43 kDa in lane 2 that correspond to the estimated molecular masses of XlnD and an N-terminal His6-XlnD fusion protein, respectively. (B and C) Purified recombinant XlnD separated on native 10% PAGE (B) and SDS-10% PAGE (C). The HMW native molecular mass standard from Amersham Biosciences was used for the native PAGE (lane M1) and is comprised of the proteins thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and albumin (66 kDa). For SDS-PAGE, the recombinant prestained standards from Bio-Rad was used as molecular mass standards (lane M2).
Spectrophotometric assay of 3-hydroxybenzoate 6-hydroxylase activity.
The in vitro assay for 3-hydroxybenzoate 6-hydroxylase was based on the 3-hydroxybenzoate-linked oxidation of NADH measured at 340 nm (16, 28). The end product in the sample cuvette was shown to be gentisate by the addition of the xlnE-encoded gentisate 1,2-dioxygenase (29) to the cuvette and observing for a shift in the λmax from 320 to 330 nm that corresponded to the conversion of gentisate to maleylpyruvate.
Cell extracts of IPTG-induced E. coli BL21(DE3)/pLysS harboring pET28xlnD showed 3-hydroxybenzoate 6-hydroxylase specific activity of 1.08 U/mg, similar to the specific activity of 1.01 U/mg obtained for the pET30xlnD recombinant. No enzyme activities were detected when 4-hydroxybenzoate or salicylate was used as the substrate.
Purification of recombinant XlnD and inferences of its trimeric nature.
Recombinant XlnD was purified from cell extracts of IPTG-induced E. coli BL21 harboring pET30xlnD by using a nickel affinity column to bind the His6 fusion protein. Separation using native PAGE of the eluted fraction that showed 6-hydroxylase activity yielded a single protein band of ∼130 kDa (Fig. 2B), whereas using SDS-PAGE, a protein band of ∼43 kDa was observed (Fig. 2C). This infers that the active recombinant XlnD is possibly homotrimeric, and this finding was verified by gel filtration. When cell extracts of IPTG-induced E. coli BL21 harboring pET28xlnD were separated on native PAGE, an overexpressed band of ∼130 kDa was also observed, indicating that recombinant XlnD without the hexahistidine tag is similarly trimeric in nature.
Subsequent analysis of the purified recombinant XlnD by isoelectric focusing revealed the pI of the purified enzyme to be 5.3.
Substrate range, effect of metal ions, and FAD.
The activity of the purified recombinant XlnD toward several 3-substituted derivatives of benzoates was investigated. When compared to 3-hydroxybenzoate, recombinant XlnD showed higher specific activities against 3-hydroxy-4-methylbenzoate and 3-hydroxy-5-methylbenzoate, the intermediates in the degradation of 2,5-xylenol and 3,5-xylenol, respectively (Table 1) . Recombinant XlnD was also active toward the different 3-substituted benzoates tested, albeit with lower specific activities compared to 3-hydroxybenzoate. Only when 3-methylthiobenzoate was presented to XlnD did the activity rise as high as that against 3-hydroxy-5-methylbenzoate (Table 1). Thus, recombinant XlnD is able to tolerate different substitutions at the C3 position of benzoate derivatives.
TABLE 1.
Specific activities of purified recombinant XlnD against 3-substituted derivatives of benzoate
| Substrate | Mean sp act (U/mg)a ± SD |
|---|---|
| 3-Hydroxybenzoate | 0.91 ± 0.11 |
| 3-Hydroxy-4-methylbenzoate | 1.25 ± 0.33 |
| 3-Hydroxy-5-methylbenzoate | 1.63 ± 0.16 |
| 3-Aminobenzoate | 0.32 ± 0.04 |
| 3-Fluorobenzoate | 0.25 ± 0.05 |
| 3-Bromobenzoate | 0.71 ± 0.05 |
| 3-Chlorobenzoate | 0.39 ± 0.01 |
| 3-Methylthiobenzoate | 1.57 ± 0.28 |
One unit of enzyme activity is defined as the conversion of 1 μmol of NADH to NAD+ per min at 23°C. Values presented are the means of results from triplicate experiments.
