Abstract
Two-component signal transduction systems play an important role in the ability of bacteria to adapt to various environments by sensing changes in their habitat and by altering gene expression. In this study, we report a novel two-component system, YhcSR, in Staphylococcus aureus which is required for bacterial growth in vitro. We found that the down-regulation of yhcSR expression by induced yhcS antisense RNA can inhibit and terminate bacterial growth. Moreover, without complementary yhcS or yhcR, no viable yhcS or yhcR gene replacement mutant was recoverable. Collectively, these results demonstrated that the YhcSR regulatory system is indispensable for S. aureus growth in culture. Moreover, induced yhcS antisense RNA selectively increased bacterial susceptibility to phosphomycin. These data suggest that YhcSR probably modulates the expression of genes critical for bacterial survival and may be a potential target for the development of novel antibacterial agents.
Staphylococcus aureus is an important community- and hospital-acquired pathogen that causes superficial skin and life-threatening infections worldwide (25, 33). The pathogenesis of S. aureus is partially due to the coordinated regulation of virulence factors allowing the bacterium to evade the host immune system and/or to promote survival during infection. This organism has developed a series of two-component signal transduction systems (TCSs) in order to sense its immediate surroundings and to modulate specific cellular responses and the expression of virulence genes (14, 32). Therefore, TCSs are being explored as potential targets for new antimicrobials (2, 17, 28).
A typical TCS is composed of a membrane-associated histidine kinase, which acts as a sensor protein extending through the cytoplasmic membrane to monitor environmental changes and to activate a response regulator existing in the cytoplasm modulating gene expression (15, 34). The well-studied TCS Agr is a positive regulator of exoproteins, including proteases, hemolysins, and toxins (32). Additionally, the TCS Agr is a repressor of the transcription of protein A, coagulase, and some adhesins in late-exponential-phase growth in vitro (32). Other two-component systems, such as saeRS, arlRS, and srrAB, also influence the expression of some virulence factors (1, 13, 11, 35, 39). Another system, LytSR, controls bacterial autolysis by positively regulating the transcription of genes responsible for the synthesis and transport of cell wall murein hydrolase (4). The vraSR loci are homologous to the yvqEC loci of B. subtilis and positively modulate cell wall biosynthesis in S. aureus (23).
The yycFG system, which has orthologs in Bacillus subtilis (7, 8) and Streptococcus pneumoniae (36), is the only known regulatory system essential for cell viability in S. aureus (26). It was reported that in B. subtilis the YycFG system controls the ftsAZ operon, which is involved in the process of cell wall division (12). However, there is no such evidence in S. aureus and S. pneumoniae, while it has been reported that YycF regulates SsaA and IsaA antigens in S. aureus (6) and cell wall synthesis in S. pneumoniae (29-31) and other essential genes (16). In this study, we report another essential two-component signal transduction system, YhcSR, in S. aureus, which may be involved in the modulation of genes critical for bacterial growth.
Construction and characterization of a TetR-regulated yhcS antisense RNA isogenic strain.
