Abstract
Twenty putative aminotransferase (AT) proteins of Corynebacterium glutamicum, or rather pyridoxal-5′-phosphate (PLP)-dependent enzymes, were isolated and assayed among others with l-glutamate, l-aspartate, and l-alanine as amino donors and a number of 2-oxo-acids as amino acceptors. One outstanding AT identified is AlaT, which has a broad amino donor specificity utilizing (in the order of preference) l-glutamate > 2-aminobutyrate > l-aspartate with pyruvate as acceptor. Another AT is AvtA, which utilizes l-alanine to aminate 2-oxo-isovalerate, the l-valine precursor, and 2-oxo-butyrate. A second AT active with the l-valine precursor and that of the other two branched-chain amino acids, too, is IlvE, and both enzyme activities overlap partially in vivo, as demonstrated by the analysis of deletion mutants. Also identified was AroT, the aromatic AT, and this and IlvE were shown to have comparable activities with phenylpyruvate, thus demonstrating the relevance of both ATs for l-phenylalanine synthesis. We also assessed the activity of two PLP-containing cysteine desulfurases, supplying a persulfide intermediate. One of them is SufS, which assists in the sulfur transfer pathway for the Fe-S cluster assembly. Together with the identification of further ATs and the additional analysis of deletion mutants, this results in an overview of the ATs within an organism that may not have been achieved thus far.
As inferred from the genome sequences, bacteria possess a number of aminotransferase (AT) proteins which, according to the KEGG entries, amount in the proteobacterium Escherichia coli to at least 16 and in the actinobacterium Corynebacterium glutamicum to 14 different proteins. The majority of these proteins are involved in amino acid synthesis, or amino acid interconversion, but also in the synthesis of biotin and porphyrin (10, 12). All ATs are pyridoxal-5′-phosphate (PLP)-dependent enzymes, where PLP forms the aldimine intermediate during transfer of the amino group from the incoming amino acid to an α-keto acid forming a new amino acid (20). However, the PLP-aldimine intermediate enables a wide variety of further reactions such as, for instance, C-S lyase activity by α,β-elimination or decarboxylation (13). Due to the mechanistic similarity of PLP-catalyzed reactions, the large number of AT proteins present, and their closely related structure, it is usually difficult, if not impossible, to derive the function of these proteins solely based on sequence studies.
An additional distinct feature of the ATs is their overlapping substrate specificity, which often leads to the nonexistence of a phenotype if one of them is absent. Thus, in E. coli the three ATs encoded by tyrB, aspC, and ilvE are involved in the synthesis of the aromatic amino acids, and the individual in vivo contribution of each of these ATs could only be studied when the other two respective genes were inactivated (7). Another example is the contribution of dapC and argD to l-lysine synthesis in E. coli (16) and C. glutamicum as well, with the latter organism possibly even possessing a third activity (9).
As mentioned above, a large number of bacterial ATs are involved in amino acid synthesis, and it is clear that these proteins are of special relevance for amino acid production with C. glutamicum. Functionally identified ATs of this organism include the N-succinyl-2,6-diaminopimelate AT (dapC) involved in the synthesis of l-lysine, of which 650,000 tonnes per year (t/y) are produced with C. glutamicum (5), as well as ilvE, encoding the branched-chain AT necessary for l-isoleucine production (26). Mutant studies further identified the genes (and respective enzymes) bioA (adenosylmethionine-8-amino-7-oxononanoate aminotransferase) (10), argD (N-acetyl-ornithine AT) (29), and pat and pdxR (involved in aromatic amino acid and pyridoxal-5′-phosphate synthesis, respectively) (19). Furthermore, an activity has been identified that uses l-alanine as the amino donor (17), thus resembling AvtA of E. coli (37). The function of some of these ATs was derived from a recent bioinformatic approach identifying a total of 20 sequences with similarities to ATs in the genome of C. glutamicum (19). However, bioinformatic and mutant analyses failed to identify specific ATs such as, for instance, the corresponding counterparts to avtA, aspC, or tyrB of E. coli.
Based on the recent bioinformatic study, we here isolate the AT proteins of C. glutamicum to study their activity with a variety of substrates. Together with in vivo studies, this investigation is an attempt to make a functional assignment of the ATs known from the genome analysis of a bacterium.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The strains and plasmids used are listed in Table 1. The standard medium for E. coli was Luria broth. C. glutamicum was precultivated on brain heart infusion (Difco) with subsequent cultivation on the minimal medium CGXII (5). When appropriate, chloramphenicol (25 mg liter−1) or kanamycin (15, 25, or 50 mg liter−1) was added to the medium. E. coli was grown at 30 or 37°C, and C. glutamicum was grown at 30°C.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant characteristicsa | Reference or sourceb |
|---|---|---|
| Strains | ||
| E. coli DH5αMCR | F−endA1 supE44 thi-1 λ− recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 mcrA Δ(mrr-hsdRMS-mcrBC) | 8 |
| C. glutamicum | ||
| ATCC 13032 | WT | ATCC |
| WTΔilvE | WT deleted of a 1,050-nt fragment of ilvE | This study |
| WTΔalaT | WT deleted of a 1,260-nt fragment of alaT | This study |
