Abstract
The general stress regulon of Bacillus subtilis is induced by activation of the σB transcription factor. σB activation occurs when one of two phosphatases responds to physical or nutritional stress to activate a positive σB regulator by dephosphorylation. The signal that triggers the nutritional stress phosphatase (RsbP) is unknown; however, RsbP activation occurs under culture conditions (glucose/phosphate starvation, azide or decoyinine treatment) that reduce the cell's levels of ATP and/or GTP. Variances in nucleotide levels in these instances may be coincidental rather than causal. RsbP carries a domain (PAS) that in some regulatory systems can respond directly to changes in electron transport, proton motive force, or redox potential, changes that typically precede shifts in high-energy nucleotide levels. The current work uses Bacillus subtilis with mutations in the oxidative phosphorylation and purine nucleotide biosynthetic pathways in conjunction with metabolic inhibitors to better define the inducing signal for RsbP activation. The data argue that a drop in ATP, rather than changes in GTP, proton motive force, or redox state, is the key to triggering σB activation.
The σB transcription factor controls the general stress regulon of Bacillus subtilis, a collection of over 200 genes whose products confer multiple stress resistances on the bacterium (14, 21, 22, 32). Induction of the σB regulon occurs by the activation of σB itself, a process that is triggered by exposure of B. subtilis to either nutritional (glucose/phosphate limitation) or physical (heat, acid, salt shock) stress (3, 5, 7, 35).
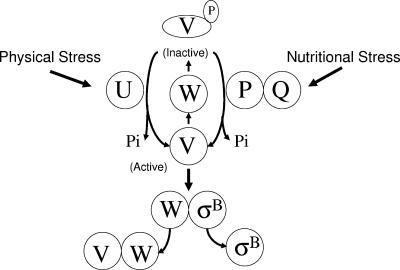
As illustrated in Fig. 1, σB is present but inactive in unstressed bacteria due to an association with an anti-σB protein (RsbW) (3, 4, 6). σB is released from RsbW when an additional protein (RsbV) binds to RsbW in place of σB (11, 12). The availability of active RsbV controls the amount of σB that is released from RsbW, with the phosphorylation state of RsbV determining its activity (12). RsbW is both an RsbV/σB binding protein and an RsbV-specific protein kinase. In the absence of stress, RsbV is phosphorylated and inactivated by the RsbW kinase (12). Exposure to stress triggers one of two stress-specific phosphatases (RsbU and RsbP) to dephosphorylate and reactivate RsbV (16, 31, 33, 34G65G). The RsbU phosphatase responds to physical stress, while RsbP is activated by nutritional stress (1, 16, 31, 35, 38). Although a number of environmental conditions that activate σB's nutritional or physical stress phosphatases have been described, the internal signals that specifically trigger their activation are unknown (28, 35).
FIG. 1.
Model of σB activation. σB is held inactive in unstressed B. subtilis as a complex with the anti-σB protein RsbW (W). σB is freed from RsbW when a release factor RsbV (V) binds to RsbW. RsbV is inactive in unstressed B. subtilis due to an RsbW-catalyzed phosphorylation (V-P). RsbV is reactivated by either of two stress-responsive phosphatases (RsbU and RsbP). Physical stress is transmitted through a series of additional proteins to the RsbV-P phosphatase, RsbU (U). Nutritional stress triggers a separate pathway in which an alternative RsbV-P phosphatase, RsbP (P) and an associated protein, RsbQ (Q) required for RsbP's activity are activated. Either phosphatase is sufficient to reactivate RsbV and allow the release of σB. The model is based on references 1, 2, 3, 4, 6, 8, 10, 12, 16, 30, 33, 34, 37, and 38.
The gene for the nutritional stress phosphatase (rsbP) is the second gene in a bicistronic operon whose promoter-proximal gene (rsbQ) encodes a product that is necessary for RsbP's activity (31). The loss of either RsbP or RsbQ prevents σB activation following carbon starvation (31). Based on results using the yeast two-hybrid system, RsbP and RsbQ appear to interact directly with each other (31). Both proteins contain recognizable domains. Determinants of the α/β hydrolase protein superfamily, including the hydrolase's catalytic triad residues, are present in RsbQ (8). In RsbQ, these residues are Ser 96, Asp 219, and His 247. Substitution of either Ser 96 or His 247 by Ala creates RsbQ variants that prevent σB activation during nutritional stress in the B. subtilis strains that carry them (8). This result has been interpreted as evidence that the catalytic function of RsbQ is needed for activation of the RsbP phosphatase.
