Abstract
The pHT plasmids pHTα (65.9 kbp), pHTβ (63.7 kbp), and pHTγ (66.5 kbp) are highly conjugative pheromone-independent pMG1-like plasmids that carry Tn1546-like transposons encoding vancomycin resistance. pHTβ is the prototype plasmid, and the pHTα and pHTγ plasmids are derivatives of the insertion into pHTβ of an IS232-like (2.2 kbp) element and a group II intron (2.8 kbp), respectively. The complete nucleotide sequence of the pHTβ plasmid was determined and, with the exception of the Tn1546-like insertion (10,851 bp), was found to be 52,890 bp. Sixty-one open reading frames (ORFs) having the same transcript orientation were identified. A homology search revealed that 22 of the pHTβ (pHT) plasmid ORFs showed similarities to the ORFs identified on the pXO2 plasmid (96.2 kbp), which is the virulence plasmid essential for capsule formation by Bacillus anthracis; however, the functions of most of the ORFs remain unknown. Most other ORFs did not show any significant homology to reported genes for which functions have been analyzed. To investigate the highly efficient transfer mechanism of the pHT plasmid, mutations with 174 unique insertions of transposon Tn917-lac insertion mutants of pHTβ were obtained. Of the 174 derivatives, 92 showed decrease or loss in transfer frequency, and 74 showed normal transfer frequency and LacZ expression. Eight derivatives showed normal transfer and no LacZ expression. Inserts within the 174 derivatives were mapped to 124 different sites on pHTβ. The Tn917-lac insertions which resulted in altered transfer frequency mapped to three separate regions designated I, II, and III, which were separated by segments in which insertions of Tn917-lac did not affect transfer. There was no region homologous to the previously reported oriT sequences in the pHT plasmid. The oriT was cloned by selection for the ability to mobilize the vector plasmid pAM401. The oriT region resided in a noncoding region (192 bp) between ORF31 and ORF32 and contained three direct repeat sequences and two inverted repeat sequences. ORF34, encoding a 506-amino-acid protein which was located downstream of the oriT region, contains the three conserved motifs (I to III) of the DNA relaxase/nickase of mobile plasmids. The transfer abilities of the Tn917-lac-insertion mutants of ORF34 or a mutant of ORF34 with an in-frame motif III deletion were completely abolished. The sequence of the oriT region and the deduced relaxase/nickase protein of ORF34 showed no significant similarity to the oriT and relaxase/nickase of other conjugative plasmids, respectively. The putative relaxase/nickase protein of ORF34 could be classified as a new member of the MOBMG family.
The isolation of vancomycin-resistant enterococci (VRE) was first reported in 1988 in the United Kingdom (43) and France (25), and shortly thereafter, VRE were detected in hospitals in the United States (34). Since then, VRE have emerged with unanticipated rapidity and are now encountered in most hospitals, especially in the United States (28).
Increased drug resistance is linked to direct selective pressure by the use of antibiotics and often the presence of a genetic transfer system for spread of resistance (5). Most VRE clinical isolates are Enterococcus faecium strains (28). Little is known about systems of efficient plasmid transfer in E. faecium. Previously, we described the isolation of the pheromone-independent gentamicin resistance conjugative plasmid pMG1 (65.1 kbp) from an E. faecium clinical strain in Japan, which was the first report describing efficient plasmid transfer in E. faecium (21). pMG1 transfers among enterococcus strains during broth mating at a frequency of about 10−4 per donor strain. Southern hybridization analysis revealed no similarity to other gram-positive conjugative plasmids, and pMG1 was categorized as a new type of conjugative plasmid. Our epidemiological study revealed that pMG1-like plasmids are widely disseminated in vancomycin-resistant E. faecium clinical isolates from a hospital in the United States, suggesting that pMG1-like plasmids may contribute to the efficient dissemination of vancomycin resistance in enterococcus strains (41).
Recently, we reported the isolation of pMG1-like vancomycin resistance pHT plasmids from clinical Enterococcus faecium and Enterococcus avium strains in Japan (42). pHT plasmids, including pHTα (65.9 kbp), pHTβ (63.7 kbp), and pHTγ (66.5 kbp), are highly conjugative plasmids carrying Tn1546-like transposons (3) that encode vancomycin resistance (VanA). The pHT plasmids are related to the gentamicin resistance conjugative plasmid pMG1 with respect to DNA hybridization, and they are thought to contain the same efficient conjugation system. The transfer gene traA, which is involved in the formation of mating aggregates, is conserved in all pMG1-like plasmids (36, 42).
In this report, sequence comparisons and genetic analysis of pHTβ obtained by generating Tn917-lac insertion mutants led to identification of a novel type of oriT region and a putative relaxase/nickase gene designated traI.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, media, and reagents.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1 (and see Table S6 in the supplemental material). E. faecalis strains were grown in Todd-Hewitt broth (THB) (Difco Laboratories) at 37°C. The following antibiotic concentrations were used for selection of E. faecalis: erythromycin, 12.5 μg ml−1; streptomycin, 250 μg ml−1; kanamycin, 250 μg ml−1; spectinomycin, 250 μg ml−1; chloramphenical, 20 μg ml−1; rifampin, 25 μg ml−1; and fusidic acid, 25 μg ml−1. Antibiotic concentrations for selection of Escherichia coli were as follows: ampicillin, 100 μg ml−1; kanamycin, 40 μg ml−1; chloramphenicol, 50 μg ml−1; and spectinomycin, 50 μg ml−1. All antibiotics were obtained from Sigma Chemical Co. X-Gal (5-bromo-4-chloro-3 indolyl-β-d-galactopyranoside) was used at 40 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant features | Reference |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| FA2-2 | rif fus | 12 |
| JH2SS | spc str | 37 |
| UV202 | fir fus, recombination-deficient mutant of JH2-2 | 46 |
| OG1X | str | 20 |
| OG1RF | rif fus | 8 |
| OG1SS | spc str | 12 |
| E. faecium | ||
| BM4105RF | rif fus | 21 |
| BM4105SS | spc str | 21 |
| E. coli DH5α | endA1 recA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(argE-lacZYA)U169 | Bethesda Research Laboratories |
| Plasmids | ||
| pHTα | pHTβ carrying IS232-like element | 42 |
| pHTβ | pMG1-like vancomycin resistance (Tn1546) conjugative plasmid | 42 |
| pHTγ | pHTβ carring group II intron | 42 |
| pAM401 | E. coli-E. faecalis shuttle; cat tet | 45 |
| pTV32Ts | Transposon delivery vector, temperature sensitive; pE194Ts(Cmr)::Tn917-lac(Emr) | 31 |
| pMW119 | E. coli cloning vector; Ampr | Nippon Gene |
| pBluescript SKII(+) | E. coli cloning vector; Ampr | Stratagene |
| pACYC184 | E. coli cloning vector; Tcr Cmr | New England Biolabs |
| pHTβ/ORF34del | pHTβ derivative mutant carrying a deletion of motif III of ORF34 (Tral) | This study |
Plasmid/DNA methodology.