| WTΔaroT | WT deleted of a 972-nt fragment of aroT | This study |
| WTΔavtA | WT deleted of a 1,107-nt fragment of avtA | This study |
| WTΔalaT ΔilvE | WT deleted of a 1,260-nt fragment of alaT and a 1,050-nt fragment of ilvE | This study |
| WTΔaroT ΔilvE | WT deleted of a 972-nt fragment of aroT and a 1,050-nt fragment of ilvE | This study |
| WTΔavtA ΔilvE | WT deleted of a 1,107-nt fragment of avtA and a 1,050-nt fragment of ilvE | This study |
| Plasmids | ||
| pASK-1BA-3C | Vector for heterologous gene expression in E. coli CmroriVEc tetR | 33 |
| pJMargD | pASK-IBA-3C with argD (1467379-1467398, 1468548-1468527) | This study |
| pJMilvE | pASK-IBA-3C with ilvE (2337049-2337028, 2335916-2335935) | This study |
| pJMavtA | pASK-IBA-3C with avtA (2766133-2766111, 2764979-2765003) | This study |
| pJMalaT | pASK-IBA-3C with alaT (3030673-3030694, 3031980-3031959) | This study |
| pJM0780 | pASK-IBA-3C with the coding sequence of NCgl0780 (861547-861569, 862752-862730) | This study |
| pJMaroT | pASK-IBA-3C with aroT (233279-233256, 232260-232279) | This study |
| pJMhisC | pASK-IBA-3C (2217588-2217565, 2216494-2216515) | This study |
| pJMhemL | pASK-IBA-3C with hemL (462560-462581, 463867-463847) | This study |
| pJM2355 | pASK-IBA-3C with the coding sequence of NCgl2355 (2584563-2584584, 2585927-2585909) | This study |
| pJM2491 | pASK-IBA-3C with the coding sequence of NCgl2491 (2742575-2742553, 2741637-2741657) | This study |
| pJMsufS | pASK-IBA-3C (1649407-1649388, 1648100-1648122) | This study |
| pJMserC | pASK-IBA-3C with serC (877121-877101, 875985-876005) | This study |
| pJMbioA | pASK-IBA-3C with bioA (2770718-2770737, 2771983-2771961) | This study |
| pJMaspT | pASK-IBA-3C with aspT (256620-256641, 257894-257874) | This study |
| pJMaecD | pASK-IBA-3C with aecD (2444610-2444632, 2445710-2445691) | This study |
| pJMdapC | pASK-IBA-3C with dapC (1149282-1149300, 1150379-1150359) | This study |
| pJMpdxR | pASK-IBA-3C with pdxR (830982-830963, 829627-829646) | This study |
| pJM0462 | pASK-IBA-3C with the coding sequence of NCgl0462 (501499-501518, 502920-502901) | This study |
| pJM1184 | pASK-IBA-3C with the coding sequence of NCgl1184 (1297215-1297238, 1298339-1298319) | This study |
| pJM1022 | pASK-IBA-3C with the coding sequence of NCgl1022 (1116902-1116881, 1115832-1115851) | This study |
| pK19mobsacB | Integration vector; KmroriVEc oriT sacB | 31 |
| pK19mobsacBΔilvE | Plasmid to delete a 1,050-nt fragment of the C. glutamicum chromosome (2336998-2335949) | This study |
| pK19mobsacBΔalaT | Plasmid to delete a 1,260-nt fragment of the C. glutamicum chromosome (3030688-3031947) | This study |
| pK19mobsacBΔaroT | Plasmid to delete a 972-nt fragment of the C. glutamicum chromosome (233264-232293) | This study |
| pK19mobsacBΔavtA | Plasmid to delete a 1,107-nt fragment of the C. glutamicum chromosome (2766118-2765012) | This study |
Kmr, kanamycin resistance; Cmr, chloramphenicol resistance. Subscripts: Ec, E. coli. The nucleotide numbers of the expression vectors refer to the genome sequence BA000036 and correspond to the specific part present in the primer used to amplify the gene. The numbers of the four deletion vectors at the bottom of the table refer to the nucleotides that are deleted in the chromosome upon use of these vectors. For details, see the text.
ATCC, American Type Culture Collection.
Construction of plasmids.
Plasmids were constructed in E. coli DH5αMCR from PCR-generated fragments (Expand High Fidelity PCR kit; Roche Diagnostics) by using C. glutamicum ATCC 13032 DNA as a template. In order to construct pJMilvE the upstream primer 5′-ATGGATGGTCTCAAATGATTCTGTCAGGATGCAGGTGAT-3′ was used. The underlined sequence is specific for ilvE and corresponds to nucleotides (nt) 2337049 to 2337028 of the C. glutamicum genome sequence BA000036. As the downstream primer, 5′-ATGGATGGTCTCAGCGCTGCCAACCAGTGGGATAAGCC-3′ was used, with the underlined sequence corresponding to nt 2335916 to 2335935. In Table 1 only the gene-specific nt numbers are given for the primers to amplify the respective ATs. The sequences common to all primers used for gene amplification are identical to those given in boldface for ilvE. The resulting fragments were BsaI digested and cloned into the BsaI site of pASK-IBA-3C (IBA GmbH, Gottingen, Germany). Accordingly, the open reading frames of the other 19 genes encoding potential ATs were cloned into pASK-IBA-3C. To enable chromosomal deletions of ilvE, alaT, aroT, and avtA, crossover PCR was applied (18) to generate a defined fragment of approximately 875 bp in size carrying upstream and downstream sequences of about equal size of the respective open reading frame to be deleted. The fragments were cloned into pK19mobsacB via their attached BamHI sites. The plasmids made eventually enabled a defined chromosomal deletion, as specified by the nucleotide numbers of the wild-type (WT) chromosome (Table 1).
Construction of strains.
C. glutamicum was transformed by electroporation (34). The AT deletion mutants were constructed by using pK19mobsacBΔilvE, pK19mobsacBΔalaT, pK19mobsacBΔaroT, and pK19mobsacBΔavtA, respectively. Clones were selected for kanamycin resistance to establish integration of the plasmid in the chromosome. In a second round of positive selection by using sucrose resistance, clones were selected for deletion of the vector (31). The deletions in the chromosome were verified by PCR analysis using primers hybridizing approximately 500 bp upstream and 500 bp downstream of the open reading frames in question.
Heterologous gene expression and protein purification.