The recently determined crystal structures of RsbQ confirm RsbQ's membership in the α/β hydrolase superfamily of proteins and reveal that its catalytic triad is buried within RsbQ. This is a site that a large molecule such as RsbP would have difficulty entering (15). The position of the catalytic triad prompted the suggestion that the substrate for RsbQ's catalytic activity is not RsbP, but rather a small hydrophobic molecule that might eventually be transferred to RsbP (15). RsbP, consistent with its role as an RsbV-P phosphatase, contains a PP2C serine phosphatase domain (31). In addition, RsbP also carries a PAS domain at its amino terminus (31). PAS domains are signaling modules that typically respond to changes in light, redox potential, oxygen, small ligands, or the overall energy level of the cell (30). Presumably, this domain on RsbP participates in sensing a nutritional stress signal that induces RsbP phosphatase activity. It is not known if the signal detected by the RsbP is generated by the catalytic activity of RsbQ or if each protein receives a separate nutritional input that is integrated in a RsbP/Q complex to generate the active phosphatase.
In previous work, we and others noted that a number of environmental conditions that induced the nutritional stress pathway shared the characteristic of likely causing a reduction in intracellular ATP levels (28, 35). This raised the possibility that changes in ATP levels might be a trigger for the nutritional stress pathway. Although changes in ATP levels are a plausible signal for the nutritional activation of σB, the observed changes might be coincidental rather than causal. Decreases in electron transport, proton motive force, and redox potential typically precede variances in ATP (30). Inasmuch as some PAS domains can respond to such changes, it is conceivable that shifts in redox rather than ATP could be RsbP's inducing signal. Additionally, if nucleotide levels and not redox state are the RsbP trigger, it is also possible that GTP rather than ATP is the nucleotide to which the nutritional stress pathway responds. Both decoyinine and mycophenolic acid, inhibitors of the GTP biosynthetic pathway, have been shown to trigger σB activation (28).
In the current work, we use mutant B. subtilis and metabolic inhibitors to revisit the question of nutritional stress induction of σB and present evidence that a decline in ATP rather than changes in GTP or redox state is the key to triggering σB activation.
MATERIALS AND METHODS
Bacterial strains.
All of the B. subtilis strains used in this study are derivatives of PY22. BSA46 carries a specialized SPβ prophage encoding a translational fusion of the σB-dependent ctc to the Escherichia coli lacZ gene (SPβ ctc::lacZ) (3). This fusion allows β-galactosidase activity to be monitored as a measure of σB activity. BSZ7 (SPβ ctc::lacZ relA::pUK-RE1) is BSA46 made RelA− by transformation with an integrating plasmid (40). BSZ131 (SPβ ctc::lacZ Δatp FHAGDC::kanr) is BSA46 transformed with chromosomal DNA from 168Δatp2 (25).
The purA null mutation in BSZ116 (SPβ ctc::lacZ ΔpurA::kanr) was created by PCR amplification of an internal (nucleotides 306 to 1290) fragment of the 1,290-bp purA gene and its cloning into pUC18 as a SacI/SalI fragment. This was followed by replacing the region from nucleotides 511 to 972 with the kanr cassette from pDG780 (13). The resulting plasmid was linearized and transformed into BSA46, allowing the purA::kanr element to replace purA in the bacterial chromosome. Kanr clones were then screened for adenine auxotrophy on a minimal medium with or without 1 mM adenine. Construction of the guaB mutation in BSZ130 (SPβ ctc::lacZ ΔguaB::erm) involved PCR amplification and cloning of a segment (nucleotides 35 to 1423) of the 1,464-nucleotide guaB gene as an EcoRI/SalI fragment into pUC18. Nucleotides 749 to 1347 of this DNA were then replaced by a BamHI/ClaI ermr cassette from pDG646 (13). The resulting plasmid was linearized, transformed into PY22, and screened for clones requiring guanine (1 mM) for growth. SPβ ctc::lacZ (ermr, cmr) was then introduced into this strain by transformation with chromosomal DNA from BSA46.