Recombinant DNA methodology, analyses of plasmid DNA with restriction enzymes, and agarose gel electrophoresis were carried out by standard methods (35). Introduction of plasmid DNA into bacterial cells was by electrotransformation, as described previously (13). Plasmid DNA was purified from E. faecalis as previously described (44). Restriction enzymes were purchased from New England Biolabs and Roche Co. The PCR was performed with a Perkin-Elmer Cetus apparatus. Specific primers were purchased from Invitrogen, and Taq DNA polymerase was obtained from Takara.
Construction of the clone sets of the pHTβ plasmid.
For determination of the physical map of the pHTβ and for DNA sequence analysis, the clone sets of the pHTβ were constructed as previously described (14, 39, 40). The partially HindIII-digested fragments of pHTβ plasmid were cloned into vector plasmid pMW119. Plasmid pACYC184 was also used for cloning of the partially BclI-digested pHTβ plasmid DNA.
DNA sequence analysis.
Sequence analysis was performed using the Dye primer and Dye terminator cycle sequencing kit (Applied Biosystems) and a 377 DNA Sequencer and 310 Gene Analyzer (ABI PRISM). To determine the DNA sequence of the pHTβ plasmid, a GPS in vitro transposition system kit (New England BioLabs) and shotgun cloning method were used (35). To determine the DNA sequences in the gap regions, PCR amplification was performed (LA-Taq; Takara, Japan) to obtain PCR products covering the gaps. The PCR products were sequenced directly using custom primers. Open reading frames (ORFs) were identified and initially analyzed using Genetyx version 5.1 computer software and the BLAST (1) database to search for putative genes.
Conjugation experiments.
Filter matings and solid surface matings were performed as previously described (7, 20). Broth matings (in THB) were for 4 h. Transfer frequencies are expressed as the number of transconjugants per donor cell (at the end of mating).
Identification and genetic analyses of the oriT region of the pHT plasmid.
Various segments of pHTβ containing sequences that were related to the consensus oriT sequence of IncP or IncQ plasmids were amplified by PCR (2, 22). Segments containing inverted repeat (IR) or direct repeat (DR) sequences were also amplified. The amplified DNAs were cloned into the pAM401 vector plasmid. Two clones, pAM401::119/113 and pAM401::119/96, were constructed as chimeric plasmids between pAM401 and the clones pMW119::113 and pMW119::96, respectively (see Fig. 1 and see Table S6 in the supplemental material). Each of the pAM401 derivatives carrying pHTβ segments to be tested for oriT activity was introduced by electrotransformation into Enterococcus faecalis UV202, which is defective in homologous recombination (46). Then, the conjugative plasmid pHTβ was introduced into each of the UV202 transformants carrying the pAM401 derivative (Cmr) by conjugation. Both broth matings and filter matings were performed using the transconjugants carrying the two plasmids as donor strains and JH2SS as the recipient strain.
FIG. 1.
Genetic map and ORFs deduced from the complete plasmid sequence of pHTβ plasmid. Boxes indicate ORFs identified on the pHT plasmid. The locations of the Tn917-lac insertions of pHTβ (vertical bar with circular head) are shown on the map. Each head color shows the transfer frequency of the derivatives (white; transfer frequency the same as parent plasmid, black; transfer frequency less than one-fifth of the frequency of the parent plasmid or no transfer by broth mating). The number on the insert (1 through 29) indicates the representative plasmids of the derivatives, which are also shown in Fig. 2 and Table 3. Horizontal bars designated as Tra regions I, II, and III on the map indicate the transfer-related regions. DR and IR indicate the direct repeat sequences and inverted repeat sequences. The asterisks under the map indicate the sequence resembling the consensus oriT sites of IncP, IncQ, or oriT of the F plasmid. Black horizontal bars designated a through q under the map show the cloned DNA segments used for the identification of the oriT region of the pHT plasmid. The clones are as follows: a, pAM401::ORF1; b, pAM401::IR4; c, pAM401::IR3; d, pAM401::119/113; e, pAM401::ORF19/21; f, pAM401::119/96; g, pAM401::IR5-1; h, pAM401::ORF39/40; i, pAM401::IR1; j, pAM401::DR1; k, pAM401::ORF51/52; l, pAM401::ORF53/56; m, pAM401::traA; n, pAM401::IR2; o, pAM401::ORF58; p, pAM401::rep5′; and q, pAM401::ORF60/61.
Transposon mutagenesis and isolation of pHTβ derivatives with Tn917-lac insertions.
The transposon delivery vector pTV32Ts (31) was used for mutagenesis (19, 44). Strain OG1X/pTV32Ts was originally constructed by protoplast formation (45), and plasmid pHTβ was introduced into this strain by conjugation. The isolation of pHTβ::Tn917-lac insertion mutants was performed as previously described (44). The locations of transposon inserts in the pHTβ plasmid were determined by PCR amplification and DNA sequencing, using primers that amplify the segment containing the junction of the inserts.
Construction of the ORF34 in-frame mutant.
The overlapping PCR technique was used to construct the ORF34 deletion mutant. The internal region of ORF34, which contained the 30-bp deletion corresponding to 10 amino acid residues (HRNTEHIHIH) of motif III, was amplified using the specific primer set (see Fig. 5 and see Table S6 in the supplemental material). The amplified DNA was cloned into the pBluescript vector plasmid, resulting in pBS::ORF34del. The 1.1-kbp DNA fragment amplified by PCR, which carries a spectinomycin resistance gene, aad(9), was cloned into pBS::ORF34del, to give pBS::ORF34del-Spc (24). pBS::ORF34del-Spc was introduced into E. faecalis FA2-2/pHTβ by electrotransformation, and recombinants occurring via double homologous recombination were selected as previously described (38). A representative recombinant carrying the 30-bp deletion of motif III of ORF34, the deduced relaxase gene, was designated as pHTβ/ORF34del.
FIG. 5.
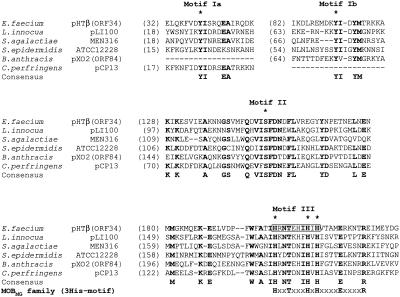
Comparison of the N-terminal region of the deduced ORF34 protein (TraI) of pHTβ with the hypothetical proteins found in sequence databases. The bold characters indicate the conserved amino acid residues in each protein. The asterisks on the sequences show the key residues, Tyr, Ser, and His3 (3His) in motifs I, II, and III, respectively. There are two motif I candidates (Ia and Ib) in most of the proteins. The gray box in motif III indicates the deleted 10-amino-acid residues of ORF34 resulting in pHTβ/ORF34del. The lowest sequence shows the putative consensus His3 motif (motif III) of these proteins (designated as MOBMG family), H(x2)T(x3)HxH(x4)E(x4)R. The accession numbers for the proteins are as follows: NP_569166 for Listeria innocua pLI100 (81,905 bp), NP_734852 for Streptococcus agalactiae MEN316, NP_765038 for Staphylococcus epidermidis ATCC 12228, NP_053238 for B. anthracis pXO2-84, and NP_150032 for Clostridium perfringens strain 13 plasmid pCP13 (54,310 bp), respectively.