The 20 E. coli DH5αMCR strains, each one harboring a different pASK-IBA-3C derivate encoding a potential AT were grown until the optical density at 550 nm reached 0.5. After induction by adding 10 μl of anhydrotetracycline (2 mg ml−1), the cultures were incubated for 3 h at 30°C. The cells were harvested by centrifugation at 6,000 rpm for 12 min at 4°C. Crude extracts were obtained by sonification using a Branson Sonifier 250 (intensity, 2; duty cycle, 20%, 4 min; Branson, Danbury, CT), while cooling on ice. After removal of the cellular debris by centrifugation (15 min, 14,000 rpm, 4°C), all preparation procedures were performed at 4°C using Strep-Tactin Sepharose and the Strep-tag Protein Purification Buffer Set (IBA GmbH, Gottingen, Germany). Purified proteins were stored in the elution buffer (100 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA) at −20°C. The identity of the potential AT proteins were confirmed by using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF-MS). Protein concentrations were determined by using a BCA Protein Assay Kit (Pierce, Rockford, IL).
MALDI-TOF-MS.
For MALDI-TOF-MS, protein spots stained with Coomassie brilliant blue were excised from the sodium dodecyl sulfate gel and washed three times with 750 μl of 30% CH3CN-100 mM NH4HCO3. After drying the gel pieces and reswelling them in 12 μl of 3 mM NH4HCO3 containing 5 ng of modified trypsin (Promega, Mannheim, Germany), digestion was carried out overnight at 37°C. Elution of the peptides from the gel was performed by the addition of 10 μl of 30% CH3CN-0.1% trifluoroacetic acid. For MALDI-TOF-MS, 2 μl of each supernatant was mixed with 0.5 μl of saturated α-cyano-4-hydroxycinnamic acid matrix (20 mg ml−1) prepared in 0.25% trifluoroacetic acid-50% acetonitrile. This mixture was spotted onto the sample probe, and MALDI mass spectra were obtained with a PerSeptive Biosystems Voyager DE STR mass spectrometer (PerSeptive Biosystems, Langen, Germany). For calibration of the mass spectrometer, the Sequazime peptide mass standard kit (Applied Bioscience, Wieterstadt, Germany) was used. Monoisotopic masses were assigned and used for in-house database searches of the C. glutamicum genome performed with the Gpmaw software (Lighthouse Data, Odense, Denmark).
Crude extracts.
C. glutamicum was grown in minimal medium until the optical density at 600 nm reached 10 (exponential phase). The cells were harvested by centrifugation for 15 min at 4,000 rpm at 4°C. All preparation procedures were performed at 4°C. The pellet was washed twice with 200 mM Tris-HCl (pH 8) and was resuspended in the same buffer. Crude extracts were obtained by sonication, and after centrifugation (15 min, 14,000 rpm, 4°C) the supernatant was desalted with PD-10 columns (Pharmacia, Uppsala, Sweden) and kept on ice until enzyme assays were performed.
Enzyme assays.
The AT assay contained 200 mM Tris-HCl (pH 8), 0.25 mM pyridoxal-5′-phosphate, 4 mM keto acid, and 50 mM l-amino acid. The reaction was started by the addition of purified protein or crude extract (in 1 ml) and was performed at 30°C. Several 50-μl samples were collected over a period of 20 min. The reaction was terminated by mixing each sample with 30 μl of 5% perchloric acid and 38% ethanol. After the sample was neutralized by the addition of 20 μl of 20 mM Tris-HCl (pH 8) buffer with 23 mM K2CO3, the precipitated salts were removed by centrifugation (10 min, 13,000 rpm). Subsequently, amino acids were quantified by high-pressure liquid chromatography as their o-phthaldehyde derivative. Assays were linear over time and proportional to the protein concentration used.
The cysteine desulfurase assays were performed with 50 mM l-cysteine and 2.5 mM pyridoxal-5′-phosphate in 20 mM Tris-HCl (pH 8). The reaction mixture was incubated at ambient temperature for 2.5 h, and samples taken at different points in time to quantify the l-alanine formed by high-pressure liquid chromatography.
RESULTS
Isolation of AT proteins and activity tests.
Application of hidden Markov models identified 20 genes in the genome of C. glutamicum putatively encoding ATs (19). We cloned all of these genes into pASK-IBA-3C, expressed them in E. coli, and isolated the proteins fused at their carboxy-terminal ends with Strep-tag II via affinity purification. In each case, 0.6 to 2.4 mg of protein was obtained from a 100-ml culture. The protein was pure as judged by SDS-PAGE analysis. Its identity was confirmed by MALDI-TOF-MS.
Due to our interest in l-isoleucine synthesis with C. glutamicum (22), we first focused on branched-chain amino acid synthesis. All 20 proteins were individually assayed with amino donor l-Glu, l-Ala, l-Asp, or l-Gln using as the amino acceptor 2-oxo-3-methylvalerate (O-Ile), 2-oxo-isocaproate (O-Leu), or 2-oxo-isovalerate (O-Val). In addition, the l-Ile intermediate 2-oxo-butyrate (O-But) was assayed, which is known to be formed during l-Ile production (38). Product formation was followed over time, and the results where detectable amino acid formation occurred are shown in Table 2. In order to avoid confusion in the nomenclature, the enzyme names resulting in the course of the studies are already given in this table. Of the 20 proteins, 5 exhibited AT activity with O-Ile, O-Leu, O-Val, and O-But, and 3 additional AT proteins had weak activities with O-But only but not with the ultimate branched-chain amino acid intermediates. With the other 12 proteins no activities were found in these assays. The highest activities were not only present for IlvE, which had been previously identified (26), but also for the AT termed AvtA. The latter AT uses l-Ala as the amino donor instead of l-Glu and thus resembles transaminase C of E. coli (37), and a corresponding activity of C. glutamicum was recently described (17).
TABLE 2.