ATP measurement.
ATP levels were measured using ATP bioluminescence assay kit CLS II (Roche Molecular Biochemicals, Palo Alto, CA). Light emission was read using the LUMAT model LB9507 (Berthold Technologies, Oak Ridge, TN). Extraction of ATP from cultures was as described by Lundin and Thore (19) using the formate-EDTA lysis buffer. Briefly, 0.5 ml of culture was added to 0.6 ml of ice-cold formate solution (0.38 M formic acid/17 mM EDTA) and rocked in ice for 15 min; 195 μl 2 N KOH was then added to the mixture to neutralize the pH. Debris was pelleted and the supernatant diluted 50-fold with an assay buffer (100 mM Tris, pH 7.75, 4 mM EDTA); 100 μl of diluted supernatant was then transferred to a clear plastic tube for ATP quantitation using the ATP bioluminescence assay reagent and LUMAT instrumentation. The LUMAT was set to inject 100 μl luciferase reagent into the tube in each assay. The light output was compared to a standard curve prepared from known amounts of ATP. The ATP concentration in each figure is given as μM ATP divided by the optical density at 540 nm of the sample.
Culture conditions.
Strains were normally grown in LB (24) and treated with azide (2 mM) or mycophenolic acid (20 μg/ml) in this medium when the culture had reached a density of 0.2 (optical density at 540 nm).
Phosphate limitation studies were performed in a synthetic medium (28) supplemented with 0.1% Casamino Acids, 0.2% glucose and either 0.6 mM or 0.15 mM PO4 as the high PO4 and PO4-limiting condition, respectively. To test the effects of adenine or guanine starvation, auxotrophic strains were grown with shaking at 37°C to an optical density of 0.2 in the abovementioned synthetic medium with 0.6 mM PO4 and either 1 mM adenine or guanosine. Cells were pelleted at room temperature, washed once with warmed (37°C) medium without adenine or guanosine and resuspended in the original volume of synthetic medium with or without adenine or guanosine. The cultures were then shaken at 37°C and samples were withdrawn at intervals for analyses of σB-dependent β-galactosidase and ATP levels.
B. subtilis treated with carboxyl cyanide m-chlorophenylhydrazone (CCCP) was grown in a Tris-based medium (0.05 M Tris pH 7.5, 0.14 mM sodium citrate, 2.5 mM K2HPO4, 0.15 mM FeCl3, 0.8 mM MgSO4, 40 μg/ml Trp, 0.004% Casamino Acids) supplemented with either 0.2% glutamine plus 0.5% glucose or 0.2% glutamate plus 0.2% succinate. CCCP (1 μM) was added to a portion of each culture at a cell density of 0.3 (optical density at 540 nm) and samples were withdrawn at intervals for β-galactosidase and ATP measurements. All of the data presented in the figures represent typical results of experiments repeated three times.
General methods.
B. subtilis transformation was carried out as described by Yasbin et al. (39). β-Galactosidase assays were performed on chloroform-permeabilized cells as described by Kenny and Moran (17).
RESULTS AND DISCUSSION
ATP decline precedes σB induction.
An indication of whether a decline in ATP or an event that precedes the ATP decline could signal σB activation was sought in an examination of the timing of ATP decline relative to σB induction. If changes in ATP levels are a trigger for σB induction, the ATP decline should precede σB induction. In contrast, if both are the result of an earlier trigger (e.g., a change in redox state) both might change in unison or σB induction may even precede the ATP drop.