Nucleotide sequence accession number.
The complete nucleotide sequence of the pHTβ plasmid has been deposited in DDBJ/EMBL/GenBank under accession no. AB183714.
RESULTS AND DISCUSSION
DNA sequence and gene organization of the pHT plasmid.
The total size of the pHTβ plasmid, excluding the Tn1546-like insertion, was 52,890 bp. The nucleotide sequence of the Tn1546-like element inserted in the pHTβ plasmid was almost identical to that of the prototype Tn1546 (10,851 bp). There was no amino acid substitution of the VanS protein found in Japanese VRE isolates that show low-level teicoplanin resistance (17). The G-C content of the core region of the pHT plasmid excluding the Tn1546-like transposon was 31.35, and that of the Tn1546-like region was 38.66. The Tn1546-like transposon is inserted at the same location in the three pHT plasmids α, β, and γ. The hexa-oligonucleotide sequence 5′-GATTAT-3′ was duplicated at the junctions. These results suggested that the pHT plasmid resulted from the transposition of the Tn1546-like element into an original plasmid without a drug resistance determinant. Sixty-one open reading frames were identified in the 52,884-bp core region of the pHT plasmid, excluding the Tn1546-like element and the 6-bp repeat (i.e., 5′-GATTAT-3′), as shown in Fig. 1 and as listed in Table 2. All the ORFs were transcribed in the same direction on the plasmid, with the exception of ORF1 of Tn1546-like transposon (Fig. 1). The nucleotide next to the right end of the Tn1546-like transposon was designated as the first base pair of the plasmid. Transcription of the genes encoded on the pHTβ plasmid was in a counterclockwise direction from the physical map described in our previous report (42).
TABLE 2.
Open reading frames identified in pHT plasmidsa
| ORF | 5′ end | 3′ end | Amino acid size | Identification [organism] | Protein family/domain | % Identity |
|---|---|---|---|---|---|---|
| 1 | 52467 | 1946 | 787 | Trsl (Tral) [L. lactis]/pXO2-77/76 [B. anthracis] | DNA topoisomerase I/III | 34/29 |
| 2 | 2264 | 2863 | 199 | AbiQ [L. lactis] | Phage abortive infection mechanism | 19 |
| 3 | 2984 | 3199 | 71 | Hypothetical protein | ||
| 4 | 3515 | 3721 | 68 | Hypothetical protein | ||
| 5 | 3801 | 4655 | 284 | ParA [E. faecalis; pTEF3]/pXO2-39 [B. anthracis] | Replication-related protein | 29/27 |
| 6 | 4668 | 5105 | 145 | YvfU [B. sutilis] | Two-component sensor histidine kinase | 19 |
| 7 | 5123 | 5422 | 99 | Hypothetical protein | ||
| 8 | 5425 | 5868 | 147 | Hypothetical protein | ||
| 9 | 6014 | 6337 | 107 | Hypothetical protein | ||
| 10 | 6755 | 10384 | 1,209 | pXO2-28 [B. anthracis] | Putative cell surfrace protein | 10 |
| 11 | 10444 | 10806 | 120 | Hypothetical protein | ||
| 12 | 10820 | 10972 | 50 | Hypothetical protein | ||
| 13 | 11079 | 11462 | 127 | pXO2-26 [B. anthracis] | 26 | |
| 14 | 11474 | 12889 | 471 | pXO2-25 [B. anthracis] | VirB11 family, secretory protein | 40 |
| 15 | 12927 | 13721 | 264 | pXO2-24/23 [B. anthracis] | 29/12 | |
| 16 | 13721 | 14518 | 265 | pXO2-22 [B. anthracis] | 43 | |
| 17 | 14536 | 14898 | 120 | pXO2-21 [B. anthracis] | 51 | |
| 18 | 14912 | 15058 | 48 | Hypothetical protein | ||
| 19 | 15310 | 15585 | 91 | pXO2-18 [B. anthracis] | 47 | |
| 20 | 15694 | 17520 | 608 | pXO2-17 [B. anthracis] | Chromosome segregation ATPases | 10 |
| 21 | 17533 | 18249 | 238 | Hypothetical protein | ||
| 22 | 18262 | 21120 | 952 | VirD4 [E. faecium]/pXO2-16/15 [B. anthracis] | VirD4/TraG/TraD family (coupling protein) | 33/33/10 |
| 23 | 21138 | 23912 | 924 | AidA [E. faecium]/pXO2-14 [B. anthracis] | Amino acid transporters | 71/25 |
| 24 | 23960 | 24271 | 103 | pXO2-11 [B. anthracis] | 35 | |
| 25 | 24268 | 24885 | 205 | pXO2-10 [B. anthracis] | 35 | |
| 26 | 24902 | 25369 | 155 | Hypothetical protein | ||
| 27 | 25385 | 27340 | 651 | TrsE (TraE) [L. lactis; pMRC01] pXO2-09 [B. anthracis] | VirB4 family (transfer complex protein) | 22/48 |
| 28 | 27362 | 28498 | 378 | ORF14 [E. faecalis; Tn916]/pXO2-08 [B. anthracis] | NLP/P60 family, cell wall-associated hydrolase | 32/43 |
| 29 | 28512 | 29162 | 216 | Hypothetical protein | ||
| 30 | 29176 | 30072 | 298 | pXO2-05 [B. anthracis] | 25 | |
| 31 | 30108 | 30338 | 76 | Hypothetical protein | ||
| 32 | 30689 | 30985 | 98 | pXO2-04 [B. anthracis] | ABC transport system, permease component | 22 |
| 33 | 30982 | 31449 | 155 | Hypothetical protein | ||
| 34 | 31532 | 33052 | 506 | pXO2-84 [B. anthracis] | Three motifs conserved in DNA relaxase/nickase | 21 |
| 35 | 33074 | 33403 | 109 | Hypothetical protein | ||
| 36 | 33405 | 33668 | 87 | Hypothetical protein | ||
| 37 | 33890 | 34756 | 288 | HlyD [E.. coli] | Hemolysin secretion protein | 11 |
| 38 | 34911 | 36737 | 608 | LtrC [L. lactis; pMRC01]/pXO2-81 [B. anthracis] | Catalytic active site (HEXXH) | 15/23 |
| 39 | 36799 | 37815 | 338 | Hypothetical protein | ABC transpoter | |
| 40 | 37824 | 38081 | 85 | Hypothetical protein | ||
| 41 | 38122 | 38412 | 96 | Hemolysin-related protein [Thermotoga maritima] | 21 | |
| 42 | 38437 | 39441 | 334 | LtrC-like protein [S. epidermidis]/pXO2-78 [B. anthracis] | 26/34 | |
| 43 | 39532 | 39822 | 96 | Hypothetical protein | ||
| 44 | 39984 | 40175 | 63 | Hypothetical protein | ||
| 45 | 40284 | 40502 | 72 | Hypothetical protein | ||
| 46 | 40539 | 40856 | 105 | Hypothetical protein | ||
| 47 | 40867 | 41064 | 65 | Hypothetical protein | ||
| 48 | 41061 | 42029 | 322 | Hypothetical protein | Predicted ATPase | |
| 49 | 42255 | 43442 | 395 | Hypothetical protein | ||
| 50 | 43553 | 43846 | 97 | Hypothetical protein | ||
| 51 | 44498 | 44905 | 135 | Prophage ps3 protein 15 [L. lactis] | Cro/CI family, transcriptional regulator | 21 |
| 52 | 45057 | 45668 | 203 | PinR [E.. coli; lambdoid prophage Rac] | Site-specific recombinases, DNA invertase | 33 |
| 53 | 46046 | 46219 | 57 | Hypothetical protein | ||
| 54 | 46387 | 46695 | 102 | Hypothetical protein | ||
| 55 | 46707 | 47024 | 105 | Hypothetical protein | ||
| 56 | 47191 | 47754 | 187 | 71ORF1 [E. faecium; pMG1] | 100 | |
| 57 | 47765 | 48625 | 286 | traA [E. faecium; pMG1] | Related to formation of mating aggregate | 98 |
| 58 | 49122 | 49376 | 84 | Hypothetical protein | ||
| 59 | 49773 | 51374 | 533 | RepS [E. faecalis; V583]/pXO2-38 [B. anthracis] | Replication protein | 34/27 |
| 60 | 51608 | 51907 | 99 | PrgN [E. faecalis; pCF10] | Replication and negative control of conjugation | 38 |
| 61 | 52041 | 52418 | 125 | Hypothetical protein |
The colums list open reading frame number, 5′ end of the ORF, 3′ end of the ORF, deduced amino acid size, identification of similar proteins and organisms, functional protein family or conserved domain, and percentage identity of the pHT plasmid ORF to the matching protein(s).