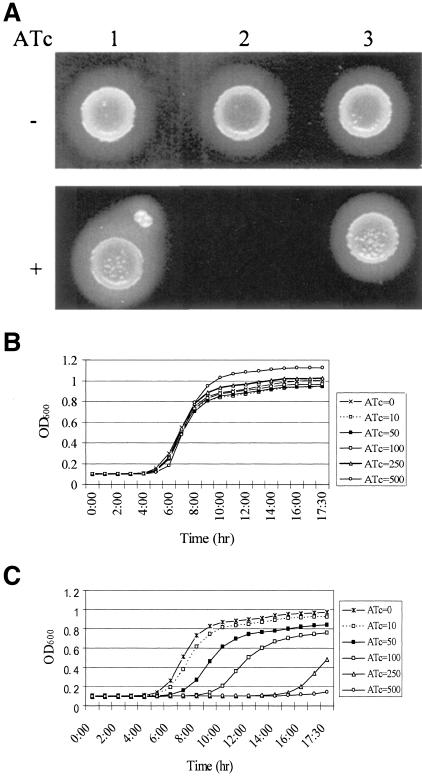
TetR-regulated antisense RNA can effectively down-regulate gene expression and has been successfully used to identify genes essential for bacterial survival in S. aureus (18, 20, 21). To examine the essentiality of the YhcSR regulatory system, a TetR-regulated yhcS (sensor) antisense strain was constructed in the clinical human S. aureus isolate WCUH29 and was denoted JSAS909. Briefly, the oligonucleotide primers YhcSfor and YhcSrev (see Table S1 in the supplemental material) were used to amplify a 353-bp yhcS fragment from S. aureus genomic DNA. The resulting DNA fragments were cloned into pYH3 DNA in an antisense orientation (40). The recombinant DNA, pJYJ909, was electroporated into RN4220 first and then was introduced into the wild-type human isolate WCUH29 and selected with erythromycin (5 μg/ml) (18, 19). The electrotransformants were confirmed by PCR and were denoted RN4220/yhcS-as and JSAS909 (WCUH29/pJYJ909). The phenotypes of the yhcS antisense strain and control strains were observed on a blood agar plate in the presence or absence of an inducer (anhydrotetracycline [ATc]) of antisense RNA. The control parent vector and a control unrelated antisense RNA (a gene encoding a nonessential hypothetical protein) grew in the presence of the inducer (Fig. 1A). In contrast, the yhcS antisense strain JSAS909 was unable to grow on the blood agar plate in the presence of the inducer. This result demonstrates that induced yhcS antisense RNA causes a lethal effect on bacterial growth.
FIG. 1.
(A) Phenotype of the S. aureus yhcS antisense strain on tryptic soy agar (TSA) plates during ATc induction. Overnight cultures of S. aureus strains were diluted and plated onto TSA-erythromycin plates with or without the inducer ATc (1 μg/ml) and incubated overnight at 37°C. Lane 1, WCUH29/pYH3 control; lane 2, yhcS antisense strain JSAS909; lane 3, unrelated antisense control. Also shown are growth curves of control strain WCUH29/pYH3 (B) and JSAS909 (C) in TSB containing 5 μg/ml of erythromycin and various concentrations of ATc (in nanograms/milliliter). The cultures were diluted to ∼104 CFU/ml with TSB containing appropriate antibiotics and different concentrations of an inducer (anhydrotetracycline [ATc], at concentrations of 0, 10, 50, 100, 250, or 500 ng/ml). Cell growth was monitored at 37°C by measuring the optical density at 600 nm (OD600) every 15 min, with 1 min of mixing before each reading using a SpectraMax plus Spectrophotometer (Molecular Devices). The grow curves represent one of three repeated experiments.
To quantitatively define the importance of yhcS, the inhibition of growth was titrated in a liquid tryptic soy broth (TSB) medium containing different concentrations of inducer. Growth curves were measured kinetically using a 96-well plate format. No significant inhibition was observed for the control strain (Fig. 1B). The yhcS antisense expression strain, JSAS909, showed inhibition of growth in a dose-dependent manner (Fig. 1C). The above experimental data indicate that yhcSR may function as a repressor or as a positive regulator to modulate the expression of genes required for staphylococcal growth.
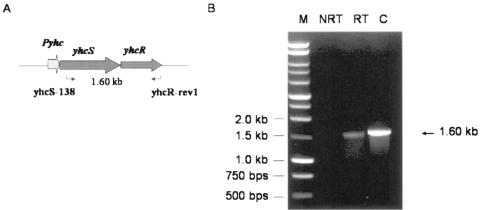
DNA sequence analyses of yhcSR indicated that yhcS and yhcR are located in the same operon and might be cotranscribed from a common promoter (Fig. 2A). To examine this possibility, reverse transcription-PCR (RT-PCR) was performed using a forward primer specifically binding to yhcS and a reverse primer specifically binding to yhcR (see Table S1 in the supplemental material). No PCR product was yielded from the negative control using total RNA as templates (Fig. 2B, lane NRT). In contrast, using the total cDNA of S. aureus as templates, a 1.6-kb PCR product was yielded at the expected size (Fig. 2B). This result indicated that both yhcS and yhcR are cotranscribed from a common promoter. Therefore, the induced yhcS antisense RNA may have an impact on yhcR expression.