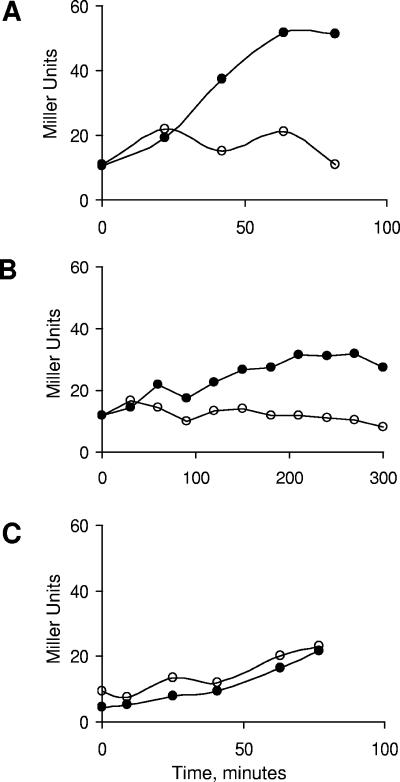
To examine the timing of σB activation relative to the ATP decline, B. subtilis with a σB-dependent reporter gene (Pctc::lacZ) was grown and allowed to enter stationary phase in PO4-limited medium. PO4 starvation is a strong inducer of σB's nutritional stress pathway (35). As illustrated in Fig. 2, there is a rapid decline in ATP levels coincident with the cessation of growth in the PO4-limited culture. This drop in ATP precedes the induction of σB by approximately 20 min. A parallel culture grown in a high-PO4 medium also induces σB, but this induction is delayed relative to the culture's entry into stationary phase (Fig. 2).
FIG. 2.

σB Activation by PO4 limitation. Wild-type B. subtilis was grown and allowed to enter stationary phase in a defined, PO4-limited (•) or high-PO4 (○) medium. Samples were taken at the indicated times for measurement of growth (top panel), σB-dependent β-galactosidase activity (middle panel), and ATP content (lower panel) as described in Materials and Methods.
The nutritional limitation that triggers σB activation in a stationary-phase culture under the high-PO4 and -glucose conditions is not known but, as with the induction initiated with PO4 limitation, there is an inverse correlation between ATP levels and σB activity. At the onset of stationary phase in the high-PO4/glucose medium, ATP levels increase, presumably due to a reduction in ATP use in growth related processes. σB remains uninduced at this time, but increases approximately 1 hour later following the eventual fall in ATP (Fig. 2). Thus, in the case of both σB induction by PO4 limitation and the induction which occurs in the high-PO4 medium, σB activation is preceded by a drop in ATP. The timing of the ATP drop relative to σB induction would allow ATP decline to be a potential trigger for σB activation.
Effects of inhibitors of nucleotide synthesis on σB induction.
If changes in nucleotide levels are the trigger for σB activation, a drop in ATP is an attractive choice for the inducing molecule; however, it is not the only possible nucleotide that might be involved. GTP levels have also been implicated in σB activation. Both mycophenolic acid and decoyinine, inhibitors of GTP synthesis, have been shown to induce σB (28). The possibility that GTP could be involved in σB activation also comes from the observation that null mutations in relA prevent σB activation following glucose or PO4 starvation (40).
RelA, the sole (p)ppGpp synthetase in B. subtilis, synthesizes ppGpp in response to amino acid, glucose, or phosphate starvation (9, 36). As a consequence of ppGpp synthesis, GTP levels fall due to its conversion into (p)ppGpp and a (p)ppGpp-dependent inhibition of inosine monophosphate dehydrogenase, an enzyme in the de novo pathway for GTP synthesis (18) (Fig. 5). Both GTP and ATP levels fall in RelA+ B. subtilis under limiting nutrient conditions. GTP levels fall under amino acid, glucose, or phosphate restriction, while ATP levels drop and σB is activated only after glucose or phosphate limitation (40). In the absence of RelA, neither GTP nor ATP levels decline when cultures are growth restricted by glucose or phosphate limitation and σB fails to be induced.
FIG. 5.
Pathways for ATP and GTP de novo synthesis. IMP is a precursor of both GTP and ATP. IMP dehydrogenase, encoded by guaB, channels IMP down a pathway for the synthesis of GTP, while adenylosuccinate synthetase (purA) generates the ATP precursor adenylosuccinate (28). GTP is required for the PurA-catalyzed synthesis of adenylosuccinate. Thus, a block in GTP synthesis ultimately inhibits ATP synthesis. IMP dehydrogenase is the target enzyme for the inhibition of GTP synthesis by either ppGpp or mycophenolic acid (18, 27).