A homology search of 61 ORFs encoded on the pHT plasmid was performed by BLAST against the protein databases, and the results are shown in Table 2. Only six ORFs (ORF1, -14, -22, -27, -57, and -60) showed significant similarity to transfer-related genes of other plasmids (Fig. 1) (16). Twenty-two ORFs that are located between ORF1 and -42 and ORF59 (gray-colored ORFs shown in Fig. 1) had significant degrees of similarity to the ORFs encoded on the Bacillus anthracis virulence plasmid pXO2 (96,231 bp; accession no. NC_002146; 94,829 bp, accession no. NC_003981) (15, 32). The pXO2 plasmid carries capsule genes and is necessary to cause the disease anthrax (27). The degree of identity between the deduced amino acid residues of the homologous ORFs ranged from about 10 to 50% (Table 2).
Although most of the functions of the homologues are not known, the putative functions of several proteins were assigned based on their similarity to other well-characterized proteins (Table 2). ORF1 encoded 787 amino acid residues, but the Tn1546-like insert at the 142nd amino acid residue (Trp) separated ORF1 into an N-terminal 142-amino-acid residue portion and a C-terminal 645-amino-acid residue portion. The intact protein was related (about 40% identity) to the trsI gene carried on the Lactococcus lactis bacteriocin plasmid pMRC01 (60.2 kbp) (6). TrsI belongs to the DNA topoisomerase family, which is frequently found in the transfer-related region of conjugative plasmids (26). Three pHT plasmids, α, β, and γ, that carried Tn1546-like insertions within ORF1 could transfer at high frequencies. This suggested that the topoisomerase homologue (ORF1) was not essential for plasmid transfer.
The sequence comparisons of the ORFs on the pHT plasmid suggested that a relatively large portion of the plasmid from ORF57 through ORF61 and from ORF1 through ORF28 (a region spanning about 33 kb) could be associated with the transfer region of the pHT plasmid (Fig. 1). However, many pHT ORFs did not have significant homology with any reported proteins, and the plasmid was therefore categorized as a new type of conjugative plasmid, as shown by our previous genetic analysis (21). There are reports describing highly efficient self-transferable large plasmids in Bacillus species and the mobilization of the anthrax toxin plasmid pXO1 (181.7 kbp) and pXO2 (4, 29, 33). The pMG1-like plasmids, including the pHT plasmids, could be closely related to these efficient conjugative plasmids.
Analysis of Tn917-lac transposon insertion mutants of pHTβ plasmid.
For genetic analysis of the transfer system of pHT plasmids, we isolated mutants which altered the transferability of the pHTβ plasmid. Transposon-insertional mutagenesis of pHTβ plasmid using Tn917-lac (Emr) was performed, and 1,000 independent insertion derivatives were obtained. Each derivative was examined for transferability in broth mating and LacZ expression in OG1X harboring the plasmid on THB plates containing X-Gal reagent. A total of 352 derivatives which showed altered transferability or expressed LacZ activity were chosen for further analysis. Agarose gel electrophoresis analysis of NdeI-digested plasmid DNA showed that 289 of 352 derivatives had a single insertion of Tn917-lac with no deletion or recombination. Of the 289 derivatives carrying a single insertion, the location of each insertion in 174 representative derivatives was determined by DNA sequencing and mapped on pHTβ, excluding the Tn1546-like region (Fig. 1). Of 174 derivatives, 92 showed altered transfer frequency and 74 showed normal transfer and positive LacZ expression. Eight derivatives showed normal transfer and no LacZ expression after repeated examinations. There were hot spots for Tn917-lac insertion on pHTβ plasmid, and the inserts were mapped to 124 different sites within the pHTβ plasmid. The locations of the Tn917-lac insertions into the ORFs within the 124 derivatives are shown in Fig. 1. The transfer frequency of each of the 124 representative derivatives was examined in broth mating. The mating experiment was repeated using different hosts: E. faecalis FA2-2 and JH2SS, E. faecalis OG1RF and OG1SS, or E. faecium BM4105RF and BM4105SS, respectively. Ninety-two derivatives showed an altered transfer frequency, which was either a reduced transfer frequency or an inability to transfer at a frequency of greater than 10−7 per donor cell. Some representative plasmids and the results of the mating experiments are shown in Table 3.
TABLE 3.
