Abstract
Beta defensins are small cationic antimicrobial peptides present in the respiratory system which have been proposed to be dysfunctional in the environment of the cystic fibrosis lung. Defb1, a murine homologue to the human beta defensins, has also been found to be expressed in the respiratory system and, in order to examine the function of beta defensins in vivo, gene targeting was used to generate Defb1-deficient (Defb1tm1Hgu/Defb1tm1Hgu [Defb1−/−]) mice. The Defb1 synthetic peptide was shown to have a salt-sensitive antimicrobial activity that was stronger against Staphylococcus aureus than against Escherichia coli or Pseudomonas aeruginosa. Defb1−/− mice were found, however, to be effective in the clearance of the cystic fibrosis relevant pathogen S. aureus from the airways after nebulization. Although no overt deleterious phenotype was evident in the Defb1−/− mice, the number of mutant mice found to harbor bacteria of the Staphylococcus species in the bladder was significantly higher (P = 0.008) than that of controls, suggesting a role for these peptides in resistance to urinary tract infection.
To maintain sterility, the mucosal surface of the airways must have an efficient innate defense system to combat the constant threat of infection from inhaled microorganisms. Defensin molecules comprise a family of small cationic peptides that exhibit a broad spectrum of antimicrobial action. Studies on the recently discovered defensin molecules expressed by the airway epithelia and present in the airway surface fluid suggest that these molecules are an important component of innate defense in the respiratory system (2, 9-12, 18). Mammals contain two classes of defensins named alpha and beta based on the arrangements of the cysteine residues, and a third class, called cyclic theta defensins, has recently been identified in the rhesus monkey (26). The first defensin isolated from a mucosal surface was from bovine tracheal mucosa and was named tracheal antimicrobial peptide (8); subsequently, four human beta defensins were identified, and all of these are expressed in the epithelia of the airways. DEFB1 (HBD-1) expression in the airways is constitutive, and inflammatory stimuli have little apparent effect on its production (10, 18, 27), whereas the expression of DEFB2 (HBD-2), DEFB3 (HBD-3), and DEFB4 (HBD-4) is increased in airway epithelia by inflammatory stimuli (4, 9, 11, 12, 16, 17, 22). In vitro analysis of the antimicrobial properties of these beta defensins revealed all to have activity against several bacteria, including Pseudomonas aeruginosa and Escherichia coli. However, some variation has been found regarding the potency of the different peptides toward the gram-positive bacterium Staphylococcus aureus. For example, Bals et al. found DEFB2 to be bactericidal against S. aureus (2), whereas Harder et al. found DEFB2 to be bacteriostatic against S. aureus and only at very high concentrations (11). The disparity in results regarding the potency of DEFB2 highlights the problems of using in vitro systems for analyzing peptide function. Interestingly, the antibacterial activities of DEFB1 and DEFB2 have been found to be greatly affected by the concentration of NaCl. Smith et al. demonstrated that the airway surface fluid from cystic fibrosis (CF) respiratory epithelial cells had a markedly reduced antimicrobial activity due to elevated concentrations of Cl− and Na+ (25), whereas Goldman et al. demonstrated that the ablation of DEFB1 expression in normal bronchial xenografts also led to a reduction in antimicrobial activity (10). Although diverse estimates have been published for the in vivo composition of airway surface liquid (ASL) (28), it is an intriguing possibility that the dysfunction of the beta defensin peptides in an abnormal salt environment of the CF lung may explain why these individuals suffer from early-onset chronic bacterial colonization of the lung. However, all experiments performed to date on beta defensins have been carried out in in vitro cell culture systems, and the creation of a model system to study defensin function and dysfunction in vivo may avoid the problems associated with these culture systems.
We have previously reported the identification and characterization of a mouse beta defensin, Defb1, whose predicted protein sequence had 57% similarity to DEFB1. Defb1 was found to be expressed in the airway epithelia, and the synthetic peptide was shown to have salt-sensitive antimicrobial activity against P. aeruginosa and E. coli (1, 14, 19). We created a mouse model deficient in Defb1 in order to gain further insights into the role of these peptides by determining the effect of impairment of defensin activity in vivo.
MATERIALS AND METHODS
Generation of Defb1 mutant embryonic stem (ES) cells and mice.
A 9-kb fragment of genomic DNA was isolated from a 129 lambda Get genomic library by using a Defb1 cDNA probe. This fragment was found to contain ca. 4 kb of DNA to either side of the EcoRV fragment that contained Defb1 exon 2. A targeting construct was made to replace the EcoRV fragment containing Defb1 exon 2 with a neo resistance gene cassette (Fig. 1A). Briefly, the lambda Get genomic clone was digested with EcoRV to remove Defb1 exon 2, and the resulting 12.2-kb fragment was purified. The plasmid pMC1NeoPolyA contains the neo resistance gene under the control of the thymidine kinase (TK) promoter, and this gene cassette was removed from pMC1NeoPolyA by digestion with XhoI and SalI. The 1.1-kb neo fragment was blunt ended and ligated into the EcoRV sites of the lambda Get 12.2-kb fragment. The orientation and copy number of the inserted neo cassette was assessed by Southern hybridizations of BamHI digested clones by using a radiolabeled oligonucleotide probe to the neo gene.