The requirement for RelA in σB induction following glucose or phosphate restriction could involve an indirect effect of RelA arising from the decline in GTP and/or ATP levels that occurs only when RelA is present or an unforeseen interaction between RelA and the σB regulators. To ask whether the RelA role in σB induction is due to its effect on nucleotide levels rather than a direct involvement in σB induction, a RelA− strain was exposed to inhibitors of ATP (azide) (Fig. 3A) or GTP (mycophenolic acid) synthesis (Fig. 3B) and monitored for σB-dependent β-galactosidase activity. The presence of either inhibitor, at concentrations that inhibited growth, was found to circumvent the need for RelA and trigger an increase in σB activity.
FIG. 3.
σB Induction by azide or mycophenolic acid. RelA− B. subtilis (BSZ7; relA::pUK-RE1) (panels A and B) or F0F1 ATPase B. subtilis (BSZ131, Δatp FHAGDC::kanr) (panel C) were grown in LB. At time zero the cultures were split and half of the cultures were treated with azide (panels A and C) or mycophenolic acid (panel B). Samples were taken at the indicated times and assayed for σB-dependent β-galactosidase activity as described in Materials and Methods. Symbols: untreated control cultures (○), inhibitor-treated cultures (•).
Azide was the more effective inducer, giving full σB activity within 60 min (Fig. 2A). In contrast, σB activity rose slowly over the course of several hours in the mycophenolic acid-treated culture (Fig. 2B). The slow growth rate and σB activation observed in the presence of mycophenolic acid is attributable to a deficiency in guanine nucleotides. Both the slowing of growth and the activation of σB fail to occur if the LB medium is supplemented with 2 mM guanosine (data not shown).
Although inhibition of GTP synthesis is the primary consequence of mycophenolic acid treatment, adenylosuccinate synthetase, the first enzyme in the ATP biosynthetic pathway, requires GTP for activity (Fig. 5) (29). Thus, GTP depletion would be expected to eventually affect ATP levels. As such, the more modest σB activation observed after mycophenolic acid treatment could be a consequence of the inhibitor's indirect effect on ATP synthesis rather than a drop in GTP per se. Overall, the data argue that the requirement for RelA in σB activation following glucose or phosphate starvation is unlikely to be due to an unforeseen interaction between RelA and the σB regulators, but rather to a RelA-dependent drop in nucleotide levels that occurs under these conditions.
Azide inhibits ATP synthesis by binding to cytochrome oxidase and arresting the transfer of electrons from the electron transport chain to O2. As a result of this block the continued movement of electrons is impeded and the oxidation state of the cytochrome pathway is altered, with the upstream portions of the pathway becoming more reduced. Given the presence of a PAS domain on RsbP, it is formally possible that azide's induction of σB might be due to changes in the cell's redox state that are directly sensed by RsbP, rather than azide's effect on ATP levels. To help distinguish between these possibilities, the experiment was repeated using a B. subtilis strain (BSZ131) with a null mutation (Δatp2) in the strain's F0F1 ATPase. F0F1 ATPase is the enzyme that couples the proton gradient developed by the electron transport system to ATP synthesis. In the absence of this enzyme, the strain must generate ATP by substrate-level phosphorylation alone. When BSZ131 was treated with azide, σB activity remained unchanged from that seen in the untreated culture (Fig. 3C). It would therefore appear that it is the failure to generate ATP and not a block in electron transport as such that triggers σB activation follow azide treatment.