Transfer of pHTβ::Tn917-lac derivatives
| No. in Fig. 1 and 2 | Representative plasmid | Location of Tn917-lac insertion | Position of insertion (bp) | Frequency of transfer (broth mating)a | Relative transfer frequency (broth mating) |
|---|---|---|---|---|---|
| Wild type | pHTβ | Wild type | Wild type | 1.1 × 10−4 | 100 |
| 1 | pHTβ::Tn917-lac/315 | ORF1 (C terminal) | 492 | 1.9 × 10−4 | 172 |
| 2 | pHTβ::Tn917-lac/261 | ORF2 | 2445 | 2.8 × 10−4 | 254 |
| 3 | pHTβ::Tn917-lac/268 | ORF8 | 5589 | <1.1 × 10−7 | <0.1 |
| 4 | pHTβ::Tn917-lac/136 | ORF10 | 8937 | 4.2 × 10−7 | 0.4 |
| 5 | pHTβ::Tn917-lac/55 | ORF10/ORF11 | 10425 | <2.0 × 10−7 | <0.2 |
| 6 | pHTβ::Tn917-lac/154 | ORF13 | 11171 | <2.1 × 10−7 | <0.2 |
| 7 | pHTβ::Tn917-lac/60 | ORF14 | 11798 | <1.8 × 10−7 | <0.2 |
| 8 | pHTβ::Tn917-lac/148 | ORF15 | 13221 | <1.5 × 10−7 | <0.2 |
| 9 | pHTβ::Tn917-lac/328 | ORF16 | 14123 | <1.5 × 10−7 | <0.2 |
| 10 | pHTβ::Tn917-lac/331 | ORF18/ORF19 | 15259 | <1.7 × 10−7 | <0.2 |
| 11 | pHTβ::Tn917-lac/57 | ORF20 | 16119 | <1.5 × 10−7 | <0.2 |
| 12 | pHTβ::Tn917-lac/274 | ORF23 | 23643 | 7.3 × 10−7 | 0.7 |
| 13 | pHTβ::Tn917-lac/230 | ORF27 | 25416 | <2.0 × 10−7 | <0.2 |
| 14 | pHTβ::Tn917-lac/321 | ORF29 | 29049 | <2.2 × 10−7 | <0.2 |
| 15 | pHTβ::Tn917-lac/142 | ORF30 | 29612 | <1.7 × 10−7 | <0.2 |
| 16 | pHTβ::Tn917-lac/323 | ORF31/ORF32 (oriT) | 30573 | <1.5 × 10−7 | <0.2 |
| 17 | pHTβ::Tn917-lac/275 | ORF32 | 30821 | <1.3 × 10−7 | <0.2 |
| 18 | pHTβ::Tn917-lac/278 | ORF33 | 31343 | <1.9 × 10−7 | <0.2 |
| 19 | pHTβ::Tn917-lac/95 | ORF34 | 32626 | <1.5 × 10−7 | <0.2 |
| 20 | pHTβ::Tn917-lac/250 | ORF48/49 | 42073 | 8.5 × 10−5 | 77 |
| 21 | pHTβ::Tn917-lac/203 | ORF50/51 | 43864 | 9.1 × 10−5 | 83 |
| 22 | pHTβ::Tn917-lac/22 | ORF51 | 44414 | 1.4 × 10−4 | 127 |
| 23 | pHTβ::Tn917-lac/309 | ORF52 | 45297 | 9.5 × 10−5 | 86 |
| 24 | pHTβ::Tn917-lac/304 | ORF54 | 46466 | 4.8 × 10−5 | 44 |
| 25 | pHTβ::Tn917-lac/82 | ORF56 | 47472 | <1.4 × 10−7 | <0.2 |
| 26 | pHTβ::Tn917-lac/10 | ORF58 | 49350 | 8.0 × 10−5 | 73 |
| 27 | pHTβ::Tn917-lac/19 | ORF60 | 51712 | 1.3 × 10−4 | 118 |
| 28 | pHTβ::Tn917-lac/210 | ORF61 | 52052 | 1.7 × 10−5 | 15 |
| 29 | pHTβ::Tn917-lac/17 | ORF1 (N terminal) | 52618 | 1.0 × 10−4 | 91 |
The mating time was 4 h. The mating experiments were performed between E. faecalis OG1RF and OG1SS when the transconjugants were obtained by solid surface mating.
Inserts that decreased in the transfer frequency of pHTβ were mapped within ORFs in three separate regions designated I, II, and III (Fig. 1). Region I could span a relatively large portion of the plasmid totaling 39.3 kb lying between 2.8 kbp and 42.1 kbp and contained 46 ORFs from ORF3 to ORF48. The precise borders of region I were not defined, since the insertions within the ORFs between ORF3 and ORF7 or between ORF35 and ORF48 could not be obtained (Fig. 1). Region II spanned a small portion of 1.7 kb between 47.0 kb and 48.7 kb and contained ORF56 and ORF57, and region III consisted of ORF61 located between 52.0 kb and 52.4 kb. Of the 22 ORFs on pHTβ which showed homology with the ORFs on the pXO2 plasmid of B. anthracis, 20 were in region I. Inserts in 11 of these 20 ORFs and insertion within the noncoding region upstream of ORF19 were obtained, and all of these inserts resulted in decreased transfer frequency.
Inserts in ORF56 of region II resulted in the inability to transfer in broth mating. ORF57 downstream of region II was almost identical to traA of pMG1, which is involved in the formation or stability of mating aggregate and is expressed in the early stage of mating (21, 36, 42). Although an insert in pHTβ ORF57 has not been isolated, insertion into traA of pMG1 resulted in the inability to transfer in broth mating (36). Insertion into ORF61 of region III resulted in a reduced transfer frequency. The predicted amino acid sequence of ORF60, which lies upstream of ORF61, was homologous with PrgN of the pheromone responsive plasmid pCF10, which is the protein involved in the negative regulation of expression of the mating aggregation substance, and insertion into ORF60 did not affect the transfer frequency (18).
We cannot exclude any potential polar effects on an adjacent gene or genes by transposon insertions in region I and region II. Research is now under way to determine the function of each ORF. Analysis of transferability and mapping of the insertion mutants implied that many ORFs in region I occupied a relatively large portion of pHTβ plasmid and could be related to transfer of the plasmid. The transfer-related ORFs in region I showed significant homology to ORFs on the pXO2 plasmid of B. anthracis. Regions I, II, and III were separated by Tn1546 or several ORFs where inserts did not affect the transfer frequency. The ORFs in region II and region III might be necessary for trans-regulating expression of the ORF(s) of region I.
Identification of the fragment containing the oriT region of pHTβ.
The transfer origin (oriT) is thought to be characteristic of the conjugative plasmid and essential for the transfer of the transferable or mobile element (11, 47). The oriT functions in cis to generate the single-stranded plasmid intermediate, after DNA relaxase cleaves a specific phosphodiester bond of the nic site. The known oriTs are classified into several groups based on sequence similarities (47). To identify the oriT region, which involves identification of the relaxase recognition sequence (i.e., direct repeat sequences) and oriT (site), the degree of homology between the DNA sequences determined for the pHT plasmid and the reported consensus sequences of the oriT region was analyzed. No sequence that was identical or similar to known oriT regions of gram-positive conjugative plasmids was found in the pHTβ sequences. It is characteristic of the oriT region that direct repeat sequences flank the oriT site and that the oriT sites are present within inverted repeat sequences. Thus, segments containing direct repeats (DR) and inverted repeats (IR) were selected as candidates for the oriT region. These candidates for the oriT region in the plasmid are indicated by DR1 and DR2 and from IR1 through IR5 in Fig. 1. Sequences which had similarities (more than 80% identity) to sequences near the oriT site (nic site) of the IncP, IncQ, or F plasmids (see Fig. 4) were also screened as candidates for the oriT site.
FIG. 4.
Comparison of the oriT consensus nucleotide sequences (nic region) of the representative mobile plasmids and the possible nick site of the pHTβ plasmid. The oriT region containing the possible nick site of the pHT plasmid found in the inverted repeat sequences is shown. The consensus oriT region found in the pheromone-responsive conjugative plasmids (PRP) and the previously reported consensus oriT sites found in mobile plasmids are shown. The consensus oriT sequences of IncP (MOBP), the pMV158 superfamily, the MOBQ family (R1162, etc.), and the F plasmid are indicated. The consensus double-stranded replication origin (dso) of RCR plasmids is also shown. The circular marks indicate the conserved nucleotides, centrally located TG site, and an A located four residues away. The black arrowheads indicate the nick sites determined in the oriT region.