FIG. 2.
Cotranscription of yhcR and yhcS genes by RT-PCR. Total RNA was extracted using the Promega mini kit from collected S. aureus cells after the appropriate cell density was reached (optical density at 600 nm, ∼0.4). RNA samples were further treated with DNase I to remove the residual genomic DNA. The cDNA was synthesized by employing RNA as a template using a Promega reverse transcription kit. Cotranscription of the yhcR and yhcS genes was demonstrated by RT-PCR using the yhcS138 and YhcRrev1 primers (Table 1). (A) Schematic representation of yhcSR genes in S. aureus. NRT, total RNA of S. aureus used as the template; RT, synthesized cDNA used as the template; C, positive control, the genomic DNA used as the template.
In order to investigate whether the inhibition of bacterial growth is caused by the downregulation of yhcR expression with yhcS antisense RNA, we characterized the production of transcripts of yhcS antisense RNA in response to ATc induction and determined the effects of this induction on YhcR expression. First, we examined the expression of yhcS antisense RNA in the TetR-regulated yhcS antisense strain before and after induction using a Northern blot assay. The results showed that the yhcS antisense strain produced much more yhcS antisense RNA product in response to ATc induction than without induction (data not shown).
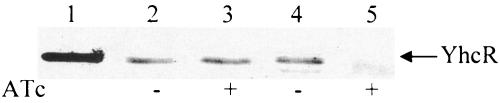
Second, we examined the effect of the induced yhcS antisense RNA on the level of the endogenous YhcR protein. The YhcR-his tag fusion protein was purified from Escherichia coli and confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown) and was used to immunize the mice for induction of YhcR antibodies, which was used for immunoblotting YhcR. The results showed that the cytoplasm fractions of JSAS909 without induction of yhcS antisense RNA reacted strongly with the anti-YhcR antisera (Fig. 3). Following an induction, the level of yhcR expression significantly decreased to an approximately 12-fold lower level of reactivity, as measured by densitometer scanning. In contrast, the reactivity of whole-cell proteins of the control strain, WCUH29/pYH3, against anti-YhcR antisera was not affected by ATc induction. This result indicated that induced yhcS antisense RNA effectively downregulated yhcR expression and that the yhcSR regulon is critical for bacterial growth. We do not know why previous studies (10, 21) did not reveal that the yhcSR regulon is critical for bacterial growth. One possible explanation is that our random library has not been exhausted for screening all open reading frame antisense RNAs. Alternatively, our results might be due to the different efficacies of different antisense RNA expression systems or to the fact that the copy number of target genes in normal cells may have an influence on a cell's sensitivity to the induced antisense RNA (data not shown).
FIG. 3.
Western blot analysis of yhcR expression during induction of yhcS antisense RNA. S. aureus strains were incubated in TSB with appropriate antibiotics in the presence of an inducer (anhydrotetracycline [ATc] or IPTG) at 37°C overnight. Cell pellets from 1 ml of culture at an optical density of 600 nm of 1.0 were collected by centrifugation and washed once with 1 ml of phosphate-buffered saline. To enrich the YhcR protein, cytoplasm fractions were prepared as described previously (5). Equal amounts of proteins samples were loaded onto a sodium dodecyl sulfate-polyacrylamide gel, and Western immunoblotting was performed as described previously (18). Proteins from S. aureus whole-cell lysis probed with mouse anti-YhcR antibodies are shown. Lane 1, standard YhcR; lanes 2 and 3, WCUH29/pYH3; lanes 4 and 5, JSAS909.