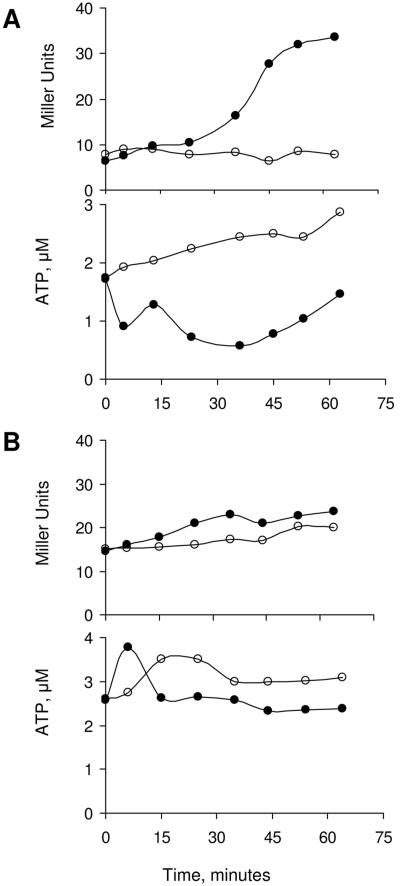
Another inhibitor of oxidative phosphorylation that triggers σB induction is CCCP (35). Unlike azide however, CCCP does not restrict electron flow. CCCP is an uncoupling agent that discharges the pH gradient and membrane potential generated by electron transport. With this reagent, respiration continues, but the resulting pH gradient is dissipated without ATP synthesis. To ask whether it is the disruption of the cell's membrane potential or the failure to generate ATP that is the CCCP-dependent inducer of σB activity, we tested the effect of CCCP on B. subtilis growing on different carbon sources. Succinate is a nonfermentable carbohydrate that requires the oxidative phosphorylation pathway to generate ATP, while glucose can generate ATP via substrate-level phosphorylation. If B. subtilis is grown in medium containing either succinate (Fig. 4A) or glucose (Fig. 4B) as the principal carbon source and exposed to CCCP, the uncoupler is found to induce σB activity only in the succinate-containing medium. As was the case with the previous σB inductions, the σB activation is preceded by a drop in ATP (Fig. 4A). The ATP reduction and σB induction is not observed when the glucose grown culture was treated with CCCP (Fig. 4B). Thus, the CCCP effect on σB activity is related to its ability to restrict ATP synthesis rather than a disruption of the bacterial cell's membrane potential.
FIG. 4.
σB induction by CCCP. Wild-type B. subtilis (BSA46) was grown in a defined medium with succinate (A) or glucose (B) as the carbon source. At time zero the cultures were split and CCCP (1 μM) was added to a portion of each culture. Samples were taken at the indicated times and assayed for σB-dependent β-galactosidase activity (upper panels) and ATP content (lower panels) as described in Materials and Methods. Symbols: untreated cultures (○), CCCP-treated cultures (•).
σB induction in adenine- or guanine-starved B. subtilis.
The data argue that a decline in nucleotide levels, particularly ATP, rather than changes in the cell's membrane potential or the redox state is important for the activation of σB during nutritional stress. A nucleotide drop is also the trigger for an alternative B. subtilis stress response entry into sporulation. In the case of sporulation, it is a drop in GTP rather than ATP that provokes the sporulation response (18, 20, 23). In a series of compelling experiments, Freese and associates used auxotrophic mutants to demonstrate that restriction of the GTP biosynthetic pathway can specifically induce sporulation (20).
If a decline in ATP and not GTP is critical to triggering σB activation, it may be possible to use a similar strategy to test which nucleotide is critical for σB activation by using mutant B. subtilis with lesions in either the GTP or ATP biosynthetic pathway. As illustrated in Fig. 5, IMP is a precursor for de novo synthesis of both GTP and ATP (29). IMP dehydrogenase, encoded by guaB, directs IMP toward formation of GTP, while adenylosuccinate synthetase, encoded by purA, sends IMP in the direction of ATP synthesis (29). B. subtilis strains with null mutations in either guaB or purA were constructed, grown in complete medium, and then resuspended in medium missing the component that each requires for growth and nucleotide synthesis. When the GuaB− strain was subjected to guanine limitation, growth slowed but σB remained uninduced (Fig. 6A). ATP levels also remained largely unchanged in this culture. In contrast, when the PurA− strain was deprived of adenine, there was a rapid rise in σB activity concomitant with a drop in ATP (Fig. 6B). Thus, unlike sporulation induction, activation of σB is largely unaffected by a reduction in guanine containing nucleotides, but is dramatically influenced by a drop in adenine containing nucleotides.
FIG. 6.