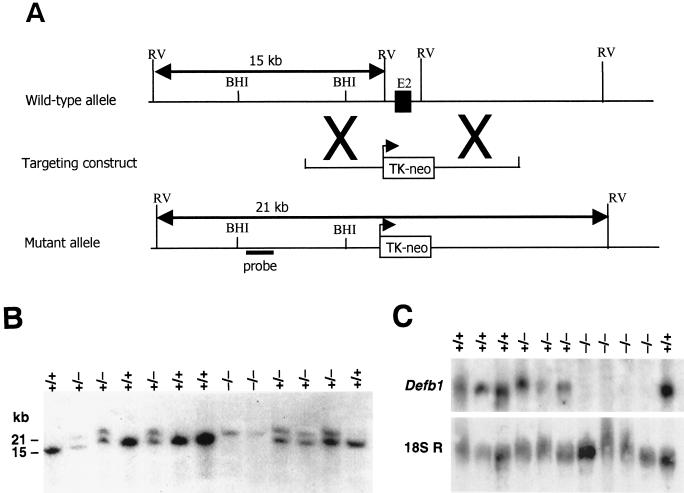
FIG. 1.
Targeted disruption of Defb1 gene. (A) Schematic representation of the Defb1 targeting strategy. A partial restriction endonuclease map of the murine Defb1 locus shows the EcoRV (RV) sites. The Defb1 targeting vector contains the neomycin resistance gene (neo) under the control of the TK promoter. The structure of the targeted mutant Defb1 allele and the location of the probe used in the Southern blot analysis is also shown. (B) Southern blot hybridization of the DNA prepared from the offspring of the Defb1+/− × Defb1+/− mating. DNA was digested with EcoRV and hybridized to the radiolabeled probe shown in Fig. 1A. The probe detects a 15-kb endogenous mouse allele and a 21-kb targeted allele. (C) Northern blot analysis of Defb1 gene expression in kidney RNA harvested from wild-type (+/+), heterozygous (+/−) and homozygous (−/−) mice. Loading and integrity of the RNA were assessed by hybridization with an 18S RNA probe.
A clone containing a single copy of the neo gene in the same orientation as Defb1 was electroporated into E14IV ES cells. G418-resistant homologous recombinants were verified by digestion with EcoRV and Southern hybridization with a 5′ probe external to the genomic sequence used in the targeting vector. Two targeted Defb1+/− ES cell lines with normal karyotypes were microinjected into C57BL/6N blastocysts and transferred to pseudopregnant foster mothers to generate chimeras, which were then mated with C57BL/6N and 129/Ola mice for germ line transmission. Genotyping of offspring was performed by Southern hybridization with the 5′ external probe mentioned above. Mice were either housed in standard (conventional) conditions or specific-pathogen-free (SPF) conditions. All experimental procedures performed with mice were in compliance with the Animals (Scientific Procedure) Act 1986 (United Kingdom).
Assessment of Defb1 gene expression by Northern blot analysis.
Total RNA was isolated from mouse kidneys by using RNAzol B reagent (Biogenesis, Ltd.) and analyzed by Northern blotting with a full-length Defb1 cDNA probe. This probe had previously been tested on Southern blots and was shown to be capable of detecting both exon 1 and exon 2 sequences. The Northern blots were subsequently probed with a control 18S ribosomal probe.
Histology.
Tissues were fixed in 10% neutral buffered formalin and processed for wax sectioning in a Tissue-Tek embedding machine. The wax blocks were sectioned on a Reichert-Jung 2030 microtome. The slides were rehydrated through graded alcohol to dH2O and stained with hematoxylin and eosin to examine the basic tissue morphology.
BAL and cell counts.
Mice were euthanized by intraperitoneal injection of 20 mg of sodium pentobarbitone (Euthatal; Rhone Merieux). The trachea was exposed by opening the skin, and a blunted 21G hypodermic needle was inserted, as a cannula, through a small incision in the upper trachea and tied in place with suture thread. Lavage was performed by introducing 1 ml of sterile phosphate-buffered saline (PBS) into the lungs via the tracheal cannula and then, after a few seconds, carefully withdrawing the fluid. This was repeated two more times with fresh PBS. The lavage fluid was placed on ice, and the total recovery volume per mouse was 2.75 ml. Samples which deviated from this volume were discarded to avoid complications from variations in dilution factors of the bronchoalveolar lavage (BAL). The BAL fluid was centrifuged for 10 min, the clear supernatant was removed, and the cell pellet was resuspended in 200 μl of PBS. Total cells were counted in a Neubauer haemocytometer chamber, and an air-dried preparation made of each sample (Cytospin; Shandon Scientific); these were then stained with Diff-Quik stain (Dade), and a differential cell count was performed.
Quantification of TNF-α.
Murine tumor necrosis factor alpha (TNF-α) was measured by using the commercially available enzyme-linked immunosorbent assay kit (R&D Systems) according to the manufacturer's instructions. Undiluted BAL fluid, in addition to a 1:10 dilution of BAL fluid, was assayed in duplicate. The sample absorbance was read at 450 nm on a Multiskan MS plate reader (Labsystems).