The activity of the purified recombinant XlnD was not affected by the addition of up to 5 mM concentration of Na+, Mg2+, and EDTA. However, partial inhibition of the 6-hydroxylase activity was observed for the following ions: Ca2+, Zn2+, and Mn2+, with 5 mM Mn2+ resulting in almost total inhibition. Recombinant XlnD was completely inactivated in the presence of 5 mM Cu2+, Hg2+, and Fe2+. For Cu2+ and Hg2+, even 50 μM was sufficient to achieve complete inhibition. K+, at 5 mM, appeared to enhance the activity of XlnD, albeit by ca. 13%.
Interestingly, the addition of 0.2 mM FAD increased the activity of the purified recombinant 6-hydroxylase by nearly 23%, suggesting the possible loss of this flavin cofactor during the purification process that was duly compensated by the addition of exogenous FAD. During purification, the recombinant XlnD protein exhibited a yellowish color, indicative of a bound flavin. The UV-visible absorption spectrum of the purified enzyme showed absorption peaks at 380 and 450 nm, with a shoulder at 480 nm, which is typical of flavoproteins (26) (data not shown).
Effects of temperature and pH on activity and storage conditions.
Optimal activity of the recombinant 3-hydroxybenzoate 6-hydroxylase was observed at 25°C, with no activity detected at temperatures lower than 15°C or higher than 50°C.
The pH dependence of the purified recombinant 6-hydroxylase was investigated by assaying for enzyme activity in 0.1 M phosphate buffers of pH values ranging from 6.0 to 9.0. Maximum enzyme activity was exhibited at pH 7.5.
For storage, the addition of 10% (vol/vol) glycerol and 0.2 mM FAD was found to stabilize the recombinant enzyme at −80°C. Under these conditions, ca. 60% of 6-hydroxylase activity was found to remain after 3 months of storage at −80°C. No differences in activity were observed when the storage buffer was 50 mM MOPS instead of 100 mM potassium phosphate. However, even with the addition of glycerol and FAD, the enzyme was completely inactivated after 12 h at room temperature and after 3 days at 4°C.
Kinetic properties.
The purified recombinant XlnD displayed typical Michaelis-Menten kinetics and Lineweaver-Burk plots of enzyme activity yielded apparent Km values of 79 μM for 3-hydroxybenzoate with NADH, and 108 μM with NADPH when the reduced pyridine nucleotide levels were fixed at 0.5 mM and the concentration of 3-hydroxybenzoate were varied. At a constant level of 3-hydroxybenzoate (0.5 mM) and variable amounts of NADH or NADPH, XlnD exhibited smaller apparent Km values for NADH (112 μM) compared to NADPH (225 μM), indicating a preference for NADH over NADPH.
For 3-hydroxy-4-methylbenzoate, the apparent Km values are 95 μM with NADH and 71 μM with NADPH.
Effects of an xlnD knockout in P. alcaligenes P25X.
Previous biochemical studies on P25X and its mutant derivatives had indicated the presence of two isofunctional 3-hydroxybenzoate 6-hydroxylases, as well as other enzymes of the gentisate pathway: one set that was constitutively expressed and another set that was strictly inducible by specific aromatic substrates (20). To investigate the role of xlnD in its native P. alcaligenes P25X host, an xlnD knockout mutant, designated G50, was constructed by transposon mutagenesis as detailed in Materials and Methods. When grown on lactate, no 3-hydroxybenzoate 6-hydroxylase activity could be detected in G50 cells, indicating the loss of the constitutive 3-hydroxybenzoate 6-hydroxylase I. 6-Hydroxylase activity could only be detected in G50 cells that were induced with 3-hydroxybenzoate and, to a lesser extent, gentisate (Table 2). In both cases, 6-hydroxylase activities against 3-hydroxy-4-methylbenzoate could be detected, although the specific activities were much less compared to 3-hydroxybenzoate as the substrate. This indicated the presence of a functional copy of the second, strictly inducible 6-hydroxylase II. Interestingly, no 6-hydroxylase activity against either 3-hydroxybenzoate or 3-hydroxy-4-methylbenzoate could be detected when G50 cells were induced with 3-hydroxy-4-methylbenzoate.