σB activity following guanine or adenine limitation. B. subtilis strains BSZ130 (guaB::erm) (A) and BSZ116 (purA::kan) (B) were grown in a defined medium supplemented with either guanosine (1 mM) or adenine (1 mM). At time zero the cultures were pelleted, washed in unsupplemented medium, and resuspended as duplicate cultures with (○) or without (•) the required base or nucleoside. Samples were taken at the indicated times and assayed for growth (upper panel), σB-dependent β-galactosidase activity (middle panel), and ATP content (lower panel) as described in Materials and Methods.
Our results reinforce the notion that changes in ATP levels are critical to the nutritional stress induction of σB. The findings that inhibitors of electron transport and uncouplers of membrane potential only induce σB when their effects can be directly channeled to ATP synthesis argue that it is ATP levels per se, rather than the cell's redox state or membrane potential, that is being conducted to the RsbP/Q proteins for the activation of their RsbV-P phosphatase activity.
In the control of sporulation, GTP interacts directly with a repressor protein (CodY) to inhibit sporulation onset (23). Recently, branched-chain amino acids have also been found to bind to CodY and activate its DNA binding (26). It is not yet known whether the effects of GTP and branched-chain amino acids on CodY's activities are independent or cooperative. It is unclear whether ATP could function in a similar fashion and interact directly with RsbP/Q to inhibit RsbP's phosphatase activity when ATP levels are high. A PAS domain on another B. subtilis protein (KinA) does, in fact, bind ATP (27). Alternatively, the ATP sensor may be distinct from RsbP. In this event, the ATP decline could trigger intermediary factors that ultimately activate RsbP/Q.
It is intriguing that B. subtilis appears to use two different purine nucleotides to communicate the cell's nutritional status to alternative stress responsive pathways and, depending on the particular purine nucleotide whose level declines (ATP or GTP), activate either σB or sporulation. This raises the interesting question of what it is about each of these nucleotides that led to its use in one or the other of these pathways. Although there may be a physiological basis for GTP decline's being a preferred signal for sporulation induction while ATP decline triggers σB activation, it is also possible that it is merely a result of evolutionary history and chance.
Acknowledgments
This study was supported by U.S. National Institutes of Health grant GM-48220.
REFERENCES
- 1.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1992. Characterization of a regulatory network that controls σB expression in Bacillus subtilis. J. Bacteriol. 174:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, A. K., and W. G. Haldenwang. 1993. The σB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J. Bacteriol. 175:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6423-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. I. Curtiss Spaceiiiqq, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. C. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 10.Chen, C.-C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supermolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 11.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of sigma B in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 14.Hecker, M., W. Schumann, and U. Voelker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, K., N. Tanaka, and T. Kumasaka. 2005. Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Sci. 14:558-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney, T. J., and C. P. Moran, Jr.1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor of Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundin, A., and A. Thore. 1975. Comparison of methods for extraction of bacterial adenine nucleotides determined by firefly assay. Appl. Microbiol. 30:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochi, K., J. Kandala, and E. Freese. 1982. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J. Bacteriol. 151:1062-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Voelker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 23.Ratnayake-Lecamwasam, M., P. Serror, K-W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early stationary phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 25.Santana, M., M. S. Ionescu, A. Vertes, R. Longin, F. Kunst, A. Danchin, and P. Glaser. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176:6802-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson, K., and J. A. Hoch. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:15251-15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stulke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 29.Switzer, R. L., H. Zalkin, and H. H. Saxild. 2002. Purine, pyrimidine, and pyridine nucleotide metabolism, p. 255-269. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 30.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 32.Voelker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Voelker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 33.Voelker, U., A. Dufour, and W. G. Haldenwang. 1995. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of σB. J. Bacteriol. 177:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist RsbV by stress or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spot homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 37.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 39.Yasbin, R. E., G. A. Wilson, and F. E. Young. 1973. Transformation and transfection of lysogenic strains of Bacillus subtilis 168. J. Bacteriol. 113:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, S., and W. G. Haldenwang. 2003. RelA is a component of the nutritional stress activation pathway of Bacillus subtilis transcription factor σB. J. Bacteriol. 185:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]