DNA segments containing potential candidates for the oriT region, which were indicated by black horizontal bars marked from a to q in Fig. 1, were cloned into pAM401. Each plasmid clone (Cmr) was tested for its ability to be mobilized by the pHTβ plasmid (Vmr). Two of the clones, pAM401::IR5-1 and pAM401::IR2, were mobilized by the pHTβ plasmid (Table 4). The issue of the mobilization of pAM401::IR2 clone will be discussed later as a possible oriV candidate. The segment containing the IR5 region between ORF31 and ORF32 (g in Fig. 1), conferred the ability to transfer the pAM401::IR5-1 chimeric plasmid at a high frequency comparable to that of the pHT plasmid (Table 4). Of the transconjugants selected on the agar plates containing vancomycin, about 40% of the transconjugants were resistant to vancomycin, and 60% were resistant to both vancomycin and chloramphenicol. The vancomycin-resistant transconjugants contained only the pHTβ plasmid, and the vancomycin- and chloramphenicol-resistant transconjugants contained both the pHTβ plasmid and the pAM401::IR5-1 chimeric plasmid. Of the transconjugants selected on agar plates containing chloramphenicol, about 10% were resistant to chloramphenicol and 90% were resistant to chloramphenicol and vancomycin. The chloramphenicol-resistant transconjugants contained only the pAM401::IR5-1 chimeric plasmid; the chloramphenicol- and vancomycin-resistant transconjugants contained both pAM401::IR5-1 and the pHTβ plasmid. These results indicated that the chimeric plasmid pAM401::IR5-1 was mobilized in trans by the coresident pHTβ plasmid. None of the other fragments of pHTβ except fragment n containing IR2 and ORF58 could mobilize transfer, and transfer of pAM401::IR5-1 required the presence of pHTβ.
TABLE 4.
Mobilization frequencies of pAM401 derivatives (Cmr) carrying the various segments of pHTβ by coresident pHTβ plasmid (Vmr)
| Segment in Fig. 1 | Plasmids | 5′/3′ end of segment on map (bp) | Length (bp) | Region | Transfer frequency (no. of conjugants/donor; broth mating)
|
|
|---|---|---|---|---|---|---|
| Vmr | Cmr | |||||
| a | pHTβ/pAM401::ORF1 | 525/1300 | 776 | ORF1 internal region | 6.8 × 10−5 | <10−7 |
| b | pHTβ/pAM401::IR4 | 1657/3060 | 1,404 | IR4 and ORF2 | 7.7 × 10−6 | <10−7 |
| c | pHTβ/pAM401::IR3 | 5618/6116 | 499 | IR3 | 5.1 × 10−5 | <10−7 |
| d | pHTβ/pAM401::119/113 | 1300/12476 | 11,177 | IR4 to ORF13 | 8.0 × 10−5 | <10−7 |
| e | pHTβ/pAM401::ORF19/21 | 15213/18648 | 3,436 | ORF19, −20, and −21 | 4.6 × 10−5 | <10−7 |
| f | pHTβ/pAM401::119/96 | 20168/25254 | 5,087 | ORF23, −24, and −25 | 5.3 × 10−5 | <10−7 |
| g | pHTβ/pAM401::IR5-1 | 29918/31161 | 1,244 | IR5, ORF31 and −32 | 6.0 × 10−5 | 4.1 × 10−5 |
| h | pHTβ/pAM401::ORF39/40 | 36696/38127 | 1,432 | ORF39 and ORF40 | 5.7 × 10−5 | <10−7 |
| i | pHTβ/pAM401::IR1 | 39726/41121 | 1,396 | IR1, ORF44 to −47 | 6.0 × 10−5 | <10−7 |
| j | pHTβ/pAM401::DR1 | 43218/44620 | 1,403 | DR1 and ORF50 | 6.5 × 10−5 | <10−7 |
| k | pHTβ/pAM401::ORF51/52 | 44171/45708 | 1,538 | ORF51 and ORF52 | 4.3 × 10−5 | <10−7 |
| l | pHTβ/pAM401::ORF53/56 | 45810/47759 | 1,950 | ORF53 to ORF56 | 8.2 × 10−5 | <10−7 |
| m | pHTβ/pAM401::traA | 47762/48505 | 744 | traA internal region | 5.2 × 10−5 | <10−7 |
| n | pHTβ/pAM401::IR2 | 48482/50108 | 1,627 | IR2 and ORF58 | 9.1 × 10−6 | 3.4 × 10−6 |
| o | pHTβ/pAM401::ORF58 | 48943/50108 | 1,166 | ORF58 | 6.5 × 10−5 | <10−7 |
| p | pHTβ/pAM401::rep5′ | 49257/50108 | 852 | Upstream region of rep | 5.0 × 10−5 | <10−7 |
| q | pHTβ/pAM401::ORF60/61 | 51124/52626 | 1,503 | ORF60 and ORF61 | 7.5 × 10−5 | <10−7 |
Cloning and genetic analysis of oriT region of pHTβ plasmid.
IR5 contained direct repeats of 13 bp and two inverted repeats (Fig. 2 and 3). The direct repeats were composed of two copies of a 13-bp sequence and 7 bp that were part of the 13-bp sequence in the same orientation (i.e., ACTATGACCAAAA [DR-a], ACTATGACCAAAA [DR-b], and TATGACC [DR-c]). The inverted repeats were composed of short and long inverted repeats, AGTTGGC/GCCAACT (IR5S) and TAGCcACCTTCCT/AGGAAGGTGCTA (IR5L), respectively. Detailed analysis of this IR5 region was performed. Deletion mutants of the IR5-1 clone were constructed (Fig. 2). Deletion mutant IR5-9, which possessed a 192-bp fragment lying between 30,409 bp and 30,600 bp of the pHTβ map, was mobilized in trans by the coresident pHTβ plasmid and was the smallest fragment retaining the ability to transfer with a frequency equivalent to that of the pHTβ plasmid. This region contained one (DR-b) of the two copies of the 13-bp direct repeat sequences and the 7 bp (DR-c) of the 13-bp sequence, as well as two inverted repeat sequences (i.e., IR5S and IR5L).
FIG. 2.
Genetic analysis and determination of oriT region of pHT plasmids. The horizontal bars indicate the cloned PCR fragments of the pAM401 derivatives, and the bar numbers show the end positions of the segments. The transfer frequency of each of the pAM401 derivatives is shown in Table 5.
FIG. 3.