TABLE 2.
Comparison of the 3-hydroxybenzoate 6-hydroxylase specific activities from P25X wild-type with the xlnD mutant G50 when grown in lactate and in the presence of aromatic substrates
| Carbon source for growth | Mean sp acta of P. alcaligenes cells ± SD assayed with:
|
|||
|---|---|---|---|---|
| P25X
|
G50
|
|||
| 3-Hydroxybenzoate | 3-Hydroxy- 4-methylbenzoate | 3-Hydroxybenzoate | 3-Hydroxy- 4-methylbenzoate | |
| Lactateb | 0.06 ± 0.01 | 0.09 ± 0.01 | NDd | ND |
| 3-Hydroxybenzoatec | 0.16 ± 0.03 | 0.14 ± 0.02 | 0.14 ± 0.02 | 0.02 ± 0.01 |
| 3-Hydroxy-4-methylbenzoatec | 0.14 ± 0.02 | 0.45 ± 0.03 | ND | ND |
| Gentisatec | 0.11 ± 0.02 | 0.26 ± 0.03 | 0.03 ± 0.01 | <0.01 |
One unit of enzyme activity is defined as the conversion of 1 μmol of NADH to NAD+ per min at 23°C. Specific activities are expressed as units per mg of protein. Values presented are the means of three separate experiments.
Lactate, at a final concentration of 20 mM, was the sole carbon source in the basal minimal medium.
Aromatic substrates were added (to a final concentration of 2.5 mM) into the cell culture grown in 20 mM lactate when the OD600 was 0.5 to 0.6.
ND, nondetectable activity.
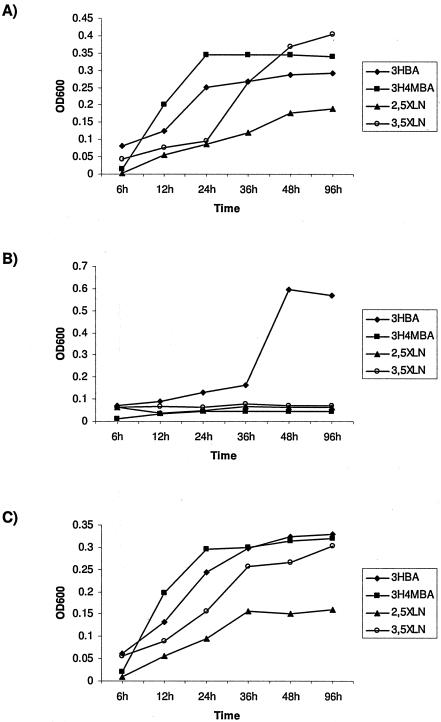
G50 was scored for growth on minimal agar plates supplemented with aromatic compounds as the sole carbon source and was found to only grow on 3-hydroxybenzoate and gentisate but not on 2,5-xylenol, 3,5-xylenol, and 3-hydroxy-4-methylbenzoate. Identical results were obtained when the growth of G50 cells in liquid minimal media containing the respective aromatic compounds was assessed by measuring OD600 over a period of 96 h (Fig. 3A and 3B). When intact xlnD was provided in trans on the pRK415 plasmid to complement the xlnD lesion in G50, restoration of the wild-type growth phenotype on 2,5-xylenol, 3,5-xylenol, and 3-hydroxy-4-methylbenzoate was observed (Fig. 3C).
FIG. 3.
Growth of P. alcaligenes P25X wild-type and its mutant derivatives in liquid minimal media containing the aromatic substrates 2,5-xylenol (2,5XLN), 3,5-xylenol (3,5XLN), 3-hydroxybenzoate (3HBA), and 3-hydroxy-4-methylbenzoate (3H4MBA) as the sole carbon source as determined by measuring OD600 over a period of 96 h. (A) Strain P25X wild type; (B) xlnD mutant G50; (C) G50 harboring the recombinant plasmid pRK415-xlnD in which xlnD is expressed from the lac promoter of the pRK415 vector.