Assessment of bacterial profile of urine.
Mice were anesthetized with halothane (Merial Animal Health, Ltd., Harlow, United Kingdom) and then given a lethal dose of sodium phenobarbitone. The abdomen was exposed, and the contents of the bladder collected by using a sterile 25G needle. The samples were divided in two and plated out on Columbia base agar (Oxoid) and MacConkey agar (Difco) and incubated overnight at 37°C, and the numbers of CFU were counted. Bacterial species were identified from colony morphology and from Gram staining.
Peptide synthesis.
The mature peptide sequence for Defb1 was predicted from the sequence of mature DEFB1 and DEFB2. The peptide was synthesized by Albachem, Ltd., as previously described (19).
Functional analysis of synthetic Defb1.
The synthetic peptide was tested against S. aureus CF clinical isolate C1705 and E.coli lab strain J2408. Fresh, overnight cultures of bacteria were grown in nutrient broth and then suspended in 0.85 mM NaCl at ∼109 CFU/ml. These samples were then centrifuged, and the resulting pellet was resuspended and diluted in 10 mM phosphate buffer containing 0.1% d-glucose and a range of concentrations of NaCl (buffer pH 7.60 at 0 mM NaCl, pH 7.40 at 30 mM NaCl, pH 7.27 at 90 mM NaCl, and pH 7.20 at 150 mM NaCl). Two sets of duplicate 500-μl reactions were prepared that contained either buffer alone or synthetic peptide rehydrated in buffer, across the same range of NaCl concentrations, to which ∼5 × 104 bacteria in the appropriate salt concentration were added. These reactions were incubated for 30 min at 37°C; duplicate sets of serial dilutions were then prepared from each sample, in buffer of the appropriate salt concentration, plated out on Columbia agar base or MacConkey agar, and incubated overnight at 37°C, and CFU counts were determined. The antimicrobial activity of the peptide was compared against buffer alone, to control for the effects on the bacteria of varying the NaCl environment.
Instillation of S. aureus by nebulization.
Mice were maintained in HEPA air-filtered isolator cabinets for the duration of the experiment. Aerosolization was carried out as previously described (6). Briefly, mice were placed in parallel tubes open to a vented cone, using a sidestream nebulizer with the addition of a baffle system (System-22 Optimist; Medic-Aid) to reduce the air particle size and aid delivery to the small airways. S. aureus was delivered with a carrier gas mixture of 95% O2 and 5% CO2 at a rate of 12 liters/min for 10 min. It was estimated that each mouse would receive ∼105 CFU from the starting culture of 2 × 108 CFU/ml and 2 × 104 CFU from the starting culture of 4 × 107 CFU/ml. Mice were euthanized 24 h after exposure by intraperitoneal injection of 20 mg of sodium pentobarbitone, and the lungs were removed and homogenized in 0.5 ml of sterile PBS. Serial dilutions of the crude homogenates were plated out on blood agar and incubated at 37°C overnight, and the numbers of colonies were determined.
LPS administration by intratracheal instillation.
Mice were anesthetized with an intraperitoneal injection of 0.2 to 0.3 ml of avertin (2.5 g of tri-bromo-ethanol, 5 ml of 2-methyl-2-butanol, 200 ml of distilled H2O)/10 g (body weight). The mice were then instilled with 40 μg of lipopolysaccharide (LPS; E. coli O55:B5; Sigma) or sterile PBS as a control. All samples were in a total volume of 20 μl. The mice were instilled by using the nonsurgical, intratracheal instillation method of Ho and Furst (13). Briefly, the anaesthetized mouse was supported in a frame so the pharynx, larynx, and trachea were arranged in a vertical straight line. The airway was illuminated externally by a lamp, the tongue was moved to one side, and a blunted Terumo spinal needle (Surgicon) was passed down the pharynx. The tip of the needle was inserted between the vocal folds at the base of the larynx, and 20 μl of the solution was instilled into the trachea. The animal was maintained in an upright position 2 min after instillation to allow the fluid to drain into the respiratory tree. Mice were euthanized 18 h after exposure by an intraperitoneal injection of 20 mg of sodium pentobarbitone, and the lungs and tracheas were removed for gene expression analysis as described below.
Semiquantitative RT-PCR.
Total RNA was isolated from a variety of tissues from C57BL/6N mice by using RNAzol B (Biogenesis, Ltd.) as described by the manufacturer. The samples were DNase treated, and then cDNA synthesis was accomplished by using a first-strand cDNA synthesis kit (Boehringer Mannheim). The resultant cDNAs were used as a template in PCRs for Defb2, Defb3, Defb4, and Defb6 as previously described (3, 15, 20, 29). The amplified products were analyzed by 2% agarose gel by electrophoresis. The sample was defined as being “positive for PCR” or to have “expression detected” (see Table 4) if 10 μl of the PCR sample gave rise to a visible product on the gel after 30 rounds of PCR. Amplification of Hprt was carried out in parallel as previously described (20). Reactions were verified for RNA amplification by including controls without reverse transcriptase.
TABLE 4.