Activities of the AT proteins of C. glutamicum with detectable activities toward branched-chain amino acid intermediates or 2-oxo-butyrate
| ATa | Amino donorb | Amino acceptor | Sp actc (μmol min−1 mg of protein−1) |
|---|---|---|---|
| ArgD | l-Glu | O-But | 0.1 |
| l-Ala | O-But | 0.1 | |
| l-Asp | O-But | 0.1 | |
| l-Gln | O-But | 0.1 | |
| IlvE | l-Glu | O-Ile | 9.6 |
| l-Glu | O-Leu | 13.9 | |
| l-Glu | O-Val | 13.7 | |
| l-Glu | O-But | 4.3 | |
| AvtA | l-Ala | O-Ile | 3.7 |
| l-Ala | O-Leu | 0.9 | |
| l-Ala | O-Val | 18.2 | |
| l-Ala | O-But | 27.5 | |
| l-Gln | O-Ile | 0.1 | |
| l-Gln | O-Leu | 0.1 | |
| l-Gln | O-Val | 0.1 | |
| l-Gln | O-But | 0.1 | |
| AlaT | l-Glu | O-But | 5.4 |
| l-Ala | O-But | 3.0 | |
| l-Asp | O-But | 2.3 | |
| l-Gln | O-But | 0.7 | |
| NCgl0780 | l-Glu | O-Leu | 0.1 |
| l-Glu | O-But | 0.2 | |
| AroT | l-Glu | O-Leu | 1.3 |
| l-Ala | O-Leu | 0.8 | |
| l-Asp | O-Leu | 0.1 | |
| l-Gln | O-Leu | 0.1 | |
| l-Glu | O-Val | 0.1 | |
| l-Ala | O-Val | 0.1 | |
| l-Asp | O-Val | 0.1 | |
| l-Gln | O-Val | 0.1 | |
| l-Glu | O-But | 1.1 | |
| l-Ala | O-But | 0.7 | |
| l-Asp | O-But | 0.1 | |
| l-Gln | O-But | 0.1 | |
| HisC | l-Glu | O-Leu | 0.8 |
| l-Ala | O-Leu | 0.1 | |
| l-Asp | O-Leu | 0.4 | |
| l-Gln | O-Leu | 0.1 | |
| l-Glu | O-But | 0.2 | |
| l-Ala | O-But | 0.1 | |
| l-Asp | O-But | 0.1 | |
| l-Gln | O-But | 0.1 | |
| NCgl0462 | l-Glu | O-But | 0.1 |
| l-Ala | O-But | 0.1 | |
| l-Gln | O-But | 0.1 |
Either names or NCgl numbers are given. See also Table 4.
Amino donors are given as their three-letter code; amino acceptors are as follows: O-Ile, 2-oxo-3-methylvalerate; O-Leu, 2-oxo-isocaproate; O-Val, 2-oxo-isovalerate; and O-But, 2-oxo butyrate.
0.1 means detectable amino acid formation compared to the other 12 ATs not included in the table. Each value represents the average of at least two independent assays.
IlvE and AvtA affinities.
As mentioned, the identified proteins, IlvE and AvtA, have the highest specific activities with the substrates assayed, but they use different amino donors. The ilvE gene was originally isolated by complementation of a mutant of C. glutamicum requiring all three branched-chain amino acids (26), and therefore an in vivo function of AvtA is not directly apparent. We therefore determined the substrate affinities for both proteins in Lineweaver-Burk plots (not shown). With the amino donor l-Glu the Km (mM) for IlvE was 0.23 (O-Ile), 0.15 (O-Leu), 0.63 (O-Val), and 1.42 (O-But). For AvtA with l-Ala as amino donor the Km values were 3.52 (O-Ile), 16.84 (O-Leu), 2.51 (O-Val), and 0.60 (O-But). This agrees with the view that the major function of IlvE is synthesis of the branched-chain amino acids and that O-But amination probably represents a side activity of this protein. Compared to this, AvtA has weak affinities for the branched-chain amino acid intermediates. The highest affinity and also activity (Table 2) was present for O-But, an activity that may not be considered to represent a housekeeping function. No activity of AvtA was detected with the substrates glycine and pyruvate.
In vivo IlvE and AvtA function.
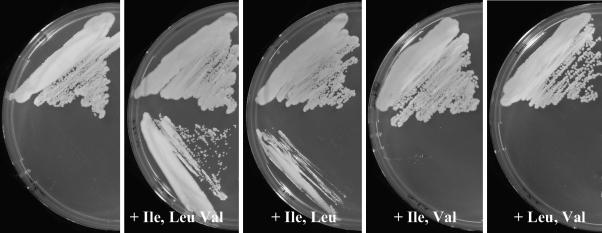
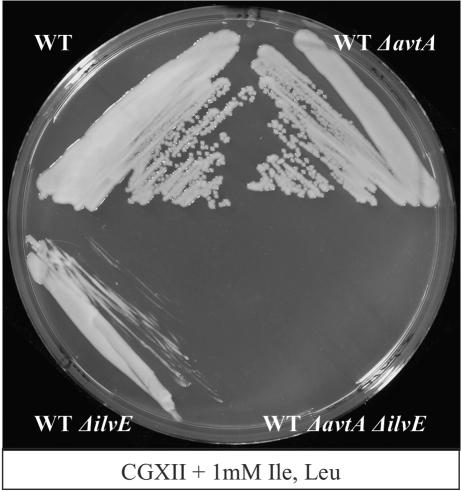
In order to analyze in vivo branched-chain amino acid synthesis, we deleted avtA and ilvE in the chromosome of the WT of C. glutamicum ATCC 13032, singly or combined, and assayed growth on mineral salts medium. WTΔilvE was fully dependent on l-Ile and l-Leu supply but not on the supply of l-Val (Fig. 1). In contrast, WTΔavtA did not exhibit a phenotype on CGXII (not shown). This illustrates that in vivo, at least for l-Ile and l-Leu synthesis, IlvE is clearly the major AT activity. The situation with l-Val is different. WTΔilvE still exhibits significant growth without l-Val addition, which is disabled in WTΔilvEΔavtA (Fig. 2). Therefore, AvtA contributes to l-Val synthesis in vivo, and the avtA deletion is silent unless ilvE is deleted as is similarly the case for E. coli (2). We did growth experiments with strain WTΔavtA to pursue the idea that AvtA might actually be necessary to catabolize externally supplied l-Ala, performed on complex medium brain heart infusion, as well as on salt medium CGXII with or without 20 mM l-Ala, but these investigations were without observable effects.