Nucleotide sequences of the oriT region of the pHT plasmid. The 420-bp noncoding DNA sequence region between ORF31 and ORF32 is shown. The horizontal arrows under the sequences indicate the direct repeats (DR-a, DR-b, and DR-c) and inverted repeats (IR5S and IR5L) in the oriT region. The names and locations of oligonucleotide primers used for the analysis of the oriT region are shown on the sequence with the angled arrows. Two downward arrowheads on the complementary sequence show the possible nick sites in the oriT region based on the sequence comparisons with other defined nick sites (see the text and Fig. 5). The vertical arrows that are numbered as 323 and 327 indicate the locations of the two Tn917-lac insertion mutants of pHTβ that abolished transfer ability. The putative promoter region (−35 and −10) and ribosome binding site (Shine-Dalgarno sequence [S.D.]) for ORF32 are shown upstream of the start codon. The asterisk marks indicate the dyad symmetric sequences overlapping the promoter region.
Deletion mutants containing only two inverted repeats, such as pAM401::IR5-3 and pAM401::IR5-10, were still mobilized at low frequencies by filter mating (Table 5). The pAM401::IR5-8 deletion mutant, which had a deletion of two inverted repeat sequences, was completely incapable of transfer, and the transfer frequency was less than 10−8 in filter mating. Two Tn917-lac mutants (numbered as 16 in Fig. 1 and 2) near the oriT region of pHTβ were obtained, and the insertion was mapped to 10 bp downstream of the long inverted repeats (IR5L) (Fig. 3). The mutants, pHTβ::Tn917-lac/323 and -327, could not transfer at all, even on a solid surface. These results indicated that the IR5 region could be the oriT region and that two inverted repeat sequences were essential for plasmid transfer. The sequence of the putative oriT region (IR5) showed no significant similarity to the reported oriT sequences.
TABLE 5.
Mobilization of pAM401 derivative plasmids by pHTβa
| Plasmid | Transfer frequency (no. of conjugants/donor)
|
|||
|---|---|---|---|---|
| Broth
|
Filter
|
|||
| Vmr | Cmr | Vmr | Cmr | |
| pAM401 | 1.2 × 10−4 | <10−7 | 4.7 × 10−2 | <10−8 |
| pAM401::IR5-1 | 6.0 × 10−5 | 4.1 × 10−5 | 5.3 × 10−2 | 1.1 × 10−1 |
| pAM401::IR5-2 | 8.0 × 10−4 | 3.8 × 10−5 | 3.2 × 10−2 | 1.0 × 10−1 |
| pAM401::IR5-3 | 1.1 × 10−4 | <10−7 | 2.5 × 10−2 | 3.2 × 10−4 |
| pAM401::IR5-4 | 1.1 × 10−4 | 5.1 × 10−5 | 4.0 × 10−2 | 1.3 × 10−1 |
| pAM401::IR5-5 | 1.4 × 10−4 | 6.9 × 10−5 | 2.5 × 10−2 | 1.2 × 10−1 |
| pAM401::IR5-6 | 9.0 × 10−5 | 3.0 × 10−5 | 4.4 × 10−2 | 1.1 × 10−1 |
| pAM401::IR5-7 | 1.6 × 10−4 | 6.7 × 10−5 | 3.1 × 10−2 | 1.2 × 10−1 |
| pAM401::IR5-8 | 7.0 × 10−5 | <10−7 | 4.8 × 10−2 | <10−8 |
| pAM401::IR5-9 | 1.4 × 10−4 | 4.2 × 10−5 | 3.0 × 10−2 | 1.2 × 10−1 |
| pAM401::IR5-10 | 1.8 × 10−4 | <10−7 | 2.7 × 10−2 | 2.5 × 10−4 |
The mating experiment was performed using E. faecalis UV202 as a donor carrying two plasmids and E. faecalis JH2SS as a recipient strain. The donor strains harbored both pHTβ (Vmr) as a mobilizer plasmid and each of the pAM401 derivatives (Cmr) containing various segments of the pHT plasmid as a tester plasmid. Each mating experiment was carried out in triplicate.
Five families of oriT core sequences have been defined through comparison of a wide range of transfer origins (Fig. 4) (9, 11, 47). There are conserved sequences within the core sequences of an oriT family. A closer inspection revealed a consensus sequence that is common even among the apparently phylogenetically remote oriT families (47). The most conserved sequence found among the families contains a centrally located TG or CG site, which is the nic site for the relaxase, and an A residue 4 bp away, which represents the highest level of conservation among the nucleotides in the nic sites of oriTs and dsos. IR5L of pHT contained a centrally located TG and an A residue 4 bp away (Fig. 4). It was possible that the nic site might be located within this region in the inverted repeat region in the pHTβ plasmid (Fig. 4).
A clone, pAM401::IR2, carrying the IR2 segment (segment n in Fig. 1) was also mobilized by the pHTβ plasmid at low frequencies, of around 10−6 per donor cell, which was about 10% of the transfer frequency of a plasmid containing the oriT region, by broth mating (Table 4). The donor strain UV202 was shown to carry the pHTβ plasmid and the cloned pAM401::IR2 plasmid by agarose gel electrophoresis of plasmid DNAs from the donor strains (data not shown). The transconjugants of JH2SS were resistant to both chloramphenicol and vancomycin and harbored one chimeric plasmid formed between pHTβ and the pAM401 derivative (data not shown), which could result from cointegration between pHTβ and pAM401::IR2. These data indicated that the mode of mobilization of pAM401::IR2 was different from that of pAM401::IR5-1. Analysis of the chimeric plasmid formed between a fragment of the F plasmid and pSC101 shows that chimeric plasmids that lack oriT but contain oriV1 of the F plasmid were mobilized in cis via cointegration with the coresident F plasmid at oriV1 in a RecA-independent recombination (23). The oriV region is essential for plasmid replication and is the start site for replication. The IR2 region of the pHTβ plasmid is located just upstream of ORF59, which is highly related to the rep genes of rolling circle replication (RCR)-type gram-positive plasmids. It was probable that IR2 was not a second oriT region but was the oriV region of the pHTβ plasmid (2, 10). Site-specific recombination could occur between the predicted oriVs of the pAM401::IR2 and pHTβ plasmids.
The putative DNA relaxase/nickase gene, ORF34.
In addition to the oriT sequence, the relaxase/nickase is an important feature of conjugative plasmids (11, 47) needed for the initiation of DNA transfer. None of the ORFs encoded on the pHT plasmid showed significant similarity to reported relaxase/nickase genes. Similarities between amino acid residues in relaxases encoded by different conjugative systems have been reported (30), and three common motifs are seen (11, 47). These suggest a shared DNA relaxation mechanism. Motif I contains the catalytic Tyr residue involved in DNA cleavage-joining activity. Motif II was reported to be involved in DNA-protein contacts through the 3′ end of the nick region, and a Ser residue is usually present. Motif III contains three conserved His residues and is known as the His3 motif. It has been suggested that the His residues aid the nucleophilic activity of the Tyr residue in motif I coordinate the required Mg2+ ions and direct activation of the active Tyr. These three motifs are thought to form part of the catalytic center of the relaxase. We examined the pHT plasmid for the presence of a relaxase by searching for the conserved motifs (residues) of the relaxase. ORF34 encoding 506 amino acid residues was found to contain the three conserved motifs I, II, and III, which contained Tyr, Ser, and three His residues, respectively, and could be the relaxase for the pHT plasmid (Fig. 5).