Expression of Defb2, Defb4, and Defb6 in mouse tracheaa
| Group | Description | Expressiona of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Defb2
|
Defb4
|
Defb6
|
||||||||
| +/+ | +/− | −/− | +/+ | +/− | −/− | +/+ | +/− | −/− | ||
| 1 | 129 × C57BL/6, conv housedb | 1/11 | 1/10 | 6/13 | 6/7 | 2/6 | 7/9 | 1/7 | 0/6 | 1/9 |
| 2 | 129 inbred, SPF housed | 2/6 | 0/2 | 2/6 | 3/6 | 1/2 | 4/6 | 0/6 | 0/2 | 0/6 |
| 3 | 129 × C57BL/6, SPF housed | 0/3 | 1/3 | 1/3 | 2/3 | 2/3 | 2/3 | 0/3 | 0/3 | 0/3 |
| 4 | 129 × C57BL/6, S. aureus treated | 0/2 | 0/2 | 0/2 | 2/2 | 1/2 | 2/2 | 0/2 | 0/2 | 0/2 |
| 5 | 129 × C57BL/6, E. coli LPS treated | 1/1 | 4/5 | 4/4 | 1/1 | 3/5 | 3/4 | 0/1 | 0/5 | 0/4 |
The numbers of mice with the indicated genotype per total number of mice tested in each genotypic group that were found to be positive by RT-PCR for defensin expression are presented. The results are separated into groups 1 to 5 based on different housing conditions and treatments. Group 4 animals were nebulized with 105 CFU of S. aureus, and group 4 animals were treated with 40 μg of E. coli LPS as described in Materials and Methods.
Conv housed, housed in conventional conditions.
Statistical analysis.
Fisher's exact test for 2 × 2 tables was used to determine the statistical difference between Defb1−/− mice and their littermates with respect to the likelihood of harboring increased numbers of bacteria in the urine. Analysis of the significance of differences between Defb1 peptide and control samples was performed and was illustrated graphically by plotting CFU means after square root transformation, with group standard errors, against the NaCl concentrations. The means of the absolute CFU counts were used to calculate the proportion of bacteria surviving in the peptide-treated samples as a percentage of the counts from the buffer-only samples to control for the effects of changes in NaCl concentration alone. These percentages were expressed as “percent killing”; the standard errors were displayed graphically.
Gene designations.
All gene designations are in accordance with the approved HUGO gene nomenclature for humans as listed in the Human Gene Nomenclature Database (http://www.gene.ucl.ac.uk/nomenclature).
RESULTS
Generation of Defb1 mutant mice.
In order to assess the loss of Defb1 gene function, homologous recombination technology was used to generate ES cells in which one copy of the Defb1 exon 2 was replaced with the neomycin resistance gene (Fig. 1A). The decision was made to target exon 2 because the entire sequence of the mature Defb1 peptide is coded for by this exon. Nine independent ES cell clones with the predicted modification were identified. Two of these clones with normal karyotypes were chosen for blastocyst injections, which produced nine chimeric males. The chimeras appeared normal and gave 100% germ line transmission of the targeted allele, with no difference observed in the phenotype of the mice derived from the two independent clones. Chimeras were crossed onto 129/Ola mice to produce an inbred population, as well as C57BL/6N mice for an outbred population. Offspring from heterozygous matings displayed a typical Mendelian pattern of inheritance of the targeted allele, indicating the absence of an embryonic lethal phenotype (Fig. 1B and Table 1). Northern hybridization with a full-length Defb1 cDNA probe gave no hybridization signal from the Defb1−/− samples, indicating the absence of the full-length Defb1 transcript or any aberrant transcripts containing the nontargeted Defb1 exon 1 (Fig. 1C). The Defb1−/− mice were of normal size and weight and had no visible abnormalities. Both sexes were fertile.
TABLE 1.
Genotype distribution of offspring from heterozygous matings
| Mouse strain and gender | No. of mice with genotype:
|
||
|---|---|---|---|
| +/+ | +/− | −/− | |
| Outbred (129 × C57BL/6) | |||
| Male | 60 | 139 | 74 |
| Female | 82 | 123 | 64 |
| Total (%) | 142 (26.2) | 160 (48.3) | 138 (25.5) |
| Inbred (129) | |||
| Male | 15 | 25 | 14 |
| Female | 19 | 24 | 15 |
| Total (%) | 34 (30.4) | 49 (43.8) | 29 (25.9) |
Histology.
There was no significant difference found between Defb1−/− mice (n = 10) and wild-type littermates (n = 12) after histologic examination of the airways, kidney, bladder, and heart (data not shown).
BAL profiles of Defb1 mutant mice do not differ significantly from wild types.