DISCUSSION
Results from the present study clearly showed that XlnD is a functional 3-hydroxybenzoate 6-hydroxylase. Very few monooxygenases that catalyze the hydroxylation of the aromatic ring at a position para to an existing hydroxyl group has been characterized. Besides mhbM from K. pneumoniae M5a1 (16), xlnD from P. alcaligenes P25X is the only other sequenced gene that has thus far been demonstrated to encode a functional 3-hydroxybenzoate 6-hydroxylase. Active recombinant XlnD was shown to be a trimeric protein, unlike the other 3-hydroxybenzoate 6-hydroxylases that have been characterized and were shown to be monomeric. The XlnD subunit is similar in size (43 kDa) to the 6-hydroxylases from K. pneumoniae (42 kDa) and B. cepacia (44 kDa), compared to the 6-hydroxylases from P. aeruginosa (85 kDa) and Micrococcus sp. (70 kDa). XlnD, along with other characterized 3-hydroxybenzoate 6-hydroxylases, was able to utilize both NADH and NADPH as cofactors. With 3-hydroxybenzoate as the substrate, XlnD has a preference for NADH over NADPH, a trait that it shares with the 3-hydroxybenzoate 6-hydroxylases from P. aeruginosa (6) and Micrococcus sp. (22). XlnD was also completely inactivated by Cu2+, Hg2+, and Fe2+, much like its counterparts from K. pneumoniae (25) and Micrococcus sp. (22).
The present study also adds to our scant knowledge of the gentisate degradative pathway in P. alcaligenes P25X and validated some of the conclusions that were earlier made based on the biochemical characterization of P25X catabolic mutants (20, 21). Different ratios of attack on 3-hydroxybenzoate and 3-hydroxy-4-methylbenzoate, as measured by the oxygen uptake of whole cells, have been previously reported to be dependent upon the carbon source utilized for the growth of P25X cells (20). The oxygen uptake studies with the wild-type and mutant strains showed that P25X synthesized two isofunctional 6-hydroxylases: 6-hydroxylase I was constitutively expressed and oxidized 3-hydroxy-4-methylbenzoate faster than 3-hydroxybenzoate, whereas 6-hydroxylase II was strictly inducible but had higher activity against 3-hydroxybenzoate than 3-hydroxy-4-methylbenzoate (20). These have been corroborated in the present study. Knocking out xlnD in mutant strain G50 led to the elimination of 6-hydroxylase I activity. The recombinant XlnD displayed higher specific activities against 3-hydroxy-4-methylbenzoate compared to 3-hydroxybenzoate. The 6-hydroxylase activity detected when G50 cells were induced with 3-hydroxybenzoate was attributed to the strictly inducible 6-hydroxylase II and its specific activity toward 3-hydroxybenzoate was higher compared to 3-hydroxy-4-methylbenzoate. 3-Hydroxy-4-methylbenzoate is the intermediate in the degradation of 2,5-xylenol and, although 6-hydroxylase II appeared to be capable of utilizing 3-hydroxy-4-methylbenzoate as a substrate, its expression is dependent on the presence of 3-hydroxybenzoate and not 3-hydroxy-4-methylbenzoate, thus indicating that 6-hydroxylase I encoded by xlnD is directly involved in 2,5-xylenol degradation.