Three Tn917-lac insertion mutants of the pHTβ plasmid, pHTβ::Tn917-lac/95, -197, and -223, were obtained in the ORF34 gene, and each of the insertions was mapped. The transposons were inserted into residues Leu366, Gln440, and Val469 of the ORF34 protein, respectively. All the insertion mutants had completely lost the ability to transfer. Each phenotype of the mutant might result from the polar effects of insertion. To confirm whether ORF34 is essential for plasmid transfer, an in-frame deletion mutant of 10 amino acids (i.e., HRNTEHIHIH) from amino acid residues 200 to 209, which is located within motif III of ORF34, was constructed in the pHTβ plasmid as described in Materials and Methods (Fig. 5). The pHTβ plasmid derivative mutant with the defective ORF34 could not transfer in broth or on a filter (data not shown). These results indicated that ORF34 was the essential transfer-related gene.
A FASTA/BLAST homology search showed that sequences homologous to the motifs of the putative relaxase ORF34 were found in ORFs of other bacterial species for which function has not been determined (Fig. 5). Most of the ORFs homologous to ORF34 were found to contain two putative motifs I, designated as motifs Ia and Ib, in the N-terminal region, and a Tyr residue was conserved in each of the motifs (Fig. 5). Although until now there has been no genetic information or characterization of the genes homologous to ORF34, the new pHT family relaxase represented by the putative relaxase ORF34 could be widespread throughout a variety of bacteria. The conserved His3 sequence in motif III of the pHT family was designated as MOBMG: i.e., H(x2)T(x3)HxH(x4)E(x4)R.
Concluding remarks.
Sequence data for the pHT plasmid revealed that the pMG1-like plasmids had little similarity to well-characterized plasmids. The ORFs encoded on pXO2, a pathogenic capsule plasmid found in B. anthracis, shared sequence homologies with the pHT plasmids. Little is known about the transfer of pXO2 and about the function of each ORF, although there are reports about the mobilization of the plasmid by other highly conjugative plasmids found in Bacillus species (4, 33). Based on the genetic analysis by transposon insertional mutagenesis, a region containing ORFs from ORF3 to ORF48 that was designated as region I could be related to the transfer of the pHT plasmid: the other two regions, region II containing ORF56 and -57 and region III consisting of ORF61, were also necessary for efficient transfer.
Both oriT and nickase/relaxase are thought to be essential and important characteristic features of the conjugative plasmid (11, 47). The oriT region of the pHT plasmid was genetically determined, and the traI gene encoding the putative DNA relaxase/nickase resided in the transfer-related region I. The biochemical activity of the product has not yet been elucidated.
Supplementary Material
Acknowledgments
This work was supported by the grants from the Japanese Ministry of Education, Culture, Sport, Science and Technology [Tokuteiryoiki (C), Kiban (B), Kiban (C)], and the Japanese Ministry of Health, Labor and Welfare (H15-Shinko-9).
We thank Koichi Tanimoto and Maria V. Francia Gil for helpful advice. We also thank Elizabeth Kamei for revising the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 1997. The origin of transfer (oriT) of the Enterococcal, pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid 37:87-94. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battisti, L., B. D. Green, and C. B. Thorne. 1985. Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J. Bacteriol. 162:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell, D. B., and G. M. Dunny. 2002. Conjugation and genetic exchange in enterococci, p. 265-300. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology Washington, D.C.
- 6.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 7.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 10.Francia, M. V., W. Haas, R. Wirth, E. Samberger, A. Muscholl-Silberhorn, M. S. Gilmore, Y. Ike, K. E. Weaver, F. Y. An, and D. B. Clewell. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46:117-127. [DOI] [PubMed] [Google Scholar]
- 11.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 12.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto, S., H. Hashimoto, and Y. Ike. 1991. Low cost device for electrotransformation and its application to the highly efficient transformation of Escherichia coli and Enterococcus faecalis. Plasmid 26:131-135. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto, S., H. Tomita, E. Wakamatsu, K. Tanimoto, and Y. Ike. 1995. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J. Bacteriol. 177:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto, Y., K. Tanimoto, Y. Ozawa, T. Murata, and Y. Ike. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185:247-254. [DOI] [PubMed] [Google Scholar]
- 18.Hedberg, P. J., B. A. Leonard, R. E. Ruhfel, and G. M. Dunny. 1996. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid 35:46-57. [DOI] [PubMed] [Google Scholar]
- 19.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 8:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ike, Y., K. Tanimoto, H. Tomita, K. Takeuchi, and S. Fujimoto. 1998. Efficient transfer of the pheromone-independent Enterococcus faecium plasmid pMG1 (Gmr) (65.1 kilobases) to Enterococcus strains during broth mating. J. Bacteriol. 180:4886-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaworski, D. D., and D. B. Clewell. 1995. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J. Bacteriol. 177:6644-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilbane, J. J., and M. H. Malamy. 1980. F factor mobilization of non-conjugative chimeric plasmids in Escherichia coli: general mechanisms and a role for site-specific recA-independent recombination at oriV1. J. Mol. Biol. 143:73-93. [DOI] [PubMed] [Google Scholar]
- 24.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 26.Li, Z., H. Hiasa, U. Kumar, and R. J. Digate. 1997. The traE gene of plasmid RP4 encodes a homologue of Escherichia coli DNA topoisomerase III. J. Biol. Chem. 272:19582-19587. [DOI] [PubMed] [Google Scholar]
- 27.Makino, S.-I., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation of Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martone, W. J. 1998. Spread of vancomycin resistant enterococci: why did it happen in the United States? Infect. Control Hosp. Epidemiol. 19:539-545. [DOI] [PubMed] [Google Scholar]
- 29.Mikesell, P., B. E. Ivins, J. D. Ristroph, and T. M. Dreier. 1983. Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infect. Immun. 39:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pansegrau, W., W. Schroder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 31.Perkins, J. B., and P. J. Youngman. 1986. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 83:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 33.Reddy, A., L. Battisti, and C. B. Thorne. 1987. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J. Bacteriol. 169:5263-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Taminoto, K., and Y. Ike. 2002. Analysis of the conjugal transfer system of the pheromone-independent highly transferable Enterococcus plasmid pMG1: identification of tra gene (traA) up-regulated during conjugation. J. Bacteriol. 184:5800-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita, H., and D. B. Clewell. 2000. A pAD1-encoded small RNA molecule, mD, negatively regulates Enterococcus faecalis pheromone response by enhancing transcription termination. J. Bacteriol. 182:1062-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomita, H., C. Pierson, S. K. Lim, D. B. Clewell, and Y. Ike. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J. Clin. Microbiol. 40:3326-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomita, H., K. Tanimoto, S. Hayakawa, K. Morinaga, K. Ezaki, H. Oshima, and Y. Ike. 2003. Highly conjugative pMG1-like plasmids carrying Tn1546-like transposons that encode vancomycin resistance in Enterococcus faecium. J. Bacteriol. 185:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 44.Weaver, K. E., and D. B. Clewell. 1988. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J. Bacteriol. 170:4343-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Bacterial plasmids and gene spread Harwood Academic Publishers, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.