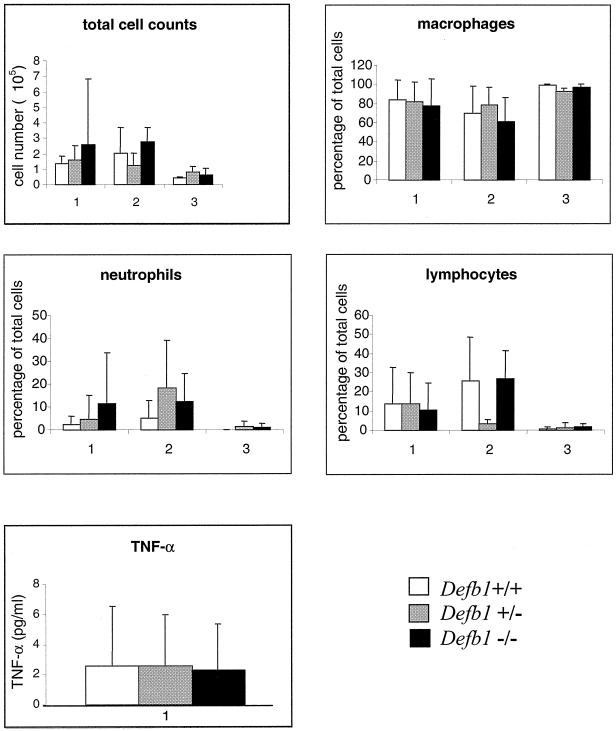
BAL fluid was obtained from Defb1−/−, Defb1+/−, and Defb1+/+ mice and analyzed as described in Materials and Methods. No significant differences in the total cell count or the percentage of neutrophils were observed between the Defb1−/− mice and wild-type littermates after the analysis of outbred conventionally housed mice, SPF-housed mice, or SPF-housed inbred 129/Ola mice (Fig. 2). Results are only shown for TNF-α levels in conventionally housed outbred animals since the levels of TNF-α in the BAL fluid from the SPF-housed sample groups were below detection.
FIG. 2.
Total and differential cell counts and TNF-α levels from bronchoalveolar lavage fluid harvested from Defb1+/+, Defb1+/−, and Defb1−/− mice. Groups: 1, mean values from conventionally housed outbred mice; 2, mean values from SPF-housed outbred mice; 3, mean values from SPF-housed inbred 129/Ola mice. TNF-α values were only obtained for conventionally housed outbred mice. Standard deviation bars have been applied to all values. Numbers of mice used: for group 1, Defb1+/+ (n = 14), Defb1+/− (n = 15), and Defb1−/− (n = 23); for group 2, Defb1+/+ (n = 5), Defb1+/− (n = 8), and Defb1−/− (n = 8); for group 3, Defb1+/+ (n = 6), Defb1+/− (n = 4), and Defb1−/− (n = 6).
Defb1−/− mice demonstrate an increased presence of bacteria in urine collected from the bladder.
Urine was collected from the mouse bladder as described and cultured for bacteria. No statistically significant differences were detected between the wild-type and heterozygous mice examined, and therefore these two genotypic classes were grouped together as the nonmutant class for the statistical analysis of mutant versus nonmutant classes. Table 2 shows that the number of Defb1−/− mice with bacteria present in the bladder was greater than that of nonmutant littermates as determined by using a one-tailed Fisher's exact test (P = 0.049). Interestingly, Staphylococcus spp. were found to be the most prominent in the bladder, with the number of Defb1−/− mice found to harbor Staphylococcus spp. in the bladder being significantly higher than the number of nonmutant mice (P = 0.008).
TABLE 2.
Bacteria retrieved from urine
| Genotype | No. of mice with:
|
Total mice |
S. aureus CFU detecteda
|
|||
|---|---|---|---|---|---|---|
| Bacteria detected | No bacteria detected | Staphylo- coccus spp. detected | Mean | SD | ||
| Defb1 +/+ | 2 | 16 | 1 | 18 | 0.22 | 0.65 |
| Defb1 +/− | 5 | 20 | 1 | 25 | 35.68 | 105.66 |
| Defb1 −/− | 12 | 22 | 9 | 34 | 103.53 | 373.06 |
CFU were counted from urine aseptically collected and cultured as detailed in Materials and Methods.
The Defb1 peptide displays antimicrobial activity against S. aureus.
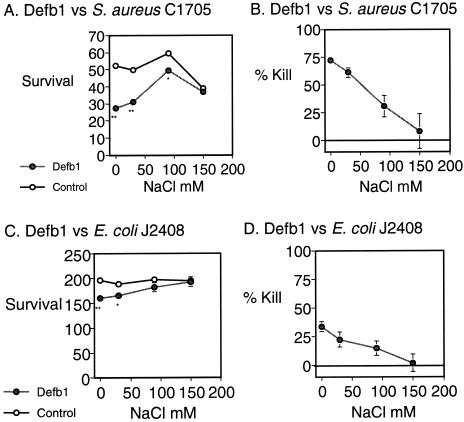
The activity of the Defb1 synthetic peptide against S. aureus and E. coli was examined in vitro according to the methods described above. The Defb1 synthetic peptide showed antimicrobial activity against S. aureus C1705 at a concentration of 50 μg/ml, killing 75% of S. aureus in 0 mM NaCl. This antimicrobial activity toward S. aureus was salt sensitive, with reduced killing at elevated (30, 90, and 150 mM) concentrations of NaCl (Fig. 3). The antimicrobial activity of Defb1 toward E. coli was less dramatic, with only 30% of E. coli being killed in 0 mM NaCl. Previously, we demonstrated Defb1's salt-sensitive antimicrobial activity against P. aeruginosa; however, this killing effect was only achieved at a Defb1 peptide concentration of 500 μg/ml (7, 19).