P25X catabolic mutants that were defective in 6-hydroxylase I synthesis were reportedly unable to use 2,5-xylenol and 3-hydroxy-4-methylbenzoate but were able to grow on m-cresol and 3,5-xylenol as the metabolism of these compounds will generate 3-hydroxybenzoate and 3-hydroxy-5-methylbenzoate, respectively, that could possibly serve as inducers for 6-hydroxylase II (21). However, in the present study, we observed that the xlnD knockout mutant, G50, was unable to utilize 3,5-xylenol besides 2,5-xylenol and 3-hydroxy-4-methylbenzoate. It should be noted that the P25X catabolic mutants in the earlier report were created by using ethylmethane sulfonate mutagenesis, followed by penicillin/cycloserine cycling with 3-hydroxybenzoate and 3-hydroxy-4-methylbenzoate as selective and counterselective carbon sources (21). Thus, the mutations that had occurred in these strains would not be as well-defined as the G50 mutant strain that was utilized in the present study where the xlnD reading frame was specifically interrupted with a custom Sm/Spr transposon. Mutations that affected either the specificity of the regulatory protein(s) or 6-hydroxylase I itself may have led to the earlier reported observations (21). Restoration of the wild-type growth phenotype on 2,5-xylenol, 3,5-xylenol and 3-hydroxy-4-methylbenzoate when xlnD was provided in trans in G50 strongly suggests that xlnD is also directly involved in 3,5-xylenol degradation.
6-Hydroxylase II activity was also detected when G50 cells were grown in the presence of gentisate, but the relatively lower specific activity indicates that gentisate is a poorer product inducer of 6-hydroxylase II, confirming our earlier findings (21). It should be noted that it is impossible to distinguish the contributions of 6-hydroxylase I and II toward the overall specific activity of P25X wild-type cells especially when grown in 3-hydroxybenzoate or gentisate since both 6-hydroxylases would be expressed. Moreover, the xln operon, which encodes 6-hydroxylase I, has been shown to be further inducible by 3-hydroxybenzoate and, to a lesser extent, by gentisate (29). Thus, the 6-hydroxylase activity in P25X cells that were grown in 3-hydroxybenzoate or gentisate would reflect the expression of the inducible 6-hydroxylase II, as well as the increased expression of 6-hydroxylase I. Whether there are any direct or indirect interactions between the two 6-hydroxylases that are competing for the same substrate and the role of the regulatory protein(s) are not known at this point and would hopefully be addressed with the cloning of the inducible 6-hydroxylase II, along with its regulatory gene(s) in the near future.
In conclusion, the results of the present study have shown that the xlnD-encoded 6-hydroxylase I, and not the strictly inducible 6-hydroxylase II, was involved in the degradation of 2,5-xylenol and 3,5-xylenol in P. alcaligenes P25X. The lower %G+C content of the xln operon compared to its host genome suggested that the xln genes are a relatively recent acquisition into the P25X genome (29). Preliminary sequence data of part of the strictly inducible gentisate 1,2-dioxygenase II gene showed a %G+C content that is similar to the host P. alcaligenes genome (∼62%), suggesting that the strictly inducible genes of the gentisate pathway have resided in the P25X genome far longer than the genes of the xln operon. The acquisition of the DNA fragment that contained the xln operon would have enabled strain P25X to expand its repertoire of aromatic substrates to include 2,5-xylenol, as well as 3,5-xylenol, thereby increasing its fitness to survive in the polluted Hull river mud from where it was originally isolated (10).
Acknowledgments
This research was supported by University Research Council grant R-182-000-069-112 to C.L.P. X.G. acknowledges the receipt of a postgraduate research scholarship from the National University of Singapore.
We thank Mark Wong for help in the construction of the mutant strain G50.
REFERENCES
- 1.Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 147:1611-1620. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, R., E. R. B. Moore, E. García-Valdéz, and D. H. Pieper. 1999. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 181:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for quantification of microgram quantities of proteins utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Eppink, M. H., H. A. Schreuder, and W. J. van Berkel. 1998. Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants. J. Biol. Chem. 273:21031-21039. [DOI] [PubMed] [Google Scholar]
- 5.Gatti, D. L., B. A. Palfey, M. S. Lah, B. Entsch, V. Massey, D. P. Ballou, and M. L. Ludwig. 1994. The mobile flavin of 4-OH benzoate hydroxylase. Science 266:110-114. [DOI] [PubMed] [Google Scholar]
- 6.Groseclose, E. E., D. W. Ribbons, and H. Hughes. 1973. 3-Hydroxybenzoate 6-hydroxylase from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 55:897-903. [DOI] [PubMed] [Google Scholar]
- 7.Harayama, S., and K. N. Timmis. 1989. Catabolism of aromatic hydrocarbons by Pseudomonas, p. 151-174. In D. A. Hopwood and K. F. Chater (ed.) Genetics of bacterial diversity. Academic Press, Ltd., London, United Kingdom.