FIG. 1.
Branched-chain amino acid requirements of C. glutamicum ΔilvE. At the top is shown the WT, and the deletion mutant is shown at the bottom. Growth was carried out on salt medium CGXII (5) with amino acids supplemented as indicated (each at 1 mM).
FIG. 2.
l-Val synthesis by AvtA. The salt medium CGXII was supplemented with l-Ile plus l-Leu. The isogenic mutants, derived from C. glutamicum WT, are as indicated.
As a further characterization of the in vivo function, activities in crude extracts of the WT and the mutants grown on CGXII were compared (Table 3). From the comparison of WT with WTΔavtA we conclude that (i) AvtA has highest activities with O-Val and O-But as has the isolated protein (Table 2), (ii) there is no further Ala-dependent activity for O-Val formation, and (iii) there are further Ala-dependent activities for O-But formation. In WTΔilvE all Glu-dependent formation of branched-chain amino acids is absent, but there is still Glu-dependent formation of aminobutyrate, which agrees with the large number of candidates identified in Table 2.
TABLE 3.
Specific activities in crude extracts of mutantsa
| Amino donor | Amino acceptor | Sp act (μmol min−1 mg of protein−1)
|
||
|---|---|---|---|---|
| WT | WTΔavtA | WTΔilvE | ||
| l-Glu | O-Ile | 38 | 35 | <1 |
| O-Leu | 44 | 45 | <1 | |
| O-Val | 24 | 21 | <1 | |
| O-But | 61 | 55 | 18 | |
| l-Ala | O-Ile | 5 | <1 | 3 |
| O-Leu | <1 | <1 | <1 | |
| O-Val | 19 | <1 | 16 | |
| O-But | 49 | 35 | 26 | |
Measurements were done twice with variations of <12%.
Identification of AroT.
In Table 2, the protein subsequently identified as aromatic AT, AroT, attracted attention since among the substrates assayed it had the highest activities with O-Leu and l-Glu as substrate. McHardy et al. (19), who termed this gene pat, observed an auxotrophy for Leu/Ile/Phe (supplied together) when inactivated in an ilvE background, suggesting that the gene under consideration encodes an aromatic AT. We therefore assayed for activity with the substrates phenylpyruvate (O-Phe) and 4-hydroxyphenylpyruvate (O-Tyr) using l-Glu as an amino donor. The specific activities (μmol min−1 mg of protein−1) were 13.6 (O-Phe) and 8.8 (O-Tyr), respectively, confirming the function of the protein as an aromatic AT. The detectable activity with O-Leu as substrate (Table 2) is not surprising since the aromatic and branched-chain amino acids share a strong hydrophobicity. Also, aromatic amino acid AT TyrB of E. coli was shown to exhibit weak activity with O-Leu (25). Based on the finding that the branched-chain AT IlvE of E. coli shows activities for the formation of O-Phe and O-Tyr (7) and the mutant study with C. glutamicum (19), we also assayed IlvE of C. glutamicum for its specificity toward aromatic substrates. A remarkably high activity (μmol min−1 mg of protein−1) of 10.7 was obtained with O-Phe as substrate and 2.4 with O-Tyr. This might explain that the single pat inactivation did not result in an aromatic amino acid requirement (19).
Identification of AspT.
In further assays with the other isolated AT proteins we searched for the aspartate AT, which is of prime interest for the synthesis of the aspartate-derived amino acids (14). These enzymes belong to the class I AT proteins (20), of which C. glutamicum possesses nine candidates (19). Since the function for three of them has been identified (9; this study), we assayed selected ATs with l-Asp and O-Glu as the substrate. With the protein encoded by NCgl0237 an activity was found. It was 10.7 μmol min−1 mg of protein−1 identifying the protein as AspT.
Identification of AlaT.
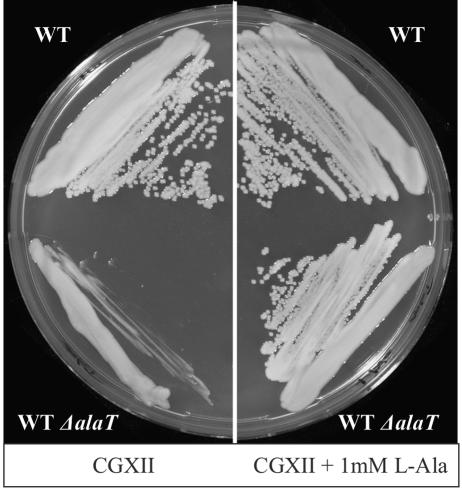
Another AT of interest is that converting pyruvate to l-alanine. The reason is that l-alanine is occasionally formed as by-product during l-lysine or l-valine production with C. glutamicum and knowledge of alanine ATs in general is scanty. For instance, the corresponding gene of E. coli has not yet been identified. In Table 2 we observed one AT encoded by NCgl2747 which uses O-But as substrate together with any of the four amino donors assayed. We therefore deleted the gene in the WT to generate WTΔalaT. Growth of this mutant was retarded on minimal medium CGXII (Fig. 3). When all 20 amino acids were supplied together, this complemented the growth defect. Further assays identified that l-Ala alone fully restores growth (Fig. 3). Interestingly, the auxotrophy was only apparent on agar plates (see Discussion). The enzyme assay subsequently performed confirmed l-alanine formation as the major activity. With pyruvate as substrate and l-Glu as amino donor the specific activity (μmol min−1 mg of protein−1) was 26.6, instead of 5.4 with O-But as substrate (Table 2). Furthermore, with pyruvate as substrate it was 1.8 with l-Asp as amino donor, 0.2 with l-Gln as amino donor, and 8.4 with aminobutyrate as amino donor. This rather broad amino donor specificity clearly distinguishes AlaT from all other ATs of C. glutamicum (see also Table 2).
FIG. 3.
l-Ala synthesis by AlaT. Only on the right side did the salt medium GCXII contain l-Ala. At the top is shown the WT, and at the bottom the WT with alaT deleted is shown.