Construction and characterization of Pspac-regulated yhcS and yhcR knockout mutants.
To further determine the essentiality of yhcSR, both Pspac-regulated yhcS and yhcR conditional mutants were created in S. aureus as described previously (9, 38). Briefly, the integration vectors pFF71-yhcS and pFF71-yhcR were created and transformed into strain RN4220 prior to knocking out yhcS and yhcR, respectively (data not shown). The plasmid DNA of pFF71-yhcS or pFF71-yhcR was inserted into the S. aureus genome via site-specific integration (geh locus, encoding lipase). This integration is a high-efficiency process mediated by the attP/attB integration sites and by an integrase encoded by the int gene (24). The transcription of the yhcS or yhcR gene is controlled by the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspac. In the presence of IPTG, both the yhcS deletion strain SASJ104 and the yhcR deletion strain SASJ204 were obtained after integrating pJS104 and pJS204 vectors into the genome, respectively, and subsequently undergoing a screening procedure (see Fig. S1A and S2A in the supplemental material). To confirm the construction of the Pspac-regulated yhcS and yhcR mutants (SASJ104 and SASJ204), Southern blot and PCR analyses were performed. The results showed that endogenous yhcS or yhcR was deleted in SASJ104 or SASJ204, respectively, compared to sequences of the control strains (see Fig. S1B and S2B in the supplemental material). To ensure a sufficient repressor level, the Lacl expression plasmid, pFF40 carrying the kanamycin resistance gene (9), was also introduced into the SASJ104 and SAS204 strains.
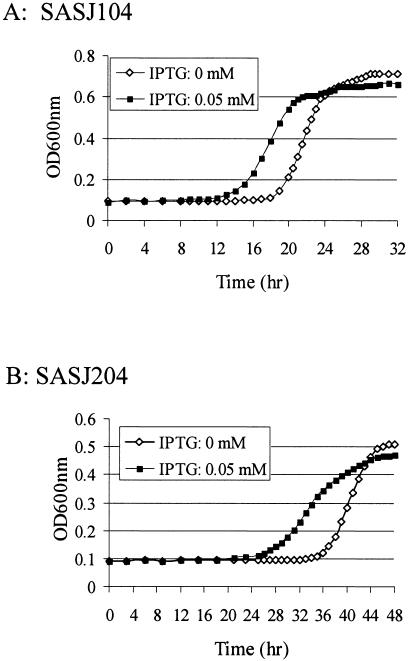
In order to determine the effect of the conditional mutations of yhcS and yhcR on growth, the bacterial cell densities were measured at an optical density of 600 nm during intervals of incubation in TSB with different IPTG concentrations. The results showed that regardless of the presence of IPTG, both Pspac-regulated yhcS and yhcR mutants showed a long lag phase of growth, which may result from the general effect of antibiotics in the medium (Fig. 4). However, it was observed that the growth of both Pspac-regulated yhcS and yhcR mutant strains reached log phase much earlier with IPTG than those without IPTG, indicating a dependency on yhcSR gene products (Fig. 4). Surprisingly, the differences of the growth of cultures disappeared after the mid-log phase of growth (Fig. 4). In addition, it has been observed that there are no significant changes of phenotypes and CFU of cells grown in medium with or without IPTG (data not shown). To explore the reason why the growth of the Pspac-regulated yhcS or yhcR mutant is not dependent on IPTG induction after early log phase, we examined whether there is leaky transcription of yhcR or yhcS without IPTG by using real-time RT-PCR. The results showed that without IPTG the leaky level of yhcS expression in SASJ104 or yhcR expression in SAS204 was almost the same level as that in wild-type cells, while the transcriptional levels of both yhcS and yhcR were significantly increased with the induction of IPTG (data not shown). These results indicated that neither SASJ104 nor SASJ204 was a tightly controlled mutant.
FIG. 4.