- 8.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-601. [DOI] [PubMed] [Google Scholar]
- 9.Hegeman, G. D. 1966. Synthesis of enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J. Bacteriol. 91:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopper, D. J., and P. J. Chapman. 1970. Gentisic acid and its 3- and 4-methyl-substituted homologues as intermediates in the bacterial degradation of m-cresol, 3,5-xylenol, and 2,5-xylenol. Biochem. J. 122:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopper, D. J., P. J. Chapman, and S. Dagley. 1971. The enzymic degradation of alkyl-substituted gentisates, maleates, and malates. Biochem. J. 122:29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, D. C. N., and R. A. Cooper. 1990. Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch. Microbiol. 154:489-495. [DOI] [PubMed] [Google Scholar]
- 13.Jones, R. M., V. Pagmantidis, and P. A. Williams. 2000. sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J. Bacteriol. 182:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 15.Kwong, S. M., C. C. Yeo, A. Suwanto, and C. L. Poh. 2000. Characterization of the endogenous plasmid from Pseudomonas alcaligenes NCIB 9867: DNA sequence and mechanism of transfer. J. Bacteriol. 182:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, D. Q., H. Liu, X. L. Gao, D. J. Leak, and N. Y. Zhou. 2005. Arg169 is essential for catalytic activity of 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae M5a1. Microbiol. Res. 160:53-59. [DOI] [PubMed] [Google Scholar]
- 17.Meganathan, R. 2001. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203:131-139. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Ortiz-Maldonado, M., L. J. Cole, S. M. Dumas, B. Entsch, and D. P. Ballou. 2004. Increased positive electrostatic potential in p-hydroxybenzoate hydroxylase accelerates hydroxylation but slows turnover. Biochemistry 43:1569-1579. [DOI] [PubMed] [Google Scholar]
- 20.Poh, C. L., and R. C. Bayly. 1980. Evidence for isofunctional enzymes used in m-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes. J. Bacteriol. 143:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poh, C. L., and R. C. Bayly. 1988. Regulation of isofunctional enzymes in Pseudomonas alcaligenes mutants defective in the gentisate pathway. J. Appl. Bacteriol. 64:451-458. [DOI] [PubMed] [Google Scholar]
- 22.Rajasekharan, S., R. Rajasekharan, and C. S. Vaidyanathan. 1990. Substrate-mediated purification and characterization of a 3-hydroxybenzoic acid-6-hydroxylase from Micrococcus. Arch. Biochem. Biophys. 278:21-25. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:37-45. [Google Scholar]
- 25.Suárez, M., E. Ferrer, A. Garrido-Pertierra, and M. Martin. 1995. Purification and characterization of the 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae. FEMS Microbiol. Lett. 126:283-290. [DOI] [PubMed] [Google Scholar]
- 26.Suh, J.-K., L. L. Poulsen, D. M. Ziegler, and J. D. Robertus. 1996. Molecular cloning and kinetic characterization of a flavin-containing monooxygenase from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 336:268-274. [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., M. Ortiz-Maldonado, B. Entsch, V. Massey, D. Ballou, and D. L. Gatti. 2002. Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase. Proc. Natl. Acad. Sci. USA 99:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, L.-H., R. Y. Hamzah, Y. Yu, and S.-C. Tu. 1987. Pseudomonas cepacia 3-hydroxybenzoate 6-hydroxylase: induction, purification, and characterization. Biochemistry 26:1099-1104. [DOI] [PubMed] [Google Scholar]
- 29.Yeo, C. C., M. V.-M. Wong, Y. Feng, K. P. Song, and C. L. Poh. 2003. Molecular characterization of an inducible gentisate 1,2-dioxygenase gene, xlnE, from Pseudomonas alcaligenes NCIMB 9867. Gene 312:239-248. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, N.-Y., J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]