Identification of cysteine desulfurases.
Three of the AT proteins (NCgl1500, NCgl1184, and NCgl1022) were isolated from E. coli as yellowish proteins. Their absorption spectrum identified them as containing pyridoxal-5′-phosphate (not shown). These proteins belong to class V of ATs containing phosphoserine ATs and cysteine desulfurases. As already concluded from the genomic context (19), the proteins might be involved in the synthesis of Fe-S complexes. In an enzyme assay we observed high alanine generation from l-cysteine accompanied by the unpleasant smell of sulfur-derived compounds with the proteins encoded by NCgl1500 and NCgl1022 identifying them as cysteine desulfurases. The determined specific activities of 0.35 and 0.04 (μmol min−1 mg of protein−1), respectively, were comparable to the cysteine desulfurase IscS of E. coli (23).
Activities of ArgD, DapC, and HemL.
The AT DapC, responsible for l-lysine synthesis, has been identified by activity determinations in crude extracts (9), and the AT ArgD, responsible for l-arginine synthesis, is known due to its clustering with arg genes in C. glutamicum (29). These enzymes aminate the structurally related substrates succinyl-diaminopimelate and acetyl-ornithine, and in E. coli both enzymes have activities with the two substrates which has led to confusion with respect to the assignment of the proteins (3, 16). Interestingly, inactivation of dapC, together with argD, in C. glutamicum still enables growth of the mutant without supplementation (9), requiring an even further AT of sufficient activity to sustain lysine-independent growth. We therefore followed the proposal of A. Tauch (University of Bielefeld, Germany) that HemL might also use succinyl-diaminopimelate as substrate and compared the activities of the proteins in question. The activities (in μmol min−1 mg of protein−1) with succinyl-diaminopimelate were 1.2 with DapC, 0.006 with ArgD, and <0.001 with HemL. With acetyl-ornithine they were <0.001 with DapC, 6.4 with ArgD, and 0.046 with HemL. It is doubtful whether the weak HemL activity contributes to l-lysine synthesis.
DISCUSSION
Table 4 provides a complete overview of the ATs plus some PLP-containing proteins as results from the various approaches based on genome information for C. glutamicum and functional studies. The PLP-containing MetC (AecD) is not an AT, but it has β-lyase activity toward cystathionine (27) or the unnatural amino acid S-(2-aminoethyl)-d,l-cysteine (35). We also did not find any AT activity with NCgl2491, which is adjacent in the genome to a putative T-protein of a glycine cleavage system. Three of the proteins were isolated as colored proteins and contain firmly bound PLP. They are likely to carry out a β-elimination, which we have demonstrated for the proteins encoded by NCgl1500 and NCgl1022. These are desulfurases that cleave l-cysteine to form alanine together with an enzyme cysteinyl persulfide intermediate (21). The mobilized sulfur is used for a number of processes, such as Fe-S cluster assembly, as well as the synthesis of thiamine, lipoic acid, or thionucleosides in tRNA. Since NCgl1500 is part of the well-conserved sufABCDS operon of bacteria and plants, assisting in the sulfur transfer pathway for Fe-S cluster assembly (23), we denote this protein SufS. SufS is an abundant protein in C. glutamicum (30), which substantiates the idea that SufS represents the major activity for Fe-S cluster generation. The gene NCgl1184 is arranged in synteny within the Corynebacterianeae with genes of lipid synthesis and electron-transferring flavoproteins, which might eventually transfer the reducing equivalents formed during the oxidation of fatty acyl-coenzyme A (CoA) to trans-Δ2-enoyl-CoA to the membrane-bound quinone pool, and the quinone oxidoreductase catalyzing this latter activity is also an Fe-S cluster protein. Thus, it appears that a more specific sulfur-providing pathway is necessary for synthesis of the Fe-S cluster in the quinone oxidoreductase. The same holds true for the third desulfurase (NCgl1022), which is clustered together with quinolinate synthetase A, an Fe-S protein required for NAD synthesis.
TABLE 4.
Overview of the ATs of C. glutamicum
| NCgl | Gene | Alias(es) | Classa
|
Enzymeb | Cellular functionb | |
|---|---|---|---|---|---|---|
| Mehta et al. | Batemann et al. | |||||
| NCgl0215 | aroT | pat, tyrB | I | I, II | Aromatic amino acid AT* | Aromatic amino acid synthesis (19) |
| NCgl0237 | aspT | I | Aspartate AT* | Aspartate synthesis | ||
| NCgl0422 | hemL | II | III | Glutamate semialdehyde AT | Uroporphyrinogen synthesis | |
| NCgl0462 | II | III | Butanoate metabolism | 4-Aminobutyrate aminotransferase | ||
| NCgl0753 | pdxR | I | Pyridoxamine-P AT | Pyridoxal-P synthesis* (19) | ||
| NCgl0780 | I | I, II | ||||
| NCgl0794 | serC | Phosphoserine AT | Serine synthesis* (24) | |||
| NCgl1022 | IV | V | Cysteine desulfurase* | Involved in NAD synthesis | ||
| NCgl1058 | dapC | I | I, II | Succinyldiaminopimelate AT* (9) | Lysine synthesis* (9) | |
| NCgl1184 | IV | V | Cysteine desulfurase | Assembly of FeS complex of electron transfer flavoprotein | ||
| NCgl1343 | argD | II | I | Acetylornithine AT* | Arginine synthesis* (29) | |
| NCgl1500 | sufS | IV | V | Cysteine desulfurase* | Assembly of FeS complexes | |
| NCgl2020 | hisC | I | I, II | Histidinol phosphate AT | Histidine synthesis* (19) | |
| NCgl2123 | ilvE | III | IV | Branched-chain amino acid AT* | BCAA synthesis* (26) | |
| NCgl2227 | metC | aecD | I | I, II | Cystathionine β-lyase* (15) | Methionine synthesis |
| NCgl2355 | II | III | ||||
| NCgl2491 | III | IV | Glycine cleavage? | |||
| NCgl2510 | avtA | I | I, II | Valine-pyruvate AT* | ||
| NCgl2515 | bioA | II | III | AdoMet-aminooxononanoate AT | Biotin synthesis* (10) | |
| NCgl2747 | alaT | I | IV | Alanine AT* | Alanine synthesis* | |
Two of the newly identified ATs are clearly separate from the others: AvtA and AlaT. Bioinformatic analyses currently recognize the AvtA structure as similar to the class I (20) or class I/II structures of ATs (1), which represent the most common types of ATs present in C. glutamicum. Nevertheless, AvtA is the only AT with exceptionally high activity toward l-alanine as an amino donor instead of preferably using l-glutamate. As 2-oxo-acid, it preferably accepts O-But with highest activity and affinity, followed by O-Val (Table 2). The enzyme corresponds to the transaminase C activity already found in extracts of C. glutamicum (17). A similar enzyme activity is present in E. coli, which has been demonstrated to use l-alanine or aminobutyrate as equivalent amino donors to aminate O-Val (28). However, the in vivo function of AvtA is difficult to assign, since in E. coli, and C. glutamicum as well, the avtA mutation has no phenotype. It could be that AvtA is involved in the adjustment of amino acid pool sizes in C. glutamicum rather than fulfilling a specific biosynthesis function.