Growth curves of the Pspac-regulated yhcS (A) and yhcR (B) mutants during the induction of yhcS or yhcR expression with IPTG. S. aureus strains were incubated at 37°C overnight in TSB with appropriate antibiotics. The cultures were diluted to ∼104 CFU/ml with TSB containing appropriate antibiotics and different concentrations of inducer (IPTG at concentrations of 0, 0.2, 50, or 4,000 μM). Cell growth was monitored at 37°C by measuring the optical density at 600 nm (OD600nm) every 15 min, with 1 min of mixing before each reading using a SpectraMax plus Spectrophotometer (Molecular Devices). The figure represents one of three repeated experiments.
It has been recently reported that insertional mutation of either yhcS or yhcR has no effect on the viability of S. aureus (37). This contradictory result may be due to the functional untruncated proteins generated from the insertional knockout mutants. To investigate this possibility, the allelic gene replacement of yhcS or yhcR in S. aureus RN4220 was performed using the strategy described above. However, no recoverable colony was obtained without IPTG-inducible complementary yhcS or yhcR, while many wild-type revertants were recovered (27/∼8,000 CFU). Collectively, the above results demonstrated that the YhcSR regulon is required for bacterial survival in vitro.
The effect of down-regulation of yhcSR expression on bacterial susceptibility to different stresses.
To explore the reason why the yhcSR regulon is required for bacterial survival, we examined the cell's susceptibility to various stresses, including pH, temperature, nutrition, ions, osmolarity, and different antibacterial agents. The results showed that the stress of pH (5.2), nutrition (diluted TSB), temperature (42°C), different ions (NO2− and NO3−), or high concentrations of sugars or NaCl had no specific impact on the growth of yhcS antisense strains (data not shown). Moreover, the results in Table 1 show that there were no significant changes in the MICs for the control strain. In contrast, the yhcS antisense-expressing strain was selectively hypersensitive to phosphomycin (the MIC decreased eightfold during the induction of yhcS antisense RNA), an inhibitor of UDP-N-glucosamine 1-carboxyvinyltransferase (MurA), which is involved in the first step in murein biosynthesis of a cell wall biosynthesis pathway (3). No significant changes were observed for the other cell wall biosynthesis inhibitors, vancomycin and bacitracin, although yhcYZ of B. subtilis was shown to be induced by bacitracin (27). It has been reported that the vraSR null mutant becomes hypersensitive to different cell wall synthesis inhibitors, including vancomycin, bacitracin, and phosphomycin (23). The yhcSR and vraSR loci are homologous to the yhcYZ and yvqEC loci of B. subtilis, respectively, and YhcYZ and YvqEC may interact with each other in B. subtilis (22). Therefore, YhcSR and VraSR may cross-talk and directly or indirectly control some overlapping genes involved in a cell wall synthesis pathway.
TABLE 1.
MICs (in micrograms/milliliter) of compounds during induction of yhcS antisense with or without ATc
| Compound and activity | WCUH29/pYH3
|
JSAS909
|
||
|---|---|---|---|---|
| Without ATc | With ATc (250 ng/ml) | Without ATc | With ATc (250 ng/ml) | |
| DNA synthesis | ||||
| Quinacrine 2 HCl | >64 | >64 | >64 | >64 |
| Novobiocin | 0.125 | 0.125 | 0.125 | 0.125 |
| RNA transcription | ||||
| Rifampin | 0.06 | 0.06 | 0.06 | 0.06 |
| Translation | ||||
| Chloramphenicol | 8 | 8 | 8 | 8 |
| Kanamycin | 64 | 64 | 64 | 64 |
| Erythromycin | >64 | >64 | >64 | >64 |
| Cell wall biosynthesis | ||||
| Phospomycin | 8 | 8 | 8 | 1 |
| Vancomycin | 0.5 | 1 | 1 | 0.5 |
| Bacitracin | >64 | >64 | >64 | >64 |
| Polymyxin B | >64 | >64 | >64 | >64 |
| Lipid biosynthesis | ||||
| Cerulenin | 64 | 64 | 64 | 64 |
| Electron transporter | ||||
| Oligomycin | >64 | >64 | >64 | >64 |
Conclusion.