FIG. 3.
Functional analysis of synthetic Defb1. S. aureus C1705 (A and B) or E. coli J2408 (C and D) were incubated with synthetic Defb1 peptide at 50 μg/ml over a range of salt concentrations. Panels A and C demonstrate survival, assessed by using the mean CFU count after square root transformation ± the standard error, for blank control samples and peptide samples. Panels B and D show the mean percent kill ± the standard error, assessed by calculating the number of bacteria surviving in the peptide-treated samples as a percentage of the counts from the buffer only control samples. ∗, P < 0.01; ∗∗, P < 0.001.
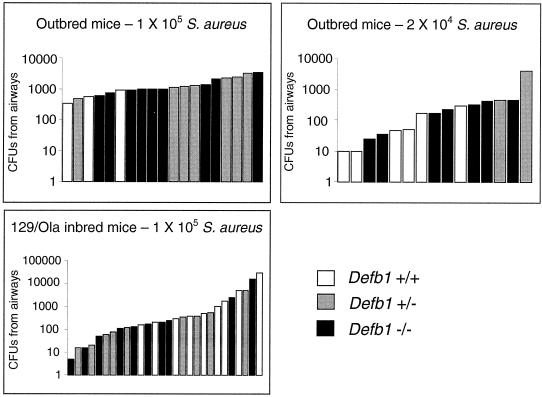
Defb1−/− mice are not impaired in their clearance of S. aureus from the airways.
Mice were exposed to S. aureus C1705 by nebulization to examine whether Defb1−/− mice demonstrated a defect in bacterial clearance in the airway. No statistically significant differences were found between Defb1−/− mice and their littermates in their ability to clear S. aureus from the airways 24 h after nebulization (Table 3 and Fig. 4). At the 105 dose, the mean number of S. aureus surviving in the outbred Defb1−/− airways was higher than that of the wild types but, due to the high variation within groups, this result could not be considered significant. In an effort to reduce the variation in the experiment, 129/Ola inbred mice were used. No significant difference was observed in the clearance of bacteria between the 129/Ola inbred Defb1−/− mice and their littermates and unfortunately, in this case, the use of inbred mice did not lead to a reduction in the values of the standard deviations of the mean within the sample groups.
TABLE 3.
Mean CFU values from mouse airways after S. aureus treatmenta
| Mouse type (CFU inoculated) | Genotype |
S. aureus CFU
|
n | |
|---|---|---|---|---|
| Mean | SD | |||
| Outbred (1 × 105) | +/+ | 61 | 29 | 4 |
| +/− | 191 | 110 | 7 | |
| −/− | 232 | 363 | 10 | |
| Outbred (2 × 104) | +/+ | 82 | 109 | 6 |
| +/− | 738 | 1,711 | 6 | |
| −/− | 310 | 121 | 10 | |
| 129 (1 × 105) | +/+ | 3,866 | 9,222 | 0 |
| +/− | 724 | 1,596 | 9 | |
| −/− | 1,854 | 4,776 | 10 | |
Mice were exposed to S. aureus at two different concentrations. n indicates the number of each genotype in each experiment, and the method was as described in Materials and Methods.
FIG. 4.
Clearance of S. aureus from the airways after nebulization. Each bar represents the number of S. aureus CFU retrieved from the airways of an individual mouse 24 h after treatment with the indicated dose of S. aureus.
Defb1−/− airways do not display upregulation of Defb2, Defb4, or Defb6
The absence of a pronounced effect on S. aureus clearance secondary to the disruption of Defb1 prompted us to examine whether any of the other published beta defensin genes with antimicrobial activity could compensate for the lack of Defb1. These genes are Defb2, Defb3, Defb4, and Defb6 (3, 15, 20, 29). Where expression of these genes was detected by RT-PCR, the level was consistently low compared to that of the control Hprt, and no significant differences were observed in the level of expression between these samples. Table 4 shows that Defb2, Defb4, and Defb6 were not upregulated in the airway of Defb1−/− mice in comparison to wild-type littermates. Defb3 expression was not observed in any of the tracheal samples. Defb6 was expressed in the tracheas of several of the conventionally housed mice but, in contrast to the findings of Yamaguchi et al. (29), which showed induction of Defb6 by P. aeruginosa LPS, we found that Defb6 expression was not upregulated in the trachea after exposure to E. coli LPS.
DISCUSSION
We have previously demonstrated that Defb1 has salt-sensitive antimicrobial activity against P. aeruginosa at a high concentration of peptide. We demonstrate here that it has a more effective activity against S. aureus and that this activity is also salt sensitive. In an effort to determine the physiological in vivo functions of Defb1, we disrupted the gene in mouse ES cells to produce a Defb1−/− mouse model.
Defb1 has previously been shown to be most highly expressed in the distal tubules and collecting ducts of the kidney (1, 19); thus, it was of interest to investigate whether the Defb1−/− mice had a compromised antimicrobial defense system in the urogenital system. Urine, like the airway surface fluid, should be sterile, and therefore we were surprised to find that 35% percent of the Defb1−/− mice tested had bacteria present in their urine. In addition, the probability of having bacteria present in the urine was significantly higher in these mice compared to the nonmutant littermates (P = 0.049). Upon further analysis it was found that the presence of Staphylococcus spp. was significantly elevated in the urine of the Defb1−/− mice compared to nonmutant littermates (P = 0.008).
Having observed a greater incidence of Staphylococcus spp. in the bladder of Defb1−/− mice and an antimicrobial profile for synthetic Defb1 peptide demonstrating more efficient killing of S. aureus than of P. aeruginosa or E. coli (7, 19), we considered that the administration of S. aureus to the airways of Defb1−/− mice would be most likely to reveal any defects in bacterial killing in the respiratory tract. Furthermore, the dysfunction of beta defensins has been proposed to play a role in the pathogenesis of CF lung disease, and we have previously demonstrated that Cftrtm1Hgu mice have a defect in bacterial clearance when exposed to S. aureus (6). However, the mean numbers of S. aureus remaining in the airways 24 h after treatment with 105 CFU were not statistically higher in the Defb1−/− mice compared to the controls. It has been proposed that defensins and other antimicrobial factors in the airway surface fluid are designed to control small numbers of bacteria to which the lungs are constantly exposed. In order to address the possibility of whether the numbers of bacteria used in these experiments were too large and thus masked any subtle effect caused by the loss of Defb1, we reduced the dose of S. aureus to 2 × 104 CFU. However, exposure of the mice to this reduced dose did not reveal any phenotypic differences between the genotypic classes.
It is possible that the outbred background of the mice may have contributed to the variability seen within the sample groups after exposure to S. aureus. The high degree of variability found in the antimicrobial activity of nasal secretions collected from different donors suggests that phenotypic modulators of antimicrobial activity may also exist in humans (5). To investigate whether the outbred background of the Defb1−/− mice contributed to the variability obtained in our results, Defb1−/− mice and littermates inbred onto a 129/Ola background were also analyzed. This did lead to a reduction in the variability in the percentages of different cell types found in the lavage fluid (samples C, Fig. 2); however, it did not reveal any significant differences between Defb1−/− mice and their wild-type littermates. The variability of results obtained with inbred 129/Ola mice in the S. aureus nebulization experiment was, however, not reduced, implying an environmental rather than a genotypic basis for the variation.
The lack of an obvious defect in the innate defense system of mice lacking Defb1 could be due to the redundant nature of the murine beta defensin molecules. Four additional family members have been identified and shown to be expressed in the airways (3, 15, 20, 29), and another beta defensin gene, Defb5, has been entered into the GenBank database under accession number AF318068. Antimicrobial activity has been demonstrated for Defb2, Defb3, and Defb6 peptides (3, 21, 29), and therefore it is possible that these peptides would be able to compensate for the lack of Defb1. However, we failed to detect the expression of Defb3 in any of the tracheal samples examined by RT-PCR, Defb6 was only expressed in two of the tracheal samples tested, and Defb2 expression was not upregulated in Defb1−/− mice compared to wild-type littermates. It is also possible that other beta defensins, uncharacterized to date, can compensate for the lack of Defb1 in the innate defense system. Indeed, 43 putative novel mature peptide-encoding beta defensin sequences present in the mouse genome have been described very recently (23). We are currently analyzing the murine beta defensin gene cluster, and we have evidence for the existence of several other sequences that may encode murine beta defensin genes. If this gene cluster has expanded in a manner similar to that observed for the alpha defensins in the mouse, then it is highly probable that the elimination of just one family member will not result in the production of an easily identifiable phenotype. It is possible that the entire family of murine beta defensin genes may need to be dysfunctional to see an effect. In addition, it is possible that direct antimicrobial activity is not the primary physiological function of the beta defensins in vivo. Recent studies of human antimicrobial peptides have demonstrated antiendotoxic and chemotactic activities (24, 30), with the latter function suggesting a possible role for antimicrobial peptides as a link between innate and adaptive immunity. In this scenario, future studies may establish a more subtle and complex phenotype in these mice. Our conclusions on beta defensin activity after our analysis of the Defb1−/− mouse therefore contrast with those of Goldman et al., who reported that the inactivation of just one of the beta defensins in human tracheal grafts is sufficient to give a reduced bacterial clearance (10).
In conclusion, Defb1−/− mice do not display any overt differences in their innate defense system and, although Defb1−/− mice are statistically more likely to harbor Staphylococcus spp. in their bladders, they are not impaired in their ability to clear S. aureus from the respiratory tract.
Acknowledgments
We thank Sheila Webb, Alison Maxwell, Peter Teague, and Andrew Carothers for help and contribution to this work and John Govan and Cathy Doherty for supplying the bacteria.
This project was supported by the Cystic Fibrosis Trust UK and the Medical Research Council UK.
Editor: E. I. Tuomanen
REFERENCES
- 1.Bals, R., M. J. Goldman, and J. M. Wilson. 1998. Mouse beta-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect. Immun. 66:1225-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bals, R., X. Wang, R. L. Meegalla, S. Wattler, D. J. Weiner, M. C. Nehls, and J. M. Wilson. 1999. Mouse beta-defensin 3 is an inducible antimicrobial peptide expressed in the epithelia of multiple organs. Infect. Immun. 67:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randell. 2000. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed]
- 5.Cole, A. M., P. Dewan, and T. Ganz. 1999. Innate antimicrobial activity of nasal secretions. Infect. Immun. 67:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson, D. J., J. R. Dorin, G. McLachlan, V. Ranaldi, D. Lamb, C. Doherty, J. Govan, and D. J. Porteous. 1995. Lung disease in the cystic fibrosis mouse exposed to bacterial pathogens. Nat. Genet. 9:351-357. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, D. J., S. Maclean, G. M. Morrison, J. Govan, D. J. Porteous, and J. R. Dorin. 1999. Contrasting antibacterial profiles on mouse and human beta defensin peptides may contribute to species specific lung phenotypes in cystic fibrosis. Pediatr. Pulm. Suppl. 19:323. [Google Scholar]
- 8.Diamond, G., M. Zasloff, H. Eck, M. Brasseur, W. L. Maloy, and C. L. Bevins. 1991. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. USA 88:3952-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia, J.-R. C., A. Krause, S. Schilz, F.-J. Rodriguez-Jimenez, E. Kluver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W.-G. Forssmann. 2001. Human β-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:1819-1821. [PubMed] [Google Scholar]
- 10.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 11.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 1997. A peptide antibiotic from human skin. Nature 387:861.. [DOI] [PubMed] [Google Scholar]
- 12.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 13.Ho, W., and A. Furst. 1973. Intratracheal instillation method for mouse lungs. Oncology 27:385-393. [DOI] [PubMed] [Google Scholar]
- 14.Huttner, K. M., C. A. Kozak, and C. L. Bevins. 1997. The mouse genome encodes a single homolog of the antimicrobial peptide human beta-defensin 1. FEBS Lett. 413:45-49. [DOI] [PubMed] [Google Scholar]
- 15.Jia, H. P., S. A. Wowk, B. C. Schutte, S. K. Lee, A. Vivado, B. F. Tack, C. L. Bevins, and P. B. McCray, Jr. 2000. A novel murine beta-defensin expressed in tongue, esophagus, and trachea. J. Biol. Chem. 275:33314-33320. [DOI] [PubMed] [Google Scholar]
- 16.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray, Jr. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 17.Liu, L., L. Wang, H. P. Jia, C. Zhao, H. H. Heng, B. C. Schutte, P. B. McCray, Jr., and T. Ganz. 1998. Structure and mapping of the human beta-defensin HBD-2 gene and its expression at sites of inflammation. Gene 222:237-244. [DOI] [PubMed] [Google Scholar]
- 18.McCray, P. B., Jr., and L. Bentley. 1997. Human airway epithelia express a beta-defensin. Am. J. Respir. Cell Mol. Biol. 16:343-349. [DOI] [PubMed] [Google Scholar]
- 19.Morrison, G. M., D. J. Davidson, F. M. Kilanowski, D. W. Borthwick, K. Crook, A. I. Maxwell, J. R. Govan, and J. R. Dorin. 1998. Mouse beta defensin-1 is a functional homolog of human beta defensin-1. Mamm. Genome 9:453-457. [DOI] [PubMed] [Google Scholar]
- 20.Morrison, G. M., D. J. Davidson, and J. R. Dorin. 1999. A novel mouse beta defensin, Defb2, which is upregulated in the airways by lipopolysaccharide. FEBS Lett. 442:112-116. [DOI] [PubMed] [Google Scholar]
- 21.Morrison, G. M., M. Rolfe, D. J. Davidson, F. M. Kilanowski, and J. R. Dorin. 2000. Genomic organisation and function of members of the murine beta defensin gene family. Pediatr. Pulm. Suppl. 20:269. [Google Scholar]
- 22.Schroder, J. M., and J. Harder. 1999. Human beta-defensin-2. Int. J. Biochem. Cell Biol. 31:645-651. [DOI] [PubMed] [Google Scholar]
- 23.Schutte, B. C., J. Mitros, J. Bartlett, J. Walters, H. P. Jia, M. Welsh, T. Casavant, and P. B. McCray. 2002. Discovery of five conserved β-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, M. G., and R. E. Hancock. 2000. Cationic antimicrobialpeptides and their multifunctional role in the immune system. Crit. Rev. Immunol. 20:407-431. [PubMed] [Google Scholar]
- 25.Smith, J. J., S. M. Travis, E. P. Greenberg, and M. J. Welsh. 1996. Cystic-fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell 85:229-236. [DOI] [PubMed] [Google Scholar]
- 26.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286:498-502. [DOI] [PubMed] [Google Scholar]
- 27.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. McCray, Jr., and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wine, J. J. 1999. The genesis of cystic fibrosis lung disease. J. Clin. Investig. 103:309-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, Y., S. Fukuhara, T. Nagase, T. Tomita, S. Hitomi, S. Kimura, H. Kurihara, and Y. Ouchi. 2001. A novel mouse β-defensin, mBD-6, predominantly expressed in skeletal muscle. J. Biol. Chem. 276:31510-31514. [DOI] [PubMed] [Google Scholar]
- 30.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defences and adaptive immunity. Cell Mol. Life Sci. 58:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]