The other AT functionally distinguished and identified in the genome of C. glutamicum is AlaT. Recent AT classifications classify AlaT into family IV, of which a total of only three proteins are present in C. glutamicum (Table 4). AlaT is characterized by a broad specificity for the amino donor, in the order Glu > But > Asp (with pyruvate as acceptor) and Glu > Ala > Asp (with O-But as acceptor). Knowledge of bacterial Ala ATs is limited, and one other AlaT is known from Pyrococcus furiosus, where it serves as an electron sink to produce l-Ala during fermentation of sugars (36). Of the C. glutamicum ATs, AlaT has the highest identity (37%) to the AlaT of P. furiosus. Using these structures as seed information, we propose that yfbQ of E. coli is an alanine transaminase. Interestingly, the alaT phenotype of C. glutamicum, which is an l-alanine requirement on plates (Fig. 3), is not present during growth of the same clone in liquid culture CGXII (not shown). This could be due to the overlapping AT activities and at the same time a different AT regulation under the two growth conditions used.
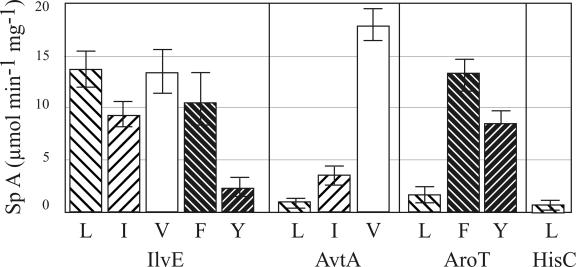
As already mentioned in the introduction, there is a strong overlap of transamination activity for the hydrophobic substrates of E. coli, which is also present in C. glutamicum (Fig. 4). An early article already reported on two separate activities in C. glutamicum for the transamination of O-Phe and O-Tyr (6), and these two activities were not considered likely to be identical to IlvE (32). This agrees with the recent observation that an ilvE, aroT double mutant (19) does not require l-Tyr for growth. Therefore, in addition to AroT and IlvE, which both have comparable activities for O-Phe (Fig. 4), a further still unknown activity for l-Tyr is required. In E. coli AspC and TyrB (together originally named transaminase A) are similar in many respects, and both have activity toward aromatic amino or oxo acids (11). We assayed AspC of C. glutamicum with aromatic amino acids and AroT with oxo acids, but in neither case was any activity detected (not shown). Therefore, the third AT active in C. glutamicum with O-Tyr still has to be identified. Also, for the branched-chain amino acids there is overlapping AT activity. However, this depends very much on the specific amino acid in question. For instance, in vivo, l-Ile appears to be exclusively synthesized via IlvE (Fig. 1), and the activity due to AvtA (Fig. 4) might be too low to sustain growth of the ilvE mutant. The situation with l-Val is different, since the high AvtA activity with this substrate (Fig. 4) is apparently sufficient to enable significant growth, and only upon deletion of both AvtA and IlvE is there an absolute requirement for l-Val (Fig. 2). Although there are three additional proteins—AvtA, AroT, and HisC—acting on 2-oxo-isocaproate to synthesize l-Leu (Fig. 4), these activities are apparently too weak to sustain significant growth. Whereas it is largely now clear which of the overlapping activities contributes to aromatic and branched-chain amino acid synthesis, this is less clear for aminobutyrate formation. The largest activities have IlvE and AvtA (Table 3). However, as is evident from the present study, there are a number of additional activities present in C. glutamicum. This is not unexpected, considering the broad substrate specificity and versatility of transaminating activities.
FIG. 4.
Comparison of the activities of the ATs showing overlapping substrate specificities for the in vitro formation of the branched-chain and aromatic amino acids. 2020 is the NCgl number of the protein (see Table 4). The amino donor for IleV, AroT, and 2020 was l-glutamate. The amino donor for AvaT was l-alanine.
Acknowledgments
We thank A. Tauch (University of Bielefeld, Germany) for the suggestion of possible NCgl0422 activity.
We thank Amino GmbH (Frellstedt, Germany), as well as the Fonds der Chemischen Industrie, for financial support.
REFERENCES
- 1.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [Online.] http://www.sanger.ac.uk/Software/Pfam/Version17.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, C. M., W. A. Whalen, and L. B. Archambault. 1983. Role of alanine-valine transaminase in Salmonella typhimurium and analysis of an avtA::Tn5 mutant. J. Bacteriol. 155:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox, R. J., and P. S. Wang. 2001. Is N-acetylornithine aminotransferase the real N-succinyl-ll-diaminopimelate aminotransferase in E. coli and M. smegmatis? J. Chem. Soc. Perkin Trans. 1:2006-2008. [Google Scholar]
- 4.Eggeling, L., S. Morbach, and H. Sahm. 1997. The fruits of molecular physiology: engineering the l-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J. Biotechnol. 56:167-182. [Google Scholar]
- 5.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Inc., Boca Raton, Fla.