YhcSR is a novel and essential two-component signal transduction system in S. aureus. Down-regulation of yhcS selectively increases its susceptibility to the cell-wall synthesis inhibitor phosphomycin. It is likely that yhcSR functions as a positive regulator and/or repressor to control expression of genes required for staphylococcal growth. The discovery of target genes regulated by yhcSR using a comprehensive microarray approach is ongoing.
Supplementary Material
Acknowledgments
We are very grateful to Martin Rosenberg for many helpful discussions and Mark Rutherford and Doug Weiss for critical reading of the manuscript.
This project was supported by grant AI057451 from the National Institute of Allergy and Infectious Disease. This work was also supported in part by AHC Faculty Research Development Grant #03-02 at the University of Minnesota.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abelnour, A., S. Arvidson, T. Bremell, C. Ryden, and A. Tarkowski. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, J. F., and J. A. Hoch. 1998. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob. Agents Chemother. 42:1529-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burts, M. L., W. Williams, K. DeBord, and D. Missiakas. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. USA 102:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis—implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, F., R. D. Lunsford, D. Sylvester, J. Fan, H. Celesnik, S. Iordanescu, M. Rosenberg, and D. McDevitt. 2001. Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 46:71-75. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth, R. A., R. J. Hselbeck, K. L. Ohlesn, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 11.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 12.Fukuchi, K., Y., Kaahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 13.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 14.Gross, R. 1993. Signal transduction and virulence regulation in human and animal pathogens. FEMS Microbiol. 10:301-326. [DOI] [PubMed] [Google Scholar]
- 15.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 16.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 17.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 176:2027-2030. [DOI] [PubMed] [Google Scholar]
- 18.Ji, Y., D. Yin, B. Fox, D. J. Holmes, D. Payne, and M. Rosenberg. 2004. Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231:177-184. [DOI] [PubMed] [Google Scholar]
- 19.Ji, Y., G. Woodnutt, M. Rosenberg, and M. K. R. Burnham. 2002. Identification of essential genes in Staphylococcus aureus using inducible antisense RNA. Methods Enzymol. 358:123-128. [DOI] [PubMed] [Google Scholar]
- 20.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated Antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji, Y., B. Zhang, S. F. Van Horn, P. Warren, G. Woodnutt, M. K. R. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 19:807-821. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of a single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 25.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascher, T., N. Margulis, T. Wang, R. Ye, and J. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 28.Matsushita, M., and K. D. Janda. 2002. Histidine kinase as targets for new antimicrobial agents. Bioorg. Med. Chem. 10:855-867. [DOI] [PubMed] [Google Scholar]
- 29.Mohedano, M., K. Overweg, A. Fuente, M. Reuter, S. Altabe, F. Mulholland, D. Mendoza, P. López, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng, W., G. Robertson, K. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 31.Ng, W., M. Krystyna, K. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 32.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 33.Steinberg, J. P., C. C. Clark, and B. O. Hackman. 1996. Nosocomial and community-acquired Staphylococcus aureus bacteremias from 1980 to 1993: impact of intravascular devices and methicillin resistance. Clin. Infect. Dis. 23:255-259. [DOI] [PubMed] [Google Scholar]
- 34.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 35.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. R. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392-10401. [DOI] [PubMed] [Google Scholar]
- 36.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 37.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Débarbouillé, J. R. Penadés, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia, M., M. R. D. Lunsford, D. McDevitt, and S. Iordanescu. 1999. Rapid method for the identification of essential genes in Staphylococcus aureus. Plasmid 42:144-149. [DOI] [PubMed] [Google Scholar]
- 39.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin, D., B. Fox, M. Lonetto, M. Etherton, D. Payne, D. Holmes, M. Rosenberg, and Y. Ji. 2004. Identification of antimicrobial targets using a comprehensive genomic approach. Pharmacogenomics 5:101-113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.