- 6.Fazel, A. M., and R. A. Jensen. 1979. Aromatic aminotransferases in coryneform bacteria. J. Bacteriol. 140:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelfand, D. H., and R. A. Steinberg. 1977. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J. Bacteriol. 130:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann, M., A. Tauch, L. Eggeling, B. Bathe, B. Möckel, A. Pühler, and J. Kalinowski. 2003. Identification and characterization of the last two unknown genes, dapC and dapF, in the succinylase branch of the l-lysine biosynthesis of Corynebacterium glutamicum. J. Biotechnol. 104:199-211. [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama, K., K. Hohama, A. A. Vertes, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1993. Genomic organization of the biotin biosynthetic genes of coryneform bacteria: cloning and sequencing of the bioA-bioD genes from Brevibacterium flavum. DNA Seq. 4:177-184. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, H., K. Inoue, T. Nagata, S. Kuramitsu, and H. Kagamiyama. 1993. Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry 32:12229-12239. [DOI] [PubMed] [Google Scholar]
- 12.Ilag, L. L., D. Jahn, G. Eggertsson, and D. Soll. 1991. The Escherichia coli hemL gene encodes glutamate 1-semialdehyde aminotransferase. J. Bacteriol. 173:3408-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John, R. A. 1995. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta 1248:81-96. [DOI] [PubMed] [Google Scholar]
- 14.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. W., H. J. Kim, Y. Kim, M. S. Lee, and H. S. Lee. 2001. Properties of the Corynebacterium glutamicum metC gene encoding cystathionine beta-lyase. Mol. Cells. 11:220-225. [PubMed] [Google Scholar]
- 16.Ledwidge, R., and J. S. Blanchard. 1999. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry 38:3019-3024. [DOI] [PubMed] [Google Scholar]
- 17.Leyval, D., D. Uy, S. Delaunay, J. L. Goergen, and J. M. Engasser. 2003. Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J. Biotechnol. 104:241-252. [DOI] [PubMed] [Google Scholar]
- 18.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McHardy, A., A. Tauch, C. Rückert, A. Pühler, and J. Kalinowski. 2003. Genome-based analysis of biosynthetic aminotransferase genes of Corynebacterium glutamicum. J. Biotechnol. 104:229-240. [DOI] [PubMed] [Google Scholar]
- 20.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 21.Mihara, H., and N. Esaki. 2002. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 60:12-23. [DOI] [PubMed] [Google Scholar]
- 22.Morbach, S. Sahm, H., and L. Eggeling. 1996. l-isoleucine production with Corynebacterium glutamicum: further flux increase and limitation of export. App. Environ. Microbiol. 62:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Outten, F. W., M. J. Wood, F. M. Munoz, and G. Storz. 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278:45713-45719. [DOI] [PubMed] [Google Scholar]
- 24.Peters-Wendisch, P., M. Stolz, H. Etterich, N. Kennerknecht, H. Sahm, and L. Eggeling. Metabolic engineering of Corynebacterium glutamicum for l-serine production. Appl. Environ. Microbiol, in press. [DOI] [PMC free article] [PubMed]
- 25.Powell, J. T., and J. F. Morrison. 1978. Role of the Escherichia coli aromatic amino acid aminotransferase in leucine biosynthesis. J. Bacteriol. 136:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radmacher, E., A. Vaitsikova, U. Burger, K. Krumbach, H. Sahm, and L. Eggeling. 2002. Linking central metabolism with increased pathway flux: l-valine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 68:2246-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossol, I., and A. Pühler. 1992. The Corynebacterium glutamicum aecD gene encodes a C-S lyase with α,β-elimination activity that degrades aminoethylcysteine. J. Bacteriol. 174:2968-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudman, D., and A. Meister. 1953. Transamination in Escherichia coli. J. Biol. Chem. 200:591-604. [PubMed] [Google Scholar]
- 29.Sakanyan, V., P. Petrosyan, M. Lecocq, A. Boyen, C. Legrain, M. J. Demarez, N. Hallet, and N. Glansdorff. 1996. Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacterium glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology 142:99-108. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer, S., B. Weil, V. D. Nguyen, G. Dongmann, K. Gunther, M. Nickolaus, T. Hermann, and M. Bott. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22:4404-4422. [DOI] [PubMed] [Google Scholar]
- 31.Schäfer, A., A. Tauch, W. Jaeger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 32.Shiio, I., and S. I. Sugimoto. 1981. Regulation at metabolic branch points of aromatic amino acid biosynthesis in Brevibacterium flavum. Agric. Biol. Chem. 45:2197-2207. [Google Scholar]
- 33.Skerra, A. 1994. Use of the production of a murine antibody fragment in Escherichia coli. Gene 151:131-135. [DOI] [PubMed] [Google Scholar]
- 34.Tauch, A., O. Kirchner, B. Löffler, S. Götker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 35.Thierbach G., J. Kalinowski, B. Bachmann, and A. Pühler. 1990. Cloning of a DNA fragment from Corynebacterium glutamicum conferring aminoethyl cysteine resistance and feedback resistance to aspartokinase. Appl. Microbiol. Biotechnol. 32:443-448. [DOI] [PubMed] [Google Scholar]
- 36.Ward, D. E., S. W. Kengen, J. van Der Oost, and W. M. de Vos. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whalen, W. A., and C. M. Berg. 1982. Analysis of an avtA::Mu d1(Ap lac) mutant: metabolic role of transaminase C. J. Bacteriol. 150:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilhelm, C., L. Eggeling, A. Nassenstein, C. Jebsen, I. Eggeling, and H. Sahm. 1989. Limitations during hydroxybutyrate conversion to isoleucine with Corynebacterium glutamicum as analysed by the formation of by-products. Appl. Microbiol. Biotechnol. 172:458-462. [Google